The K-turn is a ubiquitous RNA structural motif. K-turn-binding proteins of the L7Ae family have been structurally and biochemically characterized in archaea and eukarya. This work expands the characterization to bacteria, presenting structural and biochemical analyses of two B. subtilis proteins, YbxF and YlxQ. The authors find that these bacterial proteins are indeed L7Ae homologs, but bind K-turns with much lower affinity than L7Ae, and do not bind K-loops. Comparison of homologs from the three kingdoms of life yields a structural rationale for the divergent RNA-binding properties of members of this protein family.
Keywords: K-turn, K-loop, RNA-binding proteins, X-ray crystallography
Abstract
The archaeal protein L7Ae and eukaryotic homologs such as L30e and 15.5kD comprise the best characterized family of K-turn-binding proteins. K-turns are an RNA motif comprised of a bulge flanked by canonical and noncanonical helices. They are widespread in cellular RNAs, including bacterial gene-regulatory RNAs such as the c-di-GMP-II, lysine, and SAM-I riboswitches, and the T-box. The existence in bacteria of K-turn-binding proteins of the L7Ae family has not been proven, although two hypothetical proteins, YbxF and YlxQ, have been proposed to be L7Ae homologs based on sequence conservation. Using purified, recombinant proteins, we show that Bacillus subtilis YbxF and YlxQ bind K-turns (Kd ∼270 nM and ∼2300 nM, respectively). Crystallographic structure determination demonstrates that both YbxF and YlxQ adopt the same overall fold as L7Ae. Unlike the latter, neither bacterial protein recognizes K-loops, a structural motif that lacks the canonical helix of the K-turn. This property is shared between the bacterial and eukaryal family members. Comparison of our structure of YbxF in complex with the K-turn of the SAM-I riboswitch and previously determined structures of archaeal and eukaryal homologs bound to RNA indicates that L7Ae approaches the K-turn at a unique angle, which results in a considerably larger RNA–protein interface dominated by interactions with the noncanonical helix of the K-turn. Thus, the inability of the bacterial and eukaryal L7Ae homologs to bind K-loops probably results from their reliance on interactions with the canonical helix. The biological functions of YbxF and YlxQ remain to be determined.
INTRODUCTION
The K-turn (Klein et al. 2001) is a widespread RNA structural motif characterized by two helices flanking a trinucleotide bulge where the nucleic acid chains make a sharp ∼60° kink. One of the duplexes next to the kink (the noncanonical, or NC helix) comprises sheared G•A and A•G base pairs immediately adjacent to the bulge; the other duplex (the canonical, or C helix) is Watson-Crick-paired. The sheared base pairs of the NC helix extrude the adenine nucleobases, enabling them to make A-minor (Nissen et al. 2001) interactions with the C helix. K-turns are often the binding sites for proteins (for review, see Schroeder et al. 2010). From the structural standpoint, the best-characterized family of K-turn-binding proteins is composed of the archaeal protein L7Ae and eukaryal homologs such as 15.5kD, L30e, and Nhp2 (Koonin et al. 1994; Reuter et al. 1999; Ban et al. 2000; Vidovic et al. 2000; Chao and Williamson 2004; Koo et al. 2011). L7Ae is a versatile protein that forms part of diverse archaeal ribonucleoprotein (RNP) complexes, including the large ribosomal subunit (Bult et al. 1996), RNase P (Cho et al. 2010), the box C/D methyltransferase (Kuhn et al. 2002), and the box H/ACA pseudouridine synthase (Rozhdestvensky et al. 2003). L7Ae not only binds to RNAs with canonical K-turns, but also RNAs that have a K-loop (a stem–loop motif composed of the NC helix of a K-turn and a closing loop, but lacking the C helix) (Rozhdestvensky et al. 2003; Nolivos et al. 2005). In eukarya, it appears that L7Ae homologs have become more specialized. For instance, L30e complexes with 60S rRNA and its own coding mRNA (Eng and Warner 1991), 15.5kD with U4 snRNA (Nottrott et al. 1999) and box C/D snoRNAs (Watkins et al. 2000), and Nhp2 with box H/ACA snoRNAs (Henras et al. 2001).
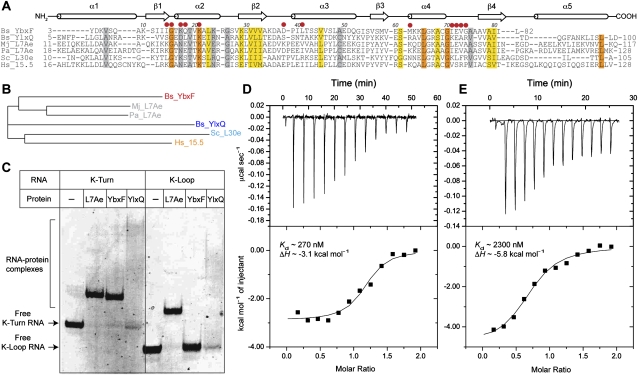
Sequence and structural characterizations reveal that many bacterial gene-regulatory RNAs contain K-turns. Conservation of the purines that form the sheared pairs of the NC helix was initially reported for T-box RNAs (Winkler et al. 2001) in addition to rRNA and eukaryal RNAs (Klein et al. 2001). Lysine and class I S-adenosylmethionine (SAM-I) riboswitches have been shown to contain K-turns that are required for the aptamer domains to fold and recognize their cognate ligands (Montange and Batey 2006; Garst et al. 2008; Serganov et al. 2008), and a K-turn is also present in the class II c-di-GMP riboswitch (Lee et al. 2010). Lafontaine and colleagues have shown that, in vitro, archaeal L7Ae can bind to the K-turns of the lysine and SAM-I riboswitches from Bacillus subtilis (Blouin and Lafontaine 2007; Heppell and Lafontaine 2008). Although these experiments suggest that K-turn-binding proteins might modulate the activity of gene-regulatory RNAs such as riboswitches and T-boxes, the heterologous interaction between the archaeal L7Ae protein and bacterial RNAs is not biologically meaningful. Sequence-based searches indicate the presence of at least two likely L7Ae homologs in Gram-positive bacteria (Koonin et al. 1994; Winkler 2002; Sojka et al. 2007), YbxF and YlxQ (also called YmxC). Alignment of the sequences of these bacterial proteins with their putative archaeal and eukaryal homologs (Fig. 1A) suggests that they might share a similar structural fold with the archaeal and eukaryal K-turn-binding proteins, since YbxF and YlxQ share 55% and 27% sequence identity with L7Ae, respectively (identity is 20% between YbxF and YlxQ). Phylogenetic analysis suggests that YbxF is more closely related to L7Ae and its eukaryal homologs than is YlxQ (Fig. 1B).
FIGURE 1.
Sequence and biochemical characterization of YbxF and YlxQ. (A) Multiple sequence alignment of B. subtilis YbxF, YlxQ, and their archaeal and eukaryal homologs. (Bs) Bacillus subtilis; (Mj) Methanococcus jannaschii; (Pa) Pyrococcus abyssi; (Sc) Saccharomyces cerevisiae; (Hs) Homo sapiens. Secondary structure elements shared by all six proteins are indicated. Colored bars denote different levels of sequence conservation among the six proteins: (gray) similar; (yellow) highly similar; (dark orange) invariant. (Red dots) YbxF residues that make RNA contacts. (B) Phylogenetic dendrogram calculated using ClustalW2 (Larkin et al. 2007) for the six proteins whose sequences are shown in A. (C) Electrophoretic mobility shift analysis of K-turn and K-loop binding by M. jannaschii L7Ae and B. subtilis YbxF and YlxQ. (D,E) Isothermal titration calorimetric analysis of K-turn binding by B. subtilis YbxF (left) and YlxQ (right).
