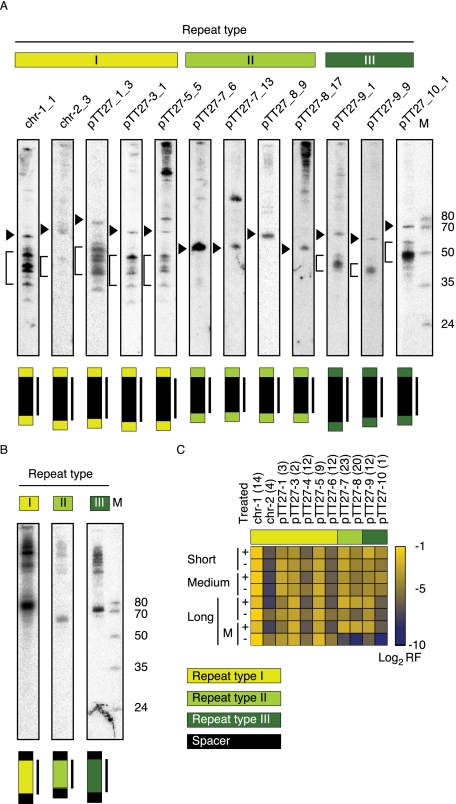
FIGURE 6.
Northern blot analyses of CRISPR spacer and repeat-sequences. Northern blots were performed with 5 μg total RNA using oligodeoxynucleotide probes designed to detect the CRISPR spacer (A) or repeat (B) sequences. The region probed against is indicated above the lane. The −8 to −9 precursor sequence is indicated by an arrowhead on the left side of each spacer panel. For type II, no other dominant band is detected, consistent with the observation that the −8 to −9 precursor comprised the mature crRNA for this type. For repeat-sequence type I and III, a bar is depicted along the mature crRNA bands. Total RNA was loaded next to a size marker cocktail (M). The sizes are indicated next to the gel. For sequences, see the Supplemental Materials and Methods. (C) Expression profile of CRISPRs in the treated (+) and untreated (−) libraries. The heatmap represents the log2 of the relative frequency of reads that were mapped to a specific CRISPR out of the total number of CRISPR reads for each corresponding library. For the long library, the values of the mature form (M) of the crRNA is also displayed. For the determination of reads corresponding to mature crRNA reads, see Materials and Methods. For an extended view of crRNA precursor and mature expression, see Supplemental Figure S4B. Interestingly, we observe a common consistent expression pattern for the three paralogous pairs of adjacent CRISPRs: chr-1:chr-2, pTT27-3:pTT27-4, and pTT27-5: pTT27-6, where the upstream CRISPR in each pair has a much higher expression level compared with the downstream CRISPR.