Although large-scale transcriptome analyses have pointed to the presence of thousands of putative long noncoding RNAs (lncRNAs) in higher eukaryotes, only a small fraction of them have been functionally characterized. To gain insight into the genomic architecture and sequence features of lncRNAs, the authors have performed an in-depth computational analysis on the functionally characterized lncRNAs. This study revealed that these RNAs not only lack large open reading frames, but also are unlikely to code even for small peptides, and thus, their cellular function is indeed RNA-mediated. Furthermore, these RNAs have fewer introns, a low GC content, and higher structural flexibility compared with protein-coding sequences, features that are also observed in the noncoding 3′ untranslated regions (3′ UTRs) of protein-coding RNAs. Together with similarities in hexamer composition between 3′ UTRs and lncRNAs, this study suggests that the regulatory RNA sequences in the cell have evolved under similar evolutionary and functional constraints.
Keywords: long noncoding RNAs, lncRNAs, database, miRNAs, splicing, peptide, 3′ UTR
Abstract
Recent transcriptome analyses have indicated that a large part of mammalian genomes are transcribed into long non-protein-coding RNAs (lncRNAs). However, only a very small fraction of them have been individually studied, and whether the majority of lncRNAs found in large-scale studies have a cellular role is debated. To gain insight into the sequence features and genomic architecture of the subset of lncRNAs that have been proven to be functional, we created a database containing studied lncRNAs manually culled from the literature along with a parallel database containing all annotated protein-coding human RNAs. The Functional lncRNA Database, which contains 204 lncRNAs and their splicing variants, is available at valadkhanlab.org/database. Analysis of the lncRNAs and their comparison to protein-coding transcripts revealed sequence features including paucity of introns and low GC content in lncRNAs, which could explain several biological characteristics of these transcripts, such as their nuclear localization and low expression level. The predicted ORFs in lncRNAs have poor start codon and ORF contexts, which would lead to activation of the nonsense-mediated decay pathways and thus make it unlikely for most lncRNAs to code for even short peptides. Interestingly, our analyses revealed significant similarities between the lncRNAs and the 3′ untranslated regions (3′ UTRs) in protein-coding RNAs in structural features and sequence composition. The presence of these intriguing parallels between the lncRNAs and 3′ UTRs, which constitute the two main components of the RNA-mediated cellular regulatory system, indicates that highly similar evolutionary constraints govern the function of regulatory RNA sequences in the cell.
INTRODUCTION
A fascinating and unanticipated outcome of large-scale transcriptome analyses has been the discovery that a significant fraction of the genome of higher eukaryotes is transcribed into thousands of long RNAs that do not seem to be translated into proteins (Carninci et al. 2005; The ENCODE Project Consortium 2007). While the biogenesis and cellular function of most long noncoding RNAs (lncRNAs) remain unknown, existing data obtained from the study of a small number of lncRNAs suggest that many of them play critical regulatory roles in diverse cellular processes including chromatin remodeling, transcription, post-transcriptional processing, and intracellular trafficking (Hannon et al. 2006; Mercer et al. 2009; Ponting et al. 2009; Wilusz et al. 2009; Chen and Carmichael 2010; Hung and Chang 2010; Valadkhan and Nilsen 2010; Pauli et al. 2011). Interestingly, the ratio of noncoding to protein-coding sequences in eukaryotic genomes increases along with the rise in the level of complexity of the organism (Taft et al. 2007). Unlike the eukaryotic proteome, which is relatively conserved among different organisms (Clamp et al. 2007), many long noncoding transcripts show a high level of sequence divergence even among closely related species and in a number of cases may even participate in generation of interspecies differences (Pang et al. 2006; Pollard et al. 2006a,b; Prabhakar et al. 2006; Ponting et al. 2009). Taken together, existing data suggest that lncRNAs constitute a highly abundant, rapidly evolving class of cellular factors with a wide range of cellular functions (Hannon et al. 2006; Clamp et al. 2007; Rymarquis et al. 2008; Mercer et al. 2009; Ponting et al. 2009; Wilusz et al. 2009).