Sojka et al. (2007) demonstrated that neither YbxF nor YlxQ is essential for B. subtilis growth in rich media, and that a YbxF-GFP fusion localizes to ribosomes (primarily the large subunits) in vivo. Although the latter result hints that YbxF might be an rRNA-binding protein, in the absence of structural and biochemical characterization, it remains unknown whether YbxF and YlxQ are authentic bacterial homologs of L7Ae, and what their RNA-binding properties might be. Using recombinant, purified YbxF and YlxQ, we now show that these bacterial proteins bind K-turns, but with affinities considerably lower (especially for YlxQ) than that of L7Ae. We were unable to detect binding to K-loops by either protein. We have determined crystal structures of the two proteins (YbxF in complex with the K-turn-containing SAM-I riboswitch aptamer domain) and find that they are indeed structural homologs of the L7Ae family of proteins. Although further in vivo research will be needed to establish what the physiological RNA targets of YbxF and YlxQ are, our work broadens the phylogenetic distribution of the L7Ae family of K-turn-binding proteins from archaea and eukarya to bacteria, and provides the molecular foundation for analyzing the cellular function of bacterial L7Ae homologs.
RESULTS AND DISCUSSION
YbxF and YlxQ bind K-turn-containing RNAs
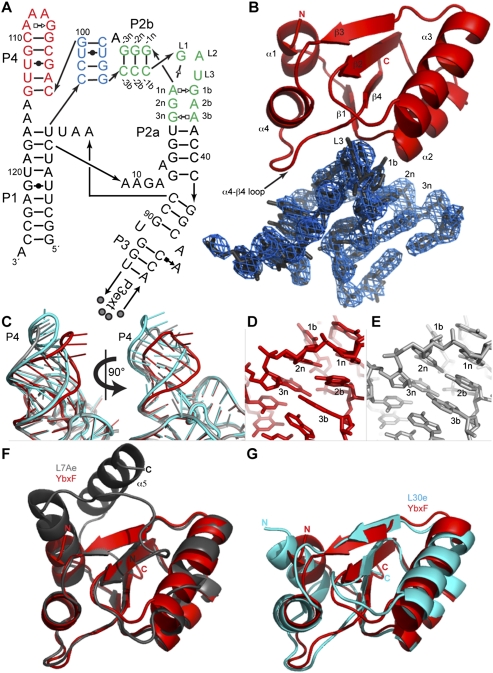
Full-length B. subtilis YbxF and YlxQ proteins were expressed in Escherichia coli and purified to near homogeneity (Materials and Methods). Electrophoretic mobility shift assays using canonical K-turn- and K-loop-containing RNAs (derived from archaeal box H/ACA guide RNAs) show that YbxF forms a stable complex with the K-turn RNA, whereas YlxQ appears to associate weakly with the K-turn RNA, retarding it into a reproducible smear (Fig. 1C). Neither protein associates detectably with K-loop-containing RNAs in this assay. In contrast, Methanococcus jannaschii L7Ae binds to both K-turn- and K-loop-containing RNAs (Fig. 1C; data not shown). Isothermal titration calorimetry (ITC) confirms K-turn binding by both YbxF and YlxQ (Fig. 1D,E). YlxQ binds weakly with Kd ∼2.3 μM, while YbxF binds with Kd ∼270 nM (Table 1). The two bacterial proteins bind K-turn-containing RNAs considerably more weakly than does L7Ae, for which Kd has been estimated to be as tight as 10 pM (Turner and Lilley 2008).
TABLE 1.
Isothermal titration calorimetry
Previously, Blouin and Lafontaine (2007) and Heppell and Lafontaine (2008) found that the archaeal protein L7Ae binds to the K-turns of the B. subtilis yitJ SAM-I and lysC lysine riboswitches in vitro. To determine if the ligand-binding affinity of the B. subtilis yitJ SAM-I riboswitch can be modulated by an endogenous K-turn-binding protein, we carried out further ITC studies. In the presence of 1 mM MgCl2, the riboswitch binds SAM with ∼265 nM affinity (Table 1). Binding of YbxF to the riboswitch before titration with SAM results in no appreciable change in affinity for the metabolite (Kd ∼247 nM) (Table 1). Many riboswitches are under kinetic, rather than thermodynamic control (for review, see Zhang et al. 2010), and our analysis does not exclude effects on the rate of aptamer domain folding or SAM binding by YbxF association with the riboswitch. For instance, YbxF association with the riboswitch may accelerate aptamer domain folding and thus kinetically facilitate metabolite binding, so that while the RNA polymerase operates at the same speed, less metabolite is necessary to activate the expression platform. Thermodynamic analysis would not detect such kinetic effects.
Crystallographic analysis of RNA binding by YbxF
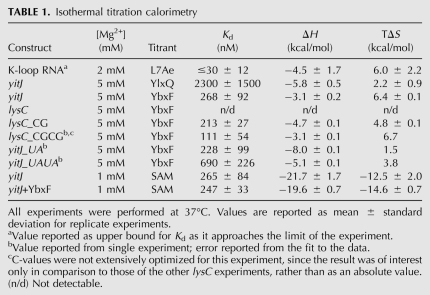
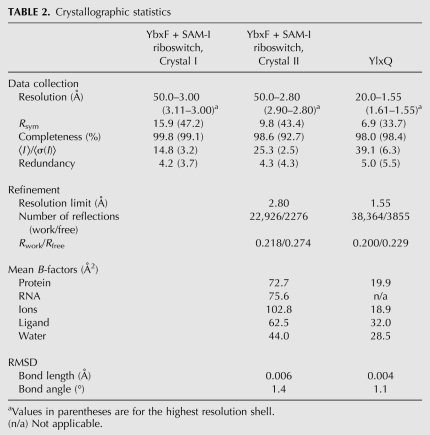
To determine how YbxF binds to the K-turn, and as a starting point for understanding how its binding might affect bacterial RNAs that contain K-turns, we cocrystallized the protein with the aptamer domain of the SAM-I riboswitch from the bacterium Thermoanaerobacter tengcongensis. To obtain well-diffracting crystals, the aptamer domain was engineered by extending the P3 helix by 16 bp and capping this helix with a UNCG tetraloop (Fig. 2A). The structure was determined at 2.8 Å resolution by molecular replacement using the previously reported structures of the SAM-I riboswitch and L7Ae (Fig. 2B; Table 2; Hamma and Ferré-D'Amaré 2004; Montange et al. 2010; Materials and Methods). The crystallographic asymmetric unit consists of two SAM-I riboswitch aptamer domains, both in complex with YbxF. The two RNA–protein complexes in the asymmetric unit are very similar, superimposing on each other with a root-mean-square deviation (RMSD) of 0.93 Å (for 126 C1′ and 77 Cα pairs).
FIGURE 2.
RNA recognition by YbxF. (A) Secondary structure of the Thermoanaerobacter tengcongensis SAM-I riboswitch construct used for cocrystallization. The distal end of P3 was engineered with the P3ext sequence described in Baird and Ferré-D'Amaré (2010). Base pairs observed in the cocrystal structure of the riboswitch bound to YbxF are denoted using Leontis and Westhof (2001) symbols. Arabic numbers indicate the numbering scheme of the riboswitch aptamer domain. K-turn residues are named using the nomenclature of Liu and Lilley (2006). (Green) K-turn; (red) P4 helix; (blue) pseudoknot. (B) Cartoon representation of YbxF bound to the SAM-I riboswitch K-turn. Secondary structure elements of the protein and several residues of the K-turn are named as in Figure 1A and part A, respectively. A portion of the 2.8 Å resolution anneal-omit 2|Fo| − |Fc| electron density map corresponding to the K-turn is shown contoured at 2σ. (C) Superposition of the SAM-I riboswitch core (red, from RNA complexed to YbxF), the protein-free T. tencongensis riboswitch (Montange et al. 2010) (PDB ID 3GX5, gray), and the B. subtilis yitJ riboswitch (Lu et al. 2010) (PDB ID 3NPB, cyan) demonstrates that P4/L4 is mobile in relation to the body of the aptamer domain. (D) Magnified view of the NC helix from the RNA complexed to YbxF reveals no interactions between G at position 2n and A at position 2b. (E) The protein-free SAM-I riboswitch (3GX5) has the same sequence as in D and indicates formation of the G•A sheared pair between 2b–2n residues of the NC helix. (F) Structural alignment of YbxF (red) and L7Ae (gray). (G) Structural alignment of YbxF (red) and L30e (cyan).