While our knowledge of the long noncoding transcriptome is still very incomplete, current estimates predict the presence of tens of thousands of lncRNAs in the mammalian transcriptome, which suggests that their number likely exceeds the approximately 20,500 protein-coding transcripts in human (Clamp et al. 2007; Pheasant and Mattick 2007; Taft et al. 2007; Mercer et al. 2009; Ponting et al. 2009). Although many lncRNAs are thousands of nucleotides long, there is no strict minimal size requirement for classifying a noncoding transcript as a “long” noncoding RNA. Rather, the term “long” and an arbitrary lower size limit of 200 used in many studies mainly serve to distinguish these transcripts from the small housekeeping or regulatory RNAs such as the snRNAs, snoRNAs, tRNAs, or the miRNAs and other Ago-associated small regulatory RNAs. Similarly, there are no clear-cut, uniformly used criteria for determining the noncoding character of an RNA (Frith et al. 2006a; Clamp et al. 2007; Dinger et al. 2008). The most widely accepted method for distinguishing protein-coding and noncoding RNAs among novel transcripts is analysis of the length of the open reading frames (ORFs) in each transcript as the primary criterion, followed by determination of the level of phylogenetic conservation of ORFs in transcripts that are classified as noncoding by the use of the first criterion. Since the majority of annotated eukaryotic proteins are longer than 100 amino acids, in most studies, RNAs that lack an ORF of 300 nt or longer have been classified as putative noncoding RNAs (Frith et al. 2006a; Clamp et al. 2007; Dinger et al. 2008). However, there are several bona fide eukaryotic proteins that are shorter than 100 amino acids, and it has been suggested that they may be much higher in number than previously thought (Frith et al. 2006b). Because the majority of annotated eukaryotic protein-coding ORFs show a high level of phylogenetic conservation, the level of conservation of the ORF and the rate of synonymous to nonsynonymous substitutions have often been used as additional criteria in distinguishing the protein-coding transcripts containing bona fide functional ORFs from noncoding transcripts among novel RNAs (Frith et al. 2006a; Clamp et al. 2007; Dinger et al. 2008). While the combination of the two sets of criteria or the use of more stringent ORF size limits (Jia et al. 2010) reduces the possibility of misclassification of protein-coding transcripts as noncoding, it is still possible that many understudied transcripts are incorrectly assigned to one or the other group, even with the use of more sophisticated algorithms (Clamp et al. 2007; Kageyama et al. 2011).
Another complicating factor is the presence of bifunctional RNAs, transcripts that function as a noncoding transcript under certain conditions and are translated into functional proteins in other situations (Dinger et al. 2008; Ulveling et al. 2011). In addition, large-scale transcriptome analyses have predicted that a significant fraction of protein-coding RNAs may have an alternatively processed isoform or one transcribed from an alternative promoter that may function as a noncoding RNA (Carninci et al. 2005; The ENCODE Project Consortium 2007; Kapranov et al. 2007; Guttman et al. 2010). These classification issues stem from and reveal a general dearth of information on the long noncoding RNAs, which highlights the need for in-depth study of their sequence and structural features and genomic architecture. Such studies will not only provide more accurate criteria for determining if an RNA is likely to be translated or not, but also they uncover important clues into the cellular biogenesis, evolution, and function of these novel transcripts and will form the basis for future mechanistic studies.
While large-scale transcriptome analyses have generated several databases of putative long noncoding RNAs (Pang et al. 2005; Dinger et al. 2009; Mituyama et al. 2009; Jia et al. 2010; among others), only a few of these transcripts have been individually studied and proven to be indeed non-protein-coding RNAs with a cellular function. A significant fraction of the identified putative lncRNAs show a developmentally regulated or tissue-specific expression pattern or are associated with functional protein complexes, which suggests that they are functional transcripts (Ravasi et al. 2006; Khalil et al. 2009). However, it is also possible that many lncRNAs result from background transcription and thus, have no functional significance (Nóbrega et al. 2004). Nonetheless, it is clear that at least a subset of lncRNAs play important functional roles in the cell and thus, they represent a novel and rapidly expanding class of functional cellular factors (Hannon et al. 2006; Rymarquis et al. 2008; Mercer et al. 2009; Ponting et al. 2009; Wilusz et al. 2009).
As a first step toward understanding the genomic architecture and sequence and structural features of this class of RNAs in higher eukaryotes, we have compiled a database of the subset of mammalian lncRNAs that have been individually studied and shown to be both noncoding and functional (see also Amaral et al. 2011). The Functional lncRNA Database (valadkhanlab.org/database) currently contains 204 lncRNAs, with lncRNAs described in human constituting the majority. Taking advantage of this resource, we have performed an in-depth in silico analysis to define the overall sequence and genomic architecture of these transcripts. To elucidate features unique to this class of functional RNAs, a parallel analysis was performed on a database containing all human protein-coding genes. The results revealed the presence of several common sequence features among lncRNAs, including the paucity or absence of introns and the low GC content, which could at least partly explain the nuclear localization and low expression levels observed in many lncRNAs (Kelly and Corbett 2009; Shabalina et al. 2010). A detailed analysis of their protein-coding capacity indicated that while the lncRNAs do contain short ORFs, their poor start codon context makes efficient translation unlikely. Furthermore, the location of ORFs within most lncRNAs was likely to trigger nonsense-mediated decay (NMD) if the short ORFs were to be translated. Together with the nuclear localization of many lncRNAs (Khalil et al. 2009), these findings make it highly unlikely for the lncRNAs to mediate their function through coding for even short functional polypeptides. Similarly, sequence analysis indicated that except in a very small number of cases, the lncRNAs did not harbor sequences that resemble miRNA precursors. Interestingly, our analyses indicated that the lncRNAs have significant similarities in structural stability, sequence composition, and architecture with the 3′ UTRs, the noncoding regulatory sequences found in protein-coding RNAs. This finding suggests that the lncRNAs, a large fraction of which play regulatory roles in the cell, and the 3′ UTRs may belong to an RNA-based cellular regulatory system and have evolved under similar evolutionary pressures and thus, have a similar sequence composition and structural flexibility that may point to a similar mode of cellular interactions.
RESULTS AND DISCUSSION
To gain insight into the features that offer clues into the evolution, biogenesis, and cellular function of lncRNAs as a group, we created a database containing the functionally validated lncRNAs in higher eukaryotes. As mentioned above, while a large number of putative lncRNAs have been identified in recent years, their designation as non-protein-coding transcripts and whether they are indeed functional entities in the cell are subject to extensive debate. Thus, to obtain insight into features of lncRNAs as functional cellular factors, we based our analysis on the small fraction of lncRNAs that have been experimentally proven to be both noncoding and have a cellular function. We included lncRNAs that had been shown to be noncoding based on experimental evidence beyond simple computational analyses commonly performed in large-scale studies, for example, through in vivo translation assays or mutational analyses. Furthermore, the included RNAs had been shown to have a clear cellular function that in many cases have been confirmed by several independent studies. Based on existing literature and available databases (Amaral et al. 2011), we included 118 mammalian lncRNAs comprising 99 and 19 lncRNAs described in human and mouse, respectively, that along with their described isoforms comprise the 204 entries in our database (Table 1). This database, which contains a range of sequence-based information for each lncRNA, is available online at www.valadkhanlab.org/database and contains both intergenic lncRNAs (lincRNAs) and lncRNAs that overlap with other transcripts in sense or antisense orientations. To be able to discern features unique to lncRNAs, we developed a parallel database containing all annotated human protein-coding RNAs in the public databases. Because many protein-coding genes have alternatively processed variants that may or may not code for a protein (Carninci et al. 2005; The ENCODE Project Consortium 2007), we only included the annotated protein-coding variants of each protein-coding gene in our database. Since many lncRNAs seem to evolve rapidly and show significant differences even among closely related mammalian species, we restricted our analysis to the lncRNAs that have been described as functional RNAs in human, which correspond to the majority of the lncRNAs in our database (Table 1).
TABLE 1.
The functional lncRNA database
Architecture of lncRNA transcripts
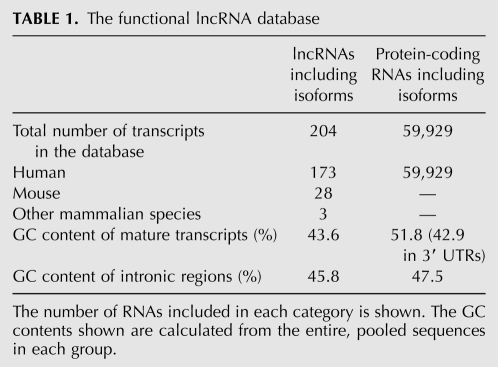
A preliminary analysis of the human functional lncRNAs and protein-coding transcripts in the database revealed significant differences in their gene architecture. The lncRNA primary, unspliced transcripts were considerably shorter than protein-coding RNAs (mRNAs), with a median length of 6 versus 24 kb, respectively, and had a narrower size distribution (Fig. 1A). This length difference largely stemmed from the smaller number of introns in lncRNAs, with a median of one versus seven introns in lncRNAs and protein-coding transcripts, respectively (Fig. 1B,C). Once the transcripts were spliced, however, the difference between the overall length of lncRNAs and protein-coding mature transcripts was not statistically significant (P-value = 0.035 based on a Student's t-test) (Fig. 1D). While lncRNAs had fewer introns, the introns were similar in length and GC content in the two groups (Fig. 1E; Table 1). Sixteen percent of lncRNAs in the database had annotated isoforms; however, due to the low expression level of these RNAs and their tissue specificity, it is likely that the true extent of alternative splicing and alternative promoter and poly(A) site usage in these transcripts is higher than suggested by current data. The individual exons in spliced transcripts were longer in lncRNAs compared with protein-coding RNAs and had a significantly lower GC content that fell within the range observed in intronic sequences (Fig. 1F,G; Table 1). Interestingly, it has been shown that a higher GC content correlates with a higher RNA steady-state level that stems from a higher rate of transcription or processing (Kudla et al. 2006). This, in turn, may at least partly explain the often much lower cellular level of lncRNAs compared with the protein-coding RNAs (Carninci et al. 2005; Mercer et al. 2009; Ponting et al. 2009; Wilusz et al. 2009).
FIGURE 1.