TABLE 2.
Crystallographic statistics
Overall, the SAM-I riboswitch aptamer domain adopts the same conformation in complex with YbxF as in isolation. SAM is bound in the compact conformation described previously (Montange and Batey 2006), with the adenine moiety of the metabolite forming a Hoogsteen to Watson-Crick base pair with U57 of the riboswitch, and also making two hydrogen bonds with the sugar edge of A45 of the RNA. Superposition of the RNA–YbxF complex with the previously determined structures of the T. tengcongensis (Montange and Batey 2006) and B. subtilis yitJ (Lu et al. 2010) SAM-I riboswitches indicates that helix P4 is the only structural element that adopts markedly different conformations between the three structures, deviating by as much as 23° between the two T. tengcongensis structures that were determined for RNAs with identical P4 sequence, and to a smaller degree with the B. subtilis riboswitch carrying several sequence differences (Fig. 2C).
The difference in disposition of P4 between the three structures might reflect the intrinsic conformational plasticity at a hinge at the junction of P4 and the core of the aptamer domain, since superposition of the core of the two T. tengcongensis structures, omitting P4-L4 and P3, yields an RMSD of 0.85 Å (for 75 C1′ atom pairs), comparable to the precision of the atomic coordinates (0.66 Å) (Materials and Methods) or the result of superimposing either T. tengcongensis riboswitch with the B. subtilis riboswitch (RMSD = 1.54 Å over 69 C1′ pairs). This would be consistent with recent solution studies suggesting that P4 is the last structural element to adopt the conformation present in the ligand-bound aptamer domain, and that the final docking of P4 enhances the affinity of the RNA for SAM (Heppell et al. 2011). On the other hand, it is possible that YbxF binding modifies the structure or flexibility of the K-turn and that this propagates across the pseudoknot (Fig. 2A, blue) to P4. Although the precise hydrogen-bonding pattern of the K-turn bound to YbxF is somewhat ambiguous in our 2.8 Å resolution structure, it is clear that while the sheared pair between positions 1b and 1n (see Fig. 2A for K-turn nomenclature) is formed, the nucleobases at 2b and 2n are severely staggered (in both RNA copies in the asymmetric unit) and probably do not hydrogen-bond with each other (Fig. 2D). This is in contrast to formation of the 2b–2n sheared pair in both the 2.9 Å and the 2.3 Å resolution structures (Montange and Batey 2006; Montange et al. 2010) of the protein-free T. tengcongensis SAM-I riboswitch (Fig. 2E) and even in a mutationally destabilized version of the riboswitch crystallized in the same crystalline environment (Schroeder et al. 2011). Interestingly, the 2b–2n sheared pair is also absent in the 3.0 Å resolution structure of the B. subtilis yitJ SAM-I riboswitch (Lu et al. 2010). Thus, it appears that the K-turn of the SAM-I riboswitch might be structurally flexible and that YbxF selects a conformation with a single sheared pair. Modulation of K-turn structure by L7Ae binding has previously been reported (Turner et al. 2005).
K-turn recognition by L7Ae, L30e, and YbxF
YbxF adopts the same fold as L7Ae and L30e, composed of a central four-stranded β-sheet flanked on each side by α-helices, with αβαβαβαβα connectivity (Figs. 1A, 2F,G). Superposition with L7Ae and L30e (Fig. 2F,G) yields an RMSD of 0.91 Å and 1.34 Å, respectively (for 81 and 80 Cα pairs). The larger discrepancy with L30e reflects differences in the orientation of the N-terminal helix and the β-strands between the bacterial and eukaryal proteins (Fig. 2G). This helix adopts a similar orientation between YbxF and L7Ae, but it is shorter by 7 residues for the bacterial homolog (Fig. 1A). However, since the helix faces away from the RNA (Fig. 2B), the K-turn interfaces of the two proteins are similar. The interface is primarily composed of two regions. First, amino acids at the C-terminal side of β1 and the N-terminal side of α2 (hereafter, the β1–α2 region) are involved in interactions with the sheared G•A pairs of the NC helix of the K-turn. The RMSD for this region is 0.19 Å between L7Ae and YbxF (five Cα pairs). Second, the loop between α4 and β4 (hereafter, the α4–β4 loop) interacts with the C-helix side of the K-turn. The RMSD for this region is 0.44 Å between L7Ae and YbxF (six Cα pairs).
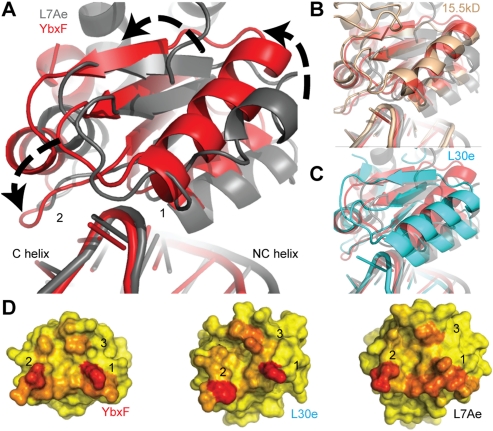
Despite the high similarity in the polypeptide backbone conformations of the RNA-binding segments of L7Ae, L30e, and YbxF, the overall orientation of these proteins with respect to the K-turn differs considerably. That this difference is primarily a rotation of the proteins with respect to the bulge of the K-turn becomes apparent when the structures of the RNP complexes are superimposed using the sheared G•A pairs of their respective K-turns as anchors (Fig. 3A–C). YbxF and the eukaryal 15.5kD protein adopt a similar orientation (Fig. 3B), and both are rotated ∼12° counterclockwise relative to L7Ae (in the view shown in Fig. 3A). L30e has an intermediate degree of rotation. That the orientation with respect to the RNA is an intrinsic feature of each particular protein and not an artifact of crystallization is supported by the observation that the orientation of L7Ae with respect to the sheared G•A pairs is the same regardless of whether it is bound to a K-turn derived from a box C/D RNA or a K-loop related to that of a box H/ACA RNA (data not shown). This analysis indicates that the orientation adopted by proteins of this family with respect to their cognate RNAs is an intrinsic property of each protein and that the orientation of L7Ae is distinctly different from those of the other family members.
FIGURE 3.
Global comparison of K-turn-binding proteins bound to K-turns. (A) Alignment of K-turn G•A sheared pairs from RNP complexes (YbxF, red; L7Ae, gray) reveal ∼12° counterclockwise rotation of YbxF relative to L7Ae. (B) Eukaryal protein 15.5kD (brown) aligns closely with YbxF in its orientation on the K-turn, while (C) eukaryal protein L30e (cyan) has an intermediate degree of rotation. (D) Analysis of the three interaction surfaces reveals that L7Ae buries more surface area (red) than YbxF and L30e and is unique in its distribution of interacting residues. The accessible surface area for YbxF was calculated subsequent to the addition of side-chain rotamers similar to L7Ae at residues K17 and K21 (α2) and E72 (α4–β4 loop).