The genomic architecture of the lncRNAs. The lengths are shown in nucleotides. When the difference between the two groups is statistically significant, the P-values obtained using the Kolmogorov-Smirnov test (KS test; see Materials and Methods) are indicated on top. For details on the box plots, see Materials and Methods. (mRNA) Protein-coding RNAs. (A) The lncRNA genes are significantly shorter than the protein-coding genes. (B,C) The lncRNAs contain fewer introns compared with protein-coding RNAs. (D) The mature, spliced transcripts of lncRNAs and protein-coding RNAs are not significantly different in size. (E) Introns in lncRNAs are similar in size to those in protein-coding RNAs. (F) Distribution of the GC content of individual RNAs in the functionally validated lncRNAs (series labeled lncRNAs), mRNAs, and lncRNAs obtained in a large-scale analysis (Guttman et al. 2009; Khalil et al. 2009). (G) In lncRNAs that contain introns, the exons are longer than those in protein-coding RNAs. (H) The length of intron-less RNAs in both lncRNAs and protein-coding RNAs.
More than 30% of the human lncRNAs in our database of functional lncRNAs did not contain any introns, compared with <3% of human protein-coding RNAs (Fig. 1C). The lack of introns in these transcripts did not necessarily stem from a very short length, since many of the intron-less transcripts are thousands of nucleotides long (Fig. 1H). Thus, the high percentage of intron-less transcripts among lncRNAs may reflect an evolutionary or functional feature of these RNAs. The low number of introns has also been noted in putative lncRNAs obtained in some but not all large-scale studies (Ravasi et al. 2006; Guttman et al. 2010; Ørom et al. 2010). It has been shown that at least among protein-coding RNAs, the intron-less transcripts as a group have a lower transcriptional expression level and a more tissue-specific expression pattern compared with spliced messages (Shabalina et al. 2010). Furthermore, the intron-less mRNAs were evolutionarily younger, showed lower interspecies sequence conservation, and seemed to code disproportionately for regulatory proteins (Shabalina et al. 2010). While the evolutionary and functional significance of splicing in lncRNAs is yet to be determined, many of the characteristics listed above for intron-less protein-coding genes are also prominent features found in the majority of lncRNAs (Pang et al. 2006; Mercer et al. 2009; Ponting et al. 2009; Wilusz et al. 2009; Guttman et al. 2010). Interestingly, many lncRNAs are nuclear transcripts and are thought to perform their cellular function through regulation of gene expression (Khalil et al. 2009). Since splicing is one of the major nuclear export signals in higher eukaryotes (Kelly and Corbett 2009), the absence or paucity of introns in lncRNAs may at least partially serve to maintain their functionally required nuclear localization.
The protein-coding capacity of functional lncRNAs
Another consequence of nuclear localization of lncRNAs is their spatial separation from the cytoplasmic translation machinery. While a strict nuclear localization automatically prohibits a transcript from being translated, several lncRNAs are cytoplasmic or are found in both compartments (Clamp et al. 2007; Khalil et al. 2009; Mercer et al. 2009; Ponting et al. 2009; Wilusz et al. 2009). Furthermore, even in the case of nuclear lncRNAs, a developmental or environmentally regulated change in subcellular localization cannot be ruled out (Chen and Carmichael 2009). Importantly, it has been suggested, although not yet proven, that even the small ORFs in putative lncRNAs may be translated into small but functional peptides (Kondo et al. 2010). Thus, a thorough analysis of the inherent protein-coding capacity of the lncRNAs using a combination of computational and experimental methods is necessary for unequivocal classification of a transcript as a noncoding RNA. As a first step in this direction, we analyzed the protein-coding potential of the studied lncRNAs in our database.
As detailed in the Introduction, in large-scale studies, the size and level of conservation of predicted ORFs in newly discovered transcripts are commonly used as rational criteria for classifying them as protein-coding or noncoding. However, a large fraction of the studied lncRNAs were not discovered in such studies but, rather, were accidentally found and were proven to be noncoding after attempts at characterizing the protein coded by them failed. Thus, computational analysis of these transcripts also provides an opportunity for validating and judging the effectiveness of the criteria used in large-scale studies for distinguishing the noncoding transcripts.