The unique orientation of L7Ae with respect to the K-turn has two consequences. First, it increases the extent of its interaction with the NC helix. While only the N-terminal end of α2 of YbxF contacts the NC helix, residues from each of the three turns of α2 of L7Ae, in addition to its β1–α2 region, contact this moiety of the RNA. Second, the angle of approach of L7Ae results in its α4–β4 loop being retracted toward the apex of the K-turn. The corresponding loops of YbxF or L30e are more extended and reach further over the K-turn toward the C helix. Analysis of burial of the solvent-accessible surface indicates that the interface between the proteins and their respective K-turn RNAs is composed of three discrete patches. The first corresponds to the β1–α2 region, the second is the α4–β4 loop, and the third is at the N terminus of α3 and the loop preceding it (Fig. 3D). As expected, the area of the first interfacial patch is considerably larger for L7Ae (298 Å2) than for YbxF (223 Å2) or L30e (179 Å2), comprising 51% of the total RNA-buried solvent-accessible surface area of L7Ae, compared with 46% and 38% for YbxF and L30e, respectively. The greater relative size of the first patch in the L7Ae interface is consistent with the ability of this protein to bind to both K-turns and K-loops (both of which have an NC helix), while YbxF, which interacts with the NC helix to a lesser degree and requires interactions with the C helix of K-turns (through its first and second patches, respectively) does not bind to K-loops (which do not have a C helix). In addition to the relative sizes of the three interfacial patches, it is also noteworthy that the absolute solvent-accessible surface area buried upon RNA binding by L7Ae is considerably larger than that buried by YbxF or L30e, and that, as expected on theoretical grounds (Eisenberg and McLachlan 1986), the affinity of the proteins for different RNAs correlates with the solvent-accessible surface area buried (589 Å2 for L7Ae vs. 487 Å2 for YbxF and 468 Å2 for L30e).
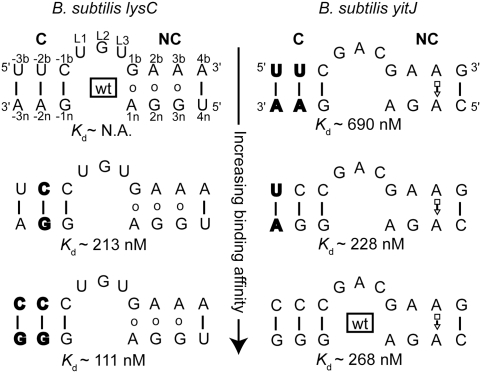
YbxF is sensitive to C-helix stability
Blouin and Lafontaine (2007) have shown that the B. subtilis lysC lysine riboswitch contains a K-turn that can be bound in vitro by L7Ae. Thus, the inability of B. subtilis YbxF to bind this RNA either in the electrophoretic mobility shift assay (data not shown) or by ITC (Table 1) was unexpected. By aligning the sequences of 32 different lysine riboswitches, Blouin and Lafontaine (2007) found that, among these RNAs, the loop positions of the K-turn do not conform to the RNN consensus proposed early on for the K-turn (Klein et al. 2001) and that U is often found in the L1 position of the K-turn, as in the case of the B. subtilis lysC riboswitch (Fig. 4). Mutation of the loop positions of the lysC riboswitch to GAU, GGU, or UAU did not confer the ability to bind to YbxF, as evaluated by ITC (data not shown).
FIGURE 4.
Binding of YbxF to the K-turn is modulated by the stability of the canonical (C) helix. Base pairs for the B. subtilis lysC K-turn are denoted as open circles because no structure is available. Base pairs for the B. subtilis yitJ K-turn are denoted with Leontis-Westhof symbols based on PDB entry 3NPB (Lu et al. 2010). Base-pair mutations (bold) in the lysC riboswitch generate a K-turn that is capable of recognition by YbxF. Mutation of the C stem of the yitJ K-turn, similar to the wild-type sequence of lysC K-turn, results in weaker binding by YbxF. Values reported for wild-type yitJ and the single-base-pair mutant do not differ given the precision of the experiments.
A second difference between the K-turn of the B. subtilis lysC riboswitch and the consensus (Klein et al. 2001) is the presence of a U•A base pair in the C helix between positions −2b and −2n. Since all other K-turns to which we detected YbxF binding have a G•C or C•G pair at this position, we prepared a double mutant of the lysC riboswitch with C at position −2b and G at position −2n. This RNA bound YbxF with a Kd ∼210 nM (Fig. 4). Further stabilizing the C helix by replacing the U•A pair at position −3 with a C•G pair resulted in even tighter binding (Kd ∼110 nM). These results suggest that YbxF is much more sensitive to the stability of the C helix than L7Ae. Conversely, we mutagenized the K-turn of the B. subtilis yitJ SAM-I riboswitch, which has three C•G pairs in the C helix and is bound by YbxF with Kd ∼270 nM to U•A pairs. Replacement of the base pair at position −3 with a U•A pair did not affect affinity for the protein, but introduction of U•A pairs at both, positions −3 and −2 decreased the binding affinity by ∼2.5-fold with a Kd ∼700 nM (Fig. 4). Overall, these experiments indicate that tight YbxF binding requires two C•G (or, presumably G•C) pairs in the −1 and −2 positions to close the C helix of the K-turn. Additional aspects of the K-turn are important for YbxF binding as evidenced by the retention of YbxF binding, albeit weakly, of a K-turn containing only a single C•G pair in the −1 position of the C helix.
YlxQ is a structural homolog of L7Ae and YbxF
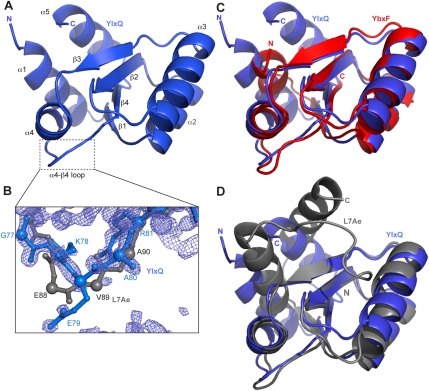
Since its low affinity for RNA (Fig. 1C,E; Table 1) precluded cocrystallization, we determined the structure of RNA-free B. subtilis YlxQ at 1.55 Å resolution to establish whether it is a structural homolog of the L7Ae family of K-turn-binding proteins (Fig. 5A; Table 2; Materials and Methods). The three-dimensional structure of YlxQ exhibits strong similarity to those of L7Ae, L30e, and YbxF (RMSD of 1.53 Å, 1.56 Å, and 1.21 Å for 85, 82, and 77 Cα pairs, respectively). A DALI (Holm and Sander 1995) search revealed that the closest structural homologs of B. subtilis YlxQ are its presumed ortholog from Bacillus halodurans, as well as L30e and L7Ae. The N- and C-terminal α-helices of YlxQ are longer than those of YbxF, and in this sense YlxQ is more similar to L7Ae or L30e (Fig. 5C,D), although the orientation of the C-terminal helix (α5) of L7Ae is unique among these proteins. The three copies of YlxQ in the asymmetric unit of our crystals superimpose closely, with an RMSD of 0.1–0.2 Å between the copies, which is comparable to the mean precision of the atomic coordinates (Materials and Methods). This suggests that YlxQ has little backbone flexibility, similar to L7Ae (Suryadi et al. 2005), but unlike Nhp2, which exhibits conformational mobility resulting from a proline that undergoes cis/trans isomerization (Koo et al. 2011). This proline (equivalent to P39 in B. subtilis YbxF) is not present in B. subtilis YlxQ.
FIGURE 5.
Structure of YlxQ. (A) Cartoon representation of the overall structure of YlxQ, with secondary structure elements labeled as in Figure 1A. (B) Portion of the 1.55 Å resolution anneal-omit 2|Fo| − |Fc| electron density map corresponding to the α4–β4 loop of YlxQ (outlined in panel A) is shown contoured at 2.5σ. The corresponding α4–β4 loop from L7Ae (only main chain atoms) is shown in gray for comparison. Cα atoms are shown as enlarged spheres to highlight the main chain trajectories of YlxQ and L7Ae. (C) Structural alignment of YlxQ (blue) and YbxF (red) and (D) of YlxQ (blue) and L7Ae (gray).
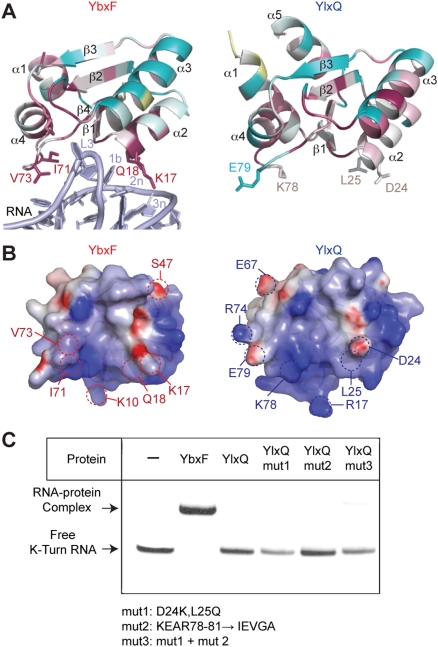
To probe the structural basis for the markedly weaker affinity for K-turn RNAs of YlxQ compared with that of YbxF, we performed sequence conservation analysis and electrostatic calculations. Position-Specific Iterated BLAST (Altschul et al. 1997) searches using YbxF and YlxQ sequences returned 65 and 150 orthologs, respectively, using a default PSI-BLAST threshold of 0.005. Alignment of these 65 YbxF orthologs shows high sequence conservation at the β1–α2 region and α4–β4 loop (Fig. 6A), consistent with the participation of these moieties in the protein–RNA interface (Fig. 3D). In contrast, the corresponding residues in 150 YlxQ orthologs are poorly conserved. This suggests that the identities of these putative RNA-interacting residues are not essential for the cellular function of YlxQ. The electrostatic properties of YlxQ, however, hint at nucleic acid binding as a possible function for this protein, since the projection of the electrostatic potential at the solvent-accessible surface of YlxQ delimits strongly positively charged patches on locations similar to the positively charged surface patches of YbxF that are involved in K-turn binding (Fig. 6B).
FIGURE 6.
Comparison of YlxQ with YbxF. (A) Sequence conservation among YbxF and YlxQ homologs calculated with the ConSurf server (Ashkenazy et al. 2010). (Dark red) Highly conserved among the top 65 YbxF homologs and 150 YlxQ homologs; (cyan) highly variable; (white) average conservation; (yellow) insufficient data. Select residues at equivalent positions at the RNA–protein interface on YbxF, YlxQ, and RNA are labeled. (B) Continuum electrostatic potentials at the solvent-accessible surfaces of YlxQ and YbxF. The view is from YbxF-bound RNA toward the YbxF–RNA interface. (Blue) Positive potential; (red) negative potential; (white) neutral. (C) Electrophoretic mobility shift assay showing that structure-based engineering of YlxQ fails to confer tighter K-turn binding.
Given the divergence of amino acid sequence between the RNA-binding surfaces of YbxF and the structurally homologous regions of YlxQ, we tested whether grafting the YbxF sequences onto YlxQ would result in tighter K-turn binding by the latter. Additionally, the α4–β4 loop of YlxQ is one residue shorter than the same loop found in YbxF, L7Ae, and L30e (Figs. 1A, 5C), and YbxF uses this loop to interact with the C helix. This prompted us to ask if the shortened α4–β4 loop is responsible for the much-reduced affinity of YlxQ for K-turns. Three site-directed mutants of YlxQ were constructed and purified to near homogeneity. Mutant 1 (D24K, L25Q) introduces a lysine and a glutamine near the N terminus of α2. YbxF uses these amino acids to make interactions with the sheared G•A pairs. Mutant 2 (KEAR78-81 → IEVGA) replaces the α4–β4 loop of YlxQ with the corresponding loop of YbxF. Mutant 3 combines both sets of mutations. When analyzed using the electrophoretic mobility shift assay (Fig. 6C) and ITC (data not shown), none of the mutants bound appreciably more strongly to K-turn-containing RNAs than did wild-type YlxQ. This implies that additional structural elements, beyond the amino acid identity at the RNA–protein interface, are important for K-turn binding by the L7Ae family of RNA-binding proteins. This also shows that the shortened α4–β4 loop on YlxQ is not principally responsible for its reduced affinity toward K-turns. This result parallels a mutational study of L7Ae in which a conserved lysine (K26 in M. jannaschii L7Ae) in the α1–β1 loop, which does not interact directly with RNA, was found to modulate RNA affinity (Gagnon et al. 2010).
Phylogenetic distribution of K-turn-binding proteins
This study extends the phylogenetic distribution of the L7Ae family of K-turn-binding proteins to bacteria from archaea and eukarya. It has been noted before that eukaryotic family members such as L30e and 15.5kD do not bind to K-loops (and Nhp2 does not even bind to K-turns specifically) (Koo et al. 2011), whereas L7Ae can bind with high affinity to both K-turns and K-loops. We now find that the bacterial homologs YbxF and YlxQ bind to K-turns with modest affinity and do not recognize K-loops. Thus, it might be expected that the bacterial and eukaryal proteins would be more similar to each other than to the archaeal L7Ae. At the overall sequence level, this is not the case, since sequence-based phylogenetic analysis places YbxF in the same branch as L7Ae (Fig. 1B, 55% sequence identity). Previously, it was hypothesized that the specific sequence of the α4–β4 loop (which differs between the archaeal and eukaryal family members) was responsible for the ability of L7Ae to bind K-loops (Hamma and Ferré-D'Amaré 2004). Gagnon et al. (2010) found that substitution of the L7Ae α4–β4 loop with the corresponding loop of 15.5kD abrogated the ability of the archaeal protein to bind to K-loops, thus showing that these amino acids are necessary for K-loop binding by L7Ae. However, those investigators did not produce the converse mutant (15.5kD protein with the L7Ae α4–β4 loop), and thus it is unclear if this loop suffices for K-loop binding. Now, our demonstration that YbxF is a bacterial K-turn-binding protein shows that the α4–β4 loop sequences do not cleanly divide among proteins that bind only K-turns and those that can bind both K-turns and K-loops, because the sequence of the loop in YbxF (IEVG) is more similar to that of L7Ae [(I/L)EVA] than that of, for instance, 15.5kD (VSRP) (Fig. 1A). Furthermore, our mutational analysis of YlxQ shows that the α4–β4 loop residues of YbxF are not the dominant determinants of the tighter K-turn binding by this protein over YlxQ. Overall, it appears that sequence similarity is not the overriding cause for the similar RNA recognition properties of the bacterial and eukaryal family members.
The special ability of L7Ae to bind to K-loops might arise from its unique orientation on its target RNAs, rather than from a particular sequence motif. Comparison of the orientation relative to the K-turn of archaeal, bacterial, and eukaryal family members (Fig. 3A–C) shows that while each of the proteins analyzed binds the K-turn from a different angle, L7Ae is an outlier. Analysis of the protein–RNA interface shows that the characteristic orientation of L7Ae results in a larger contact area between this protein and the NC helix of the K-turn (Fig. 3D). Thus, it appears that the archaeal protein places more emphasis on the NC helix than on the C helix or the bulge in binding its target, when compared with its bacterial and eukaryal homologs, thereby allowing it to recognize K-loops, which lack a C helix. In addition, the free energy of RNA binding is considerably more favorable for L7Ae than for YbxF (Kd ∼10 pM vs. Kd ∼270 nM, respectively) (Turner and Lilley 2008; this study). Our ITC analysis (Table 1) shows that the affinity of L7Ae for a K-loop RNA is about 1000-fold weaker than the 10 pM Kd that has been estimated using stopped-flow FRET by Turner and Lilley (2008) for its binding to K-turns. This difference suggests that the interactions that L7Ae makes with the C helix do contribute to its high affinity for the K-turn. Moreover, our analysis of binding by YbxF to mutant C helix RNAs (Fig. 4) shows that the bacterial protein is very sensitive to the stability of the C helix, and, by inference, the interactions it makes with this RNA element through its α4–β4 loop contribute importantly to its K-turn binding. We reason that if the magnitude of the free-energy gain that results from C-helix recognition by YbxF is comparable to that gained by L7Ae, YbxF might be able to bind to K-loops with a Kd ∼270 μM (1000-fold weaker than its affinity for K-turns) (Fig. 1D; Table 1); such low-affinity binding would not be detected in our experiments. Thus, the experimentally observed ability of L7Ae to recognize K-loops might simply reflect its extraordinarily high affinity for K-turns.