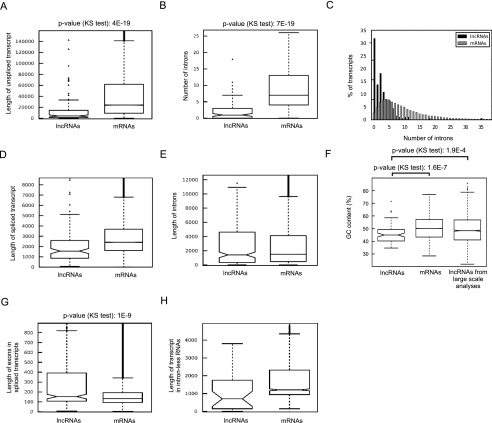
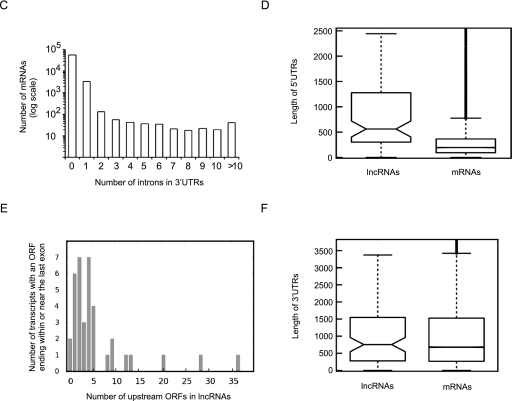
Analysis of the predicted ORFs in the lncRNAs and protein-coding RNAs indicated that transcripts in both groups contained a large number of predicted open reading frames (ORFs) (Fig. 2A). Only 17 lncRNAs, all of them shorter than 1 kb in length, did not contain any predicted ORFs longer than 100 nt (33 amino acids). Analysis of all predicted ORFs in both lncRNAs and protein-coding RNAs indicated that while the majority were shorter than 300 nt, both groups contained a large number of predicted ORFs that were longer (Fig. 2B). Thus, adopting the commonly used ORF size limit of 300 nt as the primary criterion for distinguishing protein-coding and noncoding RNAs would have led to incorrect classification of >20% of the studied human functional lncRNAs in our database as protein-coding RNAs, including such well-studied lncRNAs as HOTAIR and XIST (Fig. 2B,C). The use of ORF size limits of 400 nt and 500 nt still led to misclassification of >10% and >5% of the lncRNAs, respectively. On the other hand, analysis of the longest predicted ORF in each transcript indicated that 2% of transcripts annotated as protein-coding in public databases did not contain an ORF longer than 300 nt (Fig. 2C). Thus, nearly 1300 RNAs currently annotated as protein-coding RNAs would be classified as lncRNAs by the use of a 300-nt ORF size as the sole criterion for distinguishing mRNAs from lncRNAs (Clamp et al. 2007; Dinger et al. 2008).
FIGURE 2.
The short predicted ORFs in lncRNAs resemble the non-protein-coding ORFs in protein-coding RNAs. All lengths are shown in nucleotides. (mRNA) Protein-coding RNAs. When the difference between the two groups is statistically significant, the P-values obtained using the KS test are indicated on top. (A) Both lncRNAs and mRNAs contain a large number of predicted ORFs per transcript. (B) The distribution of length of all predicted ORFs in mRNAs and lncRNAs. (C) The longest ORFs in mRNAs are significantly larger than the longest ones in lncRNAs. (D) After excluding the longest ORF in mRNAs, the rest of the predicted ORFs in mRNAs resemble the ORFs in lncRNAs in their length. (E) Both lncRNA and mRNA sequences contain a large number of interspersed stop codons. (F) The longest stop-codon-free region in mRNAs is significantly longer than those in lncRNAs.
As detailed above, the majority of the predicted ORFs in all three reading frames in protein-coding and lncRNAs were small, with a median length of ∼150 nt in both groups (Fig. 2B), which corresponds to the size range of fortuitous ORFs in an RNA of 1 kb or longer length (Senapathy 1986; Clamp et al. 2007; Dinger et al. 2008). However, the length of the longest predicted ORF in each transcript is much larger in the protein-coding RNAs compared with lncRNAs, with a median ORF length of 1200 nt versus 250 nt, respectively (Fig. 2C). Once we omitted the longest predicted ORF in the protein-coding transcripts, which in almost all cases corresponds to the functional translated ORF (see Materials and Methods), the remaining untranslated ORFs in protein-coding RNAs were identical in length to the ORFs in noncoding transcripts (cf. Fig. 2D and 2B). This finding is consistent with the ORFs predicted in lncRNAs being fortuitous occurrences. To determine if the use of noncanonical start codons can create longer ORFs and thus an improved protein-coding capacity in lncRNAs, we analyzed the frequency of stop codons in the three reading frames in the two groups. The results indicated that other than a single long stop-codon-free region in one of the reading frames in mRNAs, the lncRNAs and mRNAs had the same high density of stop codons in the rest of their sequence, with the median size of the longest stop-codon-free region in lncRNAs being 300 nt (Fig. 2E,F). Thus, even with the use of noncanonical start codons, the protein-coding capacity of functional lncRNAs is highly limited.