In relation to the above, it should be noted that Kd values ranging from 10 pM to 8 nM have been reported for L7Ae binding to K-turns, depending on the assay used (filter binding [Kuhn et al. 2002], electrophoretic mobility shift [Blouin and Lafontaine 2007], stopped-flow FRET [Turner and Lilley 2008]) as well as the specific experimental conditions (temperature, Mg2+ concentration, RNA sequence). In our hands, the extraordinarily high affinity of L7Ae toward K-turns precluded accurate ITC measurements (data not shown), whereas the considerably weaker association (Kd ∼30 nM) of the protein to a K-loop could be characterized calorimetrically (Table 1).
What are the biological functions of YbxF and YlxQ? The pronounced phylogenetic conservation of residues on the K-turn interface of YbxF (Fig. 6A), as well as its relatively high RNA-binding affinity, suggest that its authentic cellular targets are K-turn-containing RNAs. Whether K-turns are the biological targets of YlxQ is uncertain, due to the lack of sequence conservation on the equivalent molecular surfaces. However, the electrostatic properties of YlxQ (Fig. 6B) as well as its conserved genomic location do point to an RNA-binding function. ylxQ is found in the autoregulatory nusA/infB operon consisting of ylxS-nusA-ylxRQ-infB-ylxP-rbfA (Shazand et al. 1993), where it is invariably associated (Winkler 2002) with YlxR, a putative RNA-binding protein (Osipiuk et al. 2001). The strong genetic linkage between YlxR and YlxQ and at least one possible occurrence of a YlxR–YlxQ ORF fusion in Geobacter sulfurreducens (Winkler 2002) suggests an intriguing possibility that YlxR and YlxQ associate and YlxQ may bind its cellular RNA target with high affinity once in complex with YlxR. Such regulation of RNA binding by protein–protein interaction has been previously reported, for instance, for the human spliceosomal U2B″ protein, which only binds its cognate RNA in the presence of U2A′ (Price et al. 1998). The genomic location of YlxQ between transcription factor NusA and translational initiation factor 2 (IF2) is consistent with an involvement in transcriptional or translational regulation, in line with proposed role in translation for YbxF (Sojka et al. 2007). Curiously, in the E. coli genome, the nusA/infB operon is similarly organized as in B. subtilis but the ylxR-ylxQ genes are absent. This may hint of a YlxQ function that is specific to B. subtilis such as sporulation. Neither YbxF nor YlxQ were found to be essential for B. subtilis under nonsporulating conditions (Sojka et al. 2007), but the nusA/infB operon and IF2 are known to be involved with the onset of sporulation (Milon et al. 2006). At present, then, the authentic biological targets of YbxF and YlxQ remain unknown, but our work suggests that these will likely be RNAs containing K-turns.
MATERIALS AND METHODS
Protein expression and purification
Open reading frames for B. subtilis YbxF and YlxQ were assembled by PCR from overlapping synthetic DNA oligonucleotides and cloned into pET16b (EMD chemicals) such that the resulting proteins have no purification tags but contain an extra glycine immediately C-terminal to the initiation methionine. Including the latter, the predicted translation products for YbxF and YlxQ would comprise 83 and 101 amino acids, respectively. YbxF and YlxQ were expressed for 3 h at 37°C in E. coli BL21 CodonPlus (DE3)-RIL (Agilent Technologies) by addition to cultures at OD600 ∼ 0.6 of 1 mM IPTG. Both proteins were purified in the same manner. Cells were resuspended in a solution composed of 20 mM Tris-HCl (pH 8.0), 400 mM KCl, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.5% (w/v) aprotinin, 5 μg/mL leupeptin, 5% (v/v) glycerol, and 1 mM β-mercaptoethanol; lysed using a microfluidizer; clarified by centrifugation; and the supernatant fractionated sequentially with neutralized polyethyleneimine (0.5%, v/v) (Burgess 1991) and ammonium sulfate (90% and 80% saturation, for YbxF and YlxQ, respectively). The ammonium sulfate precipitate was dissolved in Buffer A (150 mM KCl, 20 mM HEPES-KOH at pH 7.5, 5% glycerol, 0.5 mM EDTA, 0.3 mM DTT) and loaded onto HiTrap Heparin column (GE Healthcare) equilibrated in Buffer A. YbxF and YlxQ were eluted with a linear gradient to 100% Buffer B (1.5 M KCl, 20 mM HEPES-KOH at pH 7.5, 5% glycerol, 0.5 mM EDTA, 0.3 mM DTT). Peak fractions were pooled, concentrated by ultrafiltration, and loaded onto a Superdex 75 column (GE Healthcare) equilibrated in Buffer C (150 mM KCl, Tris-HCl at pH 7.5, 5% glycerol, 0.5 mM EDTA, 0.3 mM DTT). YbxF and YlxQ were concentrated to 12–14 g/L and stored at −80°C in Buffer C. Electrospray ionization mass spectrometry yields masses of 8387.5 Da and 10,979.3 Da for YbxF and YlxQ, respectively, implying that the proteins are missing their initiation methionines but are otherwise intact (calculated: 8387.8 Da and 10,979.7 Da, respectively). Coomassie Blue staining of serial dilutions analyzed by Tricine-SDS PAGE indicates that the two proteins are at least 99% pure. Nuclease contamination was assayed as described (Ferré-D'Amaré 2010). Site-directed mutagenesis of YlxQ was performed with the QuikChange Lightning kit (Agilent Technologies). YlxQ mutants were purified by the same procedure used for the wild type. The presence of desired mutations was verified by electrospray ionization mass spectrometry of the purified proteins, which yields masses of 11,007.4 Da, 10,964.3 Da, and 10,992.4 Da, for Mutant 1, Mutant 2, and Mutant 3, respectively (calculated: 11,007.8 Da, 10,964.7 Da, and 10,992.8 Da).
RNA production and purification
Six RNAs (plus a total of seven mutants of two of these) were used in this study. The B. subtilis yitJ riboswitch aptamer used in this study is identical to that used by Heppell and Lafontaine (2008) with the addition of two initial G residues. The B. subtilis lysC riboswitch aptamer domain used herein comprises nucleotides 27–201 of the construct reported by Blouin and Lafontaine (2007). RNAs used in gel shift assays (Fig. 1C) were from Box H/ACA guide RNAs containing K-loop (Hamma and Ferré-D'Amaré 2004) and K-turn (Afu-4 stem II) (Rozhdestvensky et al. 2003) motifs. The RNA used in the gel shift assay with YlxQ mutants (Fig. 6C) was derived from K-turn-containing Oceanobacillus iheyensis glyQ RNA. K-turn mutants of the lysine and SAM-I riboswitch were generated by QuikChange Lightning mutagenesis (Agilent Technologies) using wild-type plasmids as templates. The cSAMP3_2 RNA construct used for crystallization was identical to PDB ID 2GIS except that the GAAA tetraloop on L3 (nucleotides 50–53) was replaced with a 16-bp helical module capped with a UUCG tetraloop, which aided in crystallization. The sequence of the insertion is identical to that used by Baird and Ferré-D'Amaré (2010). All RNAs used in this study were transcribed in vitro from double-stranded DNA templates generated by PCR essentially as described (Xiao et al. 2008) using two 2′OMe residues at the 5′ positions of the reverse primer. Following amplification, the full 2-mL PCR reaction was added directly to make 5-mL transcription reactions. RNAs were purified under denaturing conditions, electroeluted, and washed once with 1 M KCl, followed by three washes with water; concentrated by ultrafiltration; and stored at 4°C until use.