Predicted ORFs in functional lncRNAs have poor start codon contexts
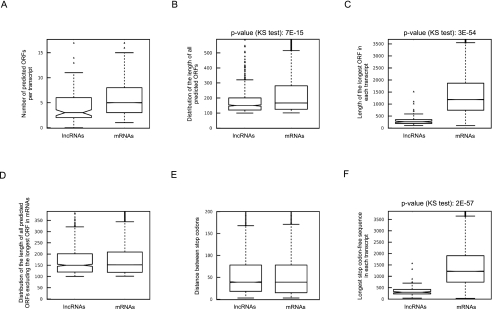
As detailed above, while the longest predicted ORFs in lncRNAs are in general smaller than those in protein-coding RNAs, there is some overlap between the ORFs in these two classes of transcripts, and thus, size cannot be a reliable criterion for defining the protein-coding capacity of an ORF (Fig. 2C). Furthermore, it has been suggested that lncRNAs may code for short peptides that mediate their cellular function, and thus, whether the function of these transcripts is truly RNA-mediated has been questioned (Kondo et al. 2010). Since analysis of the phylogenetic conservation of the short ORFs found in lncRNAs is of little use in this respect, because such coded peptides may have a high evolutionary rate and yet be functional, and considering the fundamental importance of determining the mode of function of these transcripts, it was essential to determine the inherent peptide-coding potential of the short predicted ORFs in lncRNAs. To this end, we first analyzed the context of the translation start site, which is a critical factor in determining the likelihood and efficiency of translation, in the ORFs of both protein-coding and lncRNAs. Analysis of the start codon context of the longest ORFs in the two sets of RNAs indicated the presence of a significantly higher number of start and stop codons upstream of the longest ORF in the putative “5′ UTR” in lncRNAs that can result in abortive translation initiation events (Fig. 3A,B). Many lncRNAs even have several shorter ORFs ranging in size from 100 nt to several hundred nucleotides within this region (Fig. 3C). Analysis of the sequences immediately upstream of the translation start site (nucleotides −30 to 0 relative to the start codon) further confirmed this finding by showing that the longest ORFs in protein-coding RNAs have significantly fewer start and stop codons in this region compared with the predicted ORFs in lncRNAs and the rest of the ORFs in protein-coding RNAs (Fig. 3D,E). Since in cap-dependent translation, the sequence of an RNA is scanned for a start codon beginning from the 5′ end of the transcript, the presence of start codons or upstream short ORFs in lncRNAs strongly reduces the likelihood of translation. Indeed, it has been shown that the presence of such upstream ORFs can lead to up to 80% reduction in translation efficiency and may even reduce the stability of the mRNAs (Calvo et al. 2009; Wethmar et al. 2010). Finally, these analyses indicated the presence of sequences known to be associated with efficient translation (Kozak 2005) around the start codon of the longest predicted ORF in protein-coding RNAs, but not in the rest of the predicted ORFs in these transcripts or in the ORFs predicted in lncRNAs (Fig. 3F).
FIGURE 3.
The sequence context of the predicted ORFs in lncRNAs is incompatible with translation. The numbers shown on top of the A–C panels are the P-values obtained using the KS test. (A,B) Compared with the translated ORF in mRNAs, which is almost always the longest ORF, the longest ORF in lncRNAs has a large number of start and stop codons in its putative 5′ UTR. (C) lncRNAs have a higher number of upstream long ORFs (>100 nt long) compared with mRNAs. (D,E) The predicted ORFs in lncRNAs resemble the noncoding ORFs in mRNAs in terms of the number of start and stop codons in the immediate vicinity (positions −1 to −30) of the initiation codon of each ORF. The columns labeled “Longest” correspond to the longest ORF in each transcript. The lanes labeled “All-longest” contain all the rest of the predicted ORFs that are >100 nt. (F) The ORFs in lncRNAs and untranslated ORFs in mRNAs lack the sequence context known to enhance translation at positions −3 to +4 of the initiation codon (arrows). Numbers at the bottom of each pictogram indicate the position relative to the first nucleotide of the putative start codon. The bottom two series reflect analyses performed on lncRNAs obtained in a large-scale study (Guttman et al. 2009; Khalil et al. 2009).
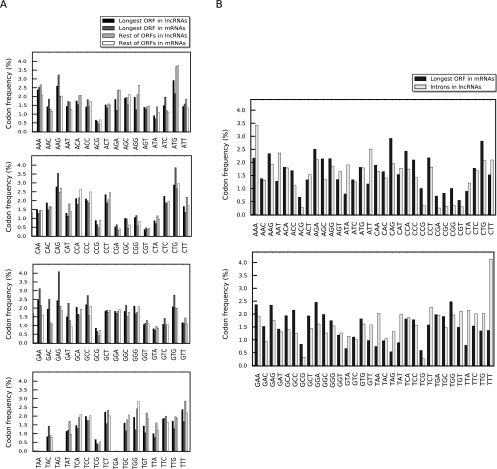
We also analyzed the codon content of the predicted ORFs in the protein-coding transcripts and lncRNAs. The codon usage in the longest predicted ORF in the protein-coding RNAs, which almost always corresponds to the annotated protein-coding ORF (see Materials and Methods), corresponded to the known codon usage bias patterns in human. However, the codon usage frequency in the rest of the predicted ORFs in protein-coding RNAs, the ORFs in the lncRNAs or introns, did not correlate with the codon usage bias pattern in human (Fig. 4A,B; Hershberg and Petrov 2008; Sharp et al. 2010). Taken together, the disparity between the codon content of the ORFs in protein-coding RNAs and functional lncRNAs indicates that the predicted ORFs in lncRNAs are not subject to the same evolutionary pressures as the protein-coding ORFs, further confirming that these transcripts are not under selective pressure for their protein-coding capacity.
FIGURE 4.
Analysis of the predicted ORFs in lncRNAs for protein-coding capacity. (A) The lncRNA ORFs and the untranslated ORFs in protein-coding genes differ from the translated ORF in their codon usage frequency. Codon frequency is shown as the percentage of all codons in an ORF. The identity of the codons is shown below each column. (B) Comparison of the codon composition of intronic sequences and protein-coding sequences in all three reading frames further points to clear distinctions between the protein-coding and noncoding sequences. (C) The vast majority of 3′ UTRs in protein-coding RNAs contain no introns. The number of mRNAs is shown in log scale. (D) The longest predicted ORFs in lncRNAs have longer 5′ UTRs compared with the protein-coding ORF in mRNAs, which increases the likelihood of inefficient translation or nonsense-mediated decay. (E) Most of the ORFs in lncRNAs that end within or <50 nt upstream of the last exon have multiple upstream ORFs, making them candidates for NMD-mediated degradation. (F) The length of 3′ UTRs of the longest predicted ORFs in lncRNAs is similar to those of the protein-coding ORFs in mRNAs. All lengths are shown in nucleotides.
Are lncRNAs substrates for NMD-mediated degradation?
In addition to mutant protein-coding messages harboring premature termination codons, nonsense-mediated decay (NMD) is known to be involved in physiological regulation of the level of many protein-coding messages. An important and interesting question is whether the presence of multiple small ORFs in mammalian lncRNAs subjects them to degradation via NMD as previously observed in plants (Kurihara et al. 2009; Nicholson et al. 2010). Extensive research has elucidated several structural features in mRNAs that help trigger NMD including a long 3′ UTR, the presence of upstream ORFs, and the presence of an intron within the 3′ UTR, although other factors such as the presence of extensive secondary structure within the 3′ UTR also affect the induction of NMD (Isken and Maquat 2007; Nicholson et al. 2010).
To determine whether the functional lncRNAs in our database are likely NMD substrates, we analyzed the relative location of the predicted ORFs in the three open reading frames and splice sites within these RNAs. Among the spliced lncRNAs that contain ORFs, in 40% of cases including HOTAIR and MEG3 RNAs, the stop codon of even the 3′-most ORF falls at least 50 nt upstream of the last exon, an arrangement that has been shown to lead to the induction of NMD and is rare among protein-coding RNAs (Fig. 4C; Isken and Maquat 2007; Kurihara et al. 2009; Nicholson et al. 2010). In the remaining cases, which did have an ORF ending within or in close vicinity of the last exon, the presence of multiple upstream ORFs in their long putative “5′ UTRs” made the translation of the terminal ORFs unlikely (Fig. 4D,E). Similarly, in the case of unspliced lncRNAs, the presence of multiple short ORFs that begin near the 5′ end of the transcript but end far from the 3′ end of the RNA will lead to the induction of NMD even in the absence of splicing (Isken and Maquat 2007; Nicholson et al. 2010). Thus, based on these analyses, the vast majority of lncRNAs will be subjected to NMD should they be recruited to the translation machinery. Analysis of the length of 3′ UTRs (based on the position of the longest predicted ORF in lncRNAs) did not indicate a significant difference between the lncRNAs and protein-coding RNAs (Fig. 4F). While large-scale analyses focusing on lncRNAs in mammalian cells are lacking, studies on a few lncRNAs suggest that many of them are subjected to NMD if recruited to the ribosomes. For example, the free cytoplasmic GAS5 (Growth arrest specific 5) RNA is known to be associated with polysomes and is rapidly degraded (Smith and Steitz 1998). GAS5 contains an ORF ending in its last exon; however, the presence of a single upstream ORF is likely the trigger for NMD in this RNA. These results also suggest that the activation of NMD pathways may at least partly explain the low steady-state level of the lncRNAs or their observed localization to the nuclei.
Taken together, the above data in aggregate indicate that based on the location and size and context of their ORFs, the lncRNAs have little, if any, inherent protein-coding capacity. Even if a lncRNA is recruited to the translation machinery, it is likely to be degraded through NMD, which prohibits the synthesis of even short peptides by most members of this class of RNAs. Thus, the vast majority of lncRNAs must indeed perform their cellular function as RNA molecules rather than templates for short peptide synthesis.
lncRNAs as potential hosts for miRNAs?
While close to 40% of all known miRNAs are found in the introns of protein-coding genes, an additional 40% are found in introns of transcripts that do not seem to harbor protein-coding capacity and thus fall into the category of lncRNAs (Carthew and Sontheimer 2009; Kim et al. 2009; Krol et al. 2010). This class of transcripts also harbors exonic miRNA precursors that account for nearly 10% of all known miRNAs (Bartel 2004; Kim 2005; Carthew and Sontheimer 2009; Kim et al. 2009; Krol et al. 2010). Large-scale transcriptome analyses suggest that while most short RNAs are part of a long RNA, only ∼10% of long RNAs host a short RNA (Carninci et al. 2005; The ENCODE Project Consortium 2007; Kapranov et al. 2007). However, due to the tissue-specific expression of many small RNAs, an accurate estimation of the fraction of the lncRNAs that harbor a small RNA requires additional comprehensive analyses. Currently several lncRNAs are known to host miRNAs in their introns or exons, including bic, 7H4, H19, Meg3, and DLEU2 RNAs (Rodriguez et al. 2004; Kim 2005; Cai and Cullen 2007; Hagan et al. 2009).
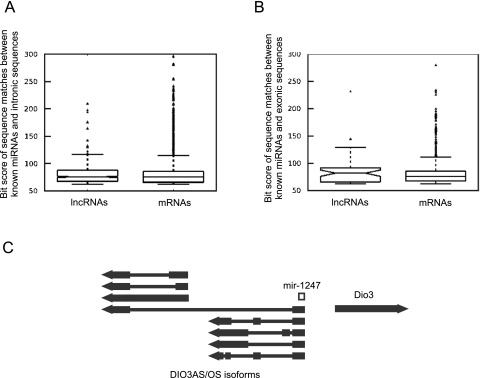
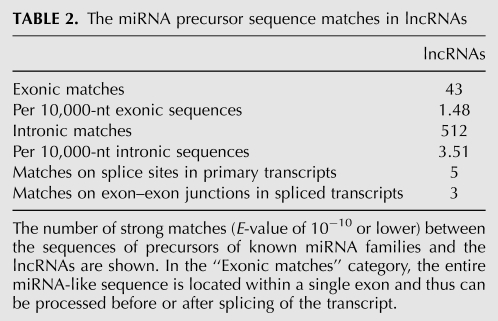
To determine if hosting an miRNA precursor is a common occurrence in functional lncRNAs, we analyzed both the primary and spliced transcripts of lncRNAs and protein-coding RNAs for the presence of sequences that showed significant similarity (E-value of 10−10 or lower) to precursors of known miRNA families. As can be seen from Figure 5 and Table 2, both lncRNAs and mRNAs contained many hairpin-like sequences with similarities to known miRNA families in both introns and exons. We also detected five miRNA-like sequences that fall on splice sites in unspliced lncRNAs or across exon–exon junctions in spliced RNAs (Table 2). For example, the H19 lncRNA hosts mir-675 (Table 3; Cai and Cullen 2007; Koerner et al. 2009), which is positioned on a splice site used in one of its alternatively spliced isomers and creates a potential regulatory switch between the processing of the miRNA and the alternatively spliced isoform. While a number of the matches between miRNA precursor sequences and lncRNAs are incomplete and lack the full-length sequence of the miRNA precursor, they contain extensive complementarity in the miRNA targeting region, suggesting that the lncRNAs may bind miRNAs either as the so-called “miRNA sponges” (Ebert and Sharp 2010a,b) or as regulatory targets.
FIGURE 5.
lncRNAs contain sequences with homology with precursors of known miRNA families. (A,B) The introns and exons of both lncRNAs and mRNAs contain a large number of sequences with homology with miRNA precursors. (C) Some isoforms of DIO3AS lncRNA host miRNA 1247. The genomic loci of some of the DIO3AS isoforms are shown (Hernandez et al. 2004), a number of which start from an alternative promoter and lack miRNA 1247. The proximity of DIO3AS and DIO3 promoter regions and the location of miRNA 1247 are shown. (Arrowheads at the end of genes) The direction of transcription. (Thick lines) Exons; (thin lines) introns.
TABLE 2.
The miRNA precursor sequence matches in lncRNAs
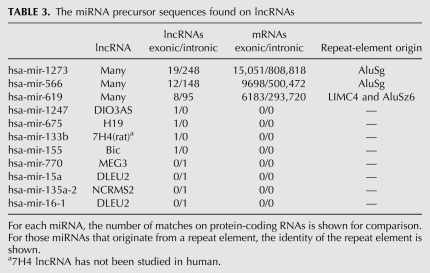
TABLE 3.
The miRNA precursor sequences found on lncRNAs
Analysis of the identity of the miRNA precursor matches indicated that the vast majority of them belong to three miRNA families—miR-1273, miR-566, and miR-