Electrophoretic mobility shift analysis
Fifty micromolar K-turn- or K-loop-containing RNA was incubated with 100 μM protein for 15 min at room temperature in Binding Buffer [20 mM Tris-HCl at pH 8.0, 20 mM NaCl, 10 mM MgCl2, and 1 mM tris(2-carboxyethyl)phosphine (TCEP)]. Samples were then mixed with equal volumes of 2× Loading Buffer [1× THE buffer (33 mM Tris, 66 mM HEPES, 0.1 mM EDTA) and 50% (v/v) glycerol] and analyzed on prerun 8%–10% total, 29:1 polyacrylamide:acrylamide gels containing 0.5× THE buffer and 10 mM MgCl2. Electrophoresis was performed adjusting the power to maintain the gel at <30°C throughout the separation.
Isothermal titration calorimetry
RNA samples for ITC experiments were prepared by annealing in 100 mM KCl and 20 mM HEPES-KOH (pH 7.5) by heating for 2 min to 85°C and holding for 5 min at room temperature. 1.5 μL of MgCl2 was added, resulting in final concentrations of 45 μM RNA, 100 mM KCl, 20 mM HEPES-KOH, and 1 mM or 5 mM MgCl2 for various samples. Samples were equilibrated with ITC buffer (100 mM KCl, 20 mM HEPES-KOH, 1 mM DTT, and 1 mM or 5 mM MgCl2) via centrifugation in Amicon Ultra centrifugal filters 10K MWCO (Millipore) three times or via dialysis. ITC experiments were performed at 37°C with 10–20 μM RNA in the cell and protein or SAM in the syringe at 10-fold higher molar concentrations than the RNA. Experiments were performed in triplicate for K-loop/L7Ae (cell contents/syringe contents) and yitJ/YbxF; in duplicate for yitJ/YlxQ, lysC_CG/YbxF, yitJ/SAM, yitJ+YbxF/SAM; and single experiments for lysC_CGCG/YbxF, yitJ_UAUA/YbxF, yitJ_UA/YbxF. Data analysis was performed using Origin 7 (OriginLab) and Sedphat v9.4 (Houtman et al. 2007).
YbxF cocrystallization, structure determination, and refinement
cSAMP3_2 RNA at 200 μM in 26.5 mM HEPES-KOH (pH 7.5) and 53 mM KCl was heated for 2 min at 85°C and then kept for 8 min at room temperature. MgCl2 and SAM were added to concentrations of 17.9 mM and 538 μM, respectively, and the solution was incubated for 15 min at 37°C. Next, YbxF at a 1.4-fold molar excess to the RNA as well as spermine hydrochloride, cobalt hexamine, and DTT were added. Final composition of the RNP mixture was 150 μM RNA, 210 μM YbxF, 10 mM MgCl2, 10 mM SAM, 40 mM KCl, 20 mM HEPES-KOH, 1 mM spermine, 1 mM cobalt hexammine, and 1 mM DTT. Crystallization was performed by vapor diffusion at 21°C by mixing 1 μL of this solution with 1.2 μL of a reservoir solution containing 100 mM potassium cacodylate (pH 6.0), 200 mM MgCl2 and 25% (v/v) PEG400. Rod-shaped crystals appeared within 2 d and grew over a week to maximum dimensions of 300 × 100 × 100 μm3. Crystals were mounted in nylon loops and flash-frozen by plunging into liquid nitrogen without any additional cryoprotection. Diffraction data were collected at 100 K at the Advanced Light Source (ALS, Lawrence Berkeley National Laboratory) beamline 5.0.2 with 1 Å X-ray radiation from two crystals (Table 2) and reduced with the HKL package (Otwinowski and Minor 1997). The two crystals were grown under the same conditions, but in separate drops. The crystals have the symmetry of space group C2, with unit cell dimensions a = 179.5 Å, b = 54.5 Å, c = 106.4 Å, β = 108.4 (Crystal I), and a = 191.8 Å, b = 54.3 Å, c = 106.4 Å, β = 116.6 (Crystal II). The structure was determined by molecular replacement using PHASER (McCoy et al. 2007) and a search model containing one copy of the SAM-I riboswitch aptamer (PDB ID 3GX5 from which the L3 GAAA tetraloop and the SAM ligand had been removed), and one copy of the M. jannaschii L7Ae protein (PDB ID 1SDS, chain A, with the N and C termini truncated to the expected full length of B. subtilis YbxF, according to the sequence alignment of Fig. 1A). The top solution obtained with data from Crystal I (translation function Z-scores of 13.6 and 12.6 for the RNA and the RNA plus protein, respectively, and an overall log-likelihood gain of 441) had Rfree = 49% after rigid body, individual isotropic B-factor, and simulated annealing refinement (Brünger et al. 1998). 2|Fo| − |Fc| Fourier syntheses calculated with phases from this partial model and Crystal I amplitudes revealed no density for P4 of the RNA. A second round of molecular replacement was performed with two copies of an RNA model from which P4 had been deleted and an L7Ae model whose side chains had been changed to correspond to the sequence of YbxF against Crystal II amplitudes. The top solution (translation function Z-scores of 11.3 and 19.9 and 10.7 for one RNA, two RNAs, and two RNAs plus the protein, respectively, and an overall log-likelihood gain of 944) had Rfree = 41.1% after rigid body, individual isotropic B-factor, and simulated annealing refinement. Iterative rounds of manual model building (Jones et al. 1991) interspersed with simulated annealing, energy minimization, and individual isotropic B-factor refinement (Brünger et al. 1998) against Crystal II amplitudes produced an essentially complete model of the two RNAs in the crystallographic asymmetric unit. One of the two YbxF molecules in the asymmetric unit is better ordered (mean B-factors of 57.8 Å2 and 90.6 Å2 for chains A and B, respectively). The bound SAM molecules were not built until near the end of the refinement, although clear residual electron density corresponding to the metabolite was present in both RNAs starting with the first difference maps. The final model comprises 126 RNA residues in chains C and D and residues 2–82 and 2–78 in YbxF chains A and B, respectively, 15 waters, 22 magnesium ions, 17 cobalt hexammine ions, and 2 SAM molecules and has a cross-validated σA coordinate precision of 0.66 Å (Table 2). Ions were assigned as either cobalt hexammine or magnesium based on distance from apparent ligands, refined individual isotropic B-factors of the presumed ion and its ligands, real-space R-factor of the ions in simulated annealing-omit 2|Fo| − |Fc| maps, and overall effect on Rfree. Ramachandran analysis shows that 80% of the amino acids are in the most favored regions, 20% in the additional allowed regions, and none in the disallowed regions. All structural analyses were performed with chains A and C. Atomic coordinates and structure factor amplitudes for Crystal II have been deposited with the PDB with accession code 3V7E.
YlxQ crystallization, structure determination, and refinement
B. subtilis YlxQ was crystallized by vapor diffusion at 4°C by mixing equal volumes of 14 g/L protein in Buffer C and reservoir solution composed of 0.2 M ammonium acetate (pH 7.0), 0.1 M sodium citrate (pH 5.6), and 30% (w/v) PEG 4000. Needle-shaped crystals grew in 3–6 d to maximum dimensions of 2000 × 20 × 20 μm3. Crystals were mounted in nylon loops and flash-frozen by plunging into liquid nitrogen without additional cryoprotection. Portions of the crystal outside of the nylon loop diffracted better than portions within the loop and were used for data collection at 100 K at the ALS beamline 5.0.1 using 0.9795 Å X-radiation, and reduced with the HKL package (Table 2; Otwinowski and Minor 1997). The crystals have the symmetry of space group P212121, with unit cell dimensions a = 50.4 Å, b = 61.3 Å, c = 94.2 Å. The structure was determined by molecular replacement using PHASER (McCoy et al. 2007) and a search model composed of three copies of the B. halodurans BH 2414 protein (identified as a hypothetical protein of unknown function, PDB ID 3ON1). The top solution (translation function Z-scores of 7.4, 9.5, and 15.4 for one, two, and three protein copies, respectively, and an overall log-likelihood gain of 429) had an Rfree = 35.3% when subjected to rigid body, simulated annealing, and individual isotropic B-factor refinement (Brünger et al. 1998) at 1.50 Å resolution. Iterative rounds of manual model building (Jones et al. 1991) interspersed with simulated annealing, energy minimization, and individual isotropic B-factor refinement (Brünger et al. 1998) against diffraction data extending to 1.55 Å resolution produced the final model, which comprises residues 3–99 of each of the three copies of YlxQ, 246 water molecules, three citrates, and two potassium ions (2532 nonhydrogen atoms). At this resolution, the identity of the citrate ions was readily apparent from residual electron density maps. Potassium ions were assigned based on their refined individual isotropic B-factors and those of their protein or water ligands. This model has a cross-validated σA coordinate precision of 0.13 Å (Fig. 5B; Table 2). Ramachandran analysis shows that 93.6% of the amino acids are in the most favored regions, 6.4% in the additional allowed regions, and none in the disallowed regions. Except where noted, structural analyses of YlxQ were performed with chain B, which has the lowest mean B-factor. Atomic coordinates and structure factor amplitudes have been deposited with the PDB with accession code 3V7Q. Continuum electrostatic potentials were calculated with DelPhi (Honig and Nicholls 1995) using default values for charges and dielectric constants. Structure figures were generated with MacPyMOL (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC).
ACKNOWLEDGMENTS
We thank the staff of Beamlines 5.0.1 and 5.0.2 of the ALS for data collection support, G. Piszczek for ITC support, D.-Y. Lee and R. Levine for generous help with mass spectrometry, and W. Winkler for discussions. This work was supported by the intramural program of the National Heart, Lung and Blood Institute, NIH.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.031518.111.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N 2010. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38: W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Ferré-D'Amaré AR 2010. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16: 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Blouin S, Lafontaine DA 2007. A loop–loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13: 1256–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, et al. 1998. Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273: 1058–1073 [DOI] [PubMed] [Google Scholar]
- Burgess RR 1991. Use of polyethyleneimine in purification of DNA-binding proteins. Methods Enzymol 208: 3–10 [DOI] [PubMed] [Google Scholar]
- Chao JA, Williamson JR 2004. Joint X-ray and NMR refinement of the yeast L30e–mRNA complex. Structure 12: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Cho I-M, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V 2010. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc Natl Acad Sci 107: 14573–14578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, McLachlan AD 1986. Solvation energy in protein folding and binding. Nature 319: 199–203 [DOI] [PubMed] [Google Scholar]
- Eng FJ, Warner JR 1991. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell 65: 797–804 [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR 2010. Use of the spliceosomal protein U1A to facilitate crystallization and structure determination of complex RNAs. Methods 52: 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Zhang X, Qu G, Biswas S, Suryadi J, Brown BA, Maxwell ES 2010. Signature amino acids enable the archaeal L7Ae box C/D RNP core protein to recognize and bind the K-loop RNA motif. RNA 16: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Héroux A, Rambo RP, Batey RT 2008. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 283: 22347–22351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma T, Ferré-D'Amaré AR 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 12: 893–903 [DOI] [PubMed] [Google Scholar]
- Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M 2001. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res 29: 2733–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppell B, Lafontaine DA 2008. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry 47: 1490–1499 [DOI] [PubMed] [Google Scholar]
- Heppell B, Blouin S, Dussault AM, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA 2011. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol 7: 384–392 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C 1995. Dali: A network tool for protein structure comparison. Trends Biochem Sci 20: 478–480 [DOI] [PubMed] [Google Scholar]
- Honig B, Nicholls A 1995. Classical electrostatics in biology and chemistry. Science 268: 1144–1149 [DOI] [PubMed] [Google Scholar]
- Houtman JC, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, Schuck P 2007. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: Application to adaptor protein complexes in cell signaling. Protein Sci 16: 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA 2001. The kink-turn: A new RNA secondary structure motif. EMBO J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B-K, Park C-J, Fernandez CF, Chim N, Ding Y, Chanfreau G, Feigon J 2011. Structure of H/ACA RNP protein Nhp2p reveals cis/trans isomerization of a conserved proline at the RNA and Nop10 binding interface. J Mol Biol 411: 927–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Bork P, Sander C 1994. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins, and a ribosome modification enzyme. Nucleic Acids Res 22: 2166–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JF, Tran EJ, Maxwell ES 2002. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res 30: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR 2010. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329: 845–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JB, Lilley DMJ 2006. The role of specific 2′-hydroxyl groups in the stabilization of the folded conformation of kink-turn RNA. RNA 13: 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ding F, Chowdhury A, Pradhan V, Tomsic J, Holmes WM, Henkin TM, Ke A 2010. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S Box/SAM-I riboswitch. J Mol Biol 404: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milon P, Tischenko E, Tomsic J, Caserta E, Folkers G, La Teana A, Rodnina MV, Pon CL, Boelens R, Gualerzi CO 2006. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci 103: 13962–13967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2006. Structure of the S-denosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Montange RK, Mondragón E, van Tyne D, Garst AD, Ceres P, Batey RT 2010. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J Mol Biol 396: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci 98: 4899–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolivos S, Carpousis AJ, Clouet-d'Orval B 2005. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res 33: 6507–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Lührmann R 1999. Functional interaction of a novel 15.5kD [U4/U6•U5] tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J 18: 6119–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk J, Górnicki P, Maj L, Dementieva I, Laskowski R, Joachimiak A 2001. Streptococcus pneumonia YlxR at 1.35 Å shows a putative new fold. Acta Crystallogr D Biol Crystallogr 57: 1747–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Price SR, Evans PR, Nagai K 1998. Crystal structure of the spliceosomal U2B″–U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature 394: 645–650 [DOI] [PubMed] [Google Scholar]
- Reuter K, Nottrott S, Fabrizio P, Lührmann R, Ficner R 1999. Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein. J Mol Biol 294: 515–525 [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in archaea. Nucleic Acids Res 31: 869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KT, McPhee SA, Ouellet J, Lilley DMJ 2010. A structural database for K-turn motifs in RNA. RNA 16: 1463–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KT, Daldrop P, Lilley DMJ 2011. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure 19: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ 2008. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455: 1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shazand K, Tucker J, Grunberg-Manago M, Rabinowitz JC, Leighton T 1993. Similar organization of the nusA-infB operon in Bacillus subtilis and Escherichia coli. J Bacteriol 175: 2880–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka L, Fucik V, Krasny L, Barvik I, Jonak J 2007. YbxF, a protein associated with exponential-phase ribosomes in Bacillus subtilis. J Bacteriol 189: 4809–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadi J, Tran EJ, Maxwell ES, Brown BA 2005. The crystal structure of the Methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry 44: 9657–9672 [DOI] [PubMed] [Google Scholar]
- Turner B, Lilley DMJ 2008. The importance of G•A hydrogen bonding in the metal ion- and protein-induced folding of a kink turn RNA. J Mol Biol 381: 431–442 [DOI] [PubMed] [Google Scholar]
- Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DMJ 2005. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA 11: 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell 6: 1331–1342 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Ségault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Lührmann R 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466 [DOI] [PubMed] [Google Scholar]
- Winkler WC. 2002 “RNA elements required for T box antitermination.” PhD thesis, Ohio State University, Columbus. [Google Scholar]
- Winkler WC, Grundy FJ, Murphy BA, Henkin TM 2001. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 7: 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Edwards TE, Ferré-D'Amaré AR 2008. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem Biol 15: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lau MW, Ferré-D'Amaré AR 2010. Ribozymes and riboswitches: Modulation of RNA function by small molecules. Biochemistry 49: 9123–9131 [DOI] [PMC free article] [PubMed] [Google Scholar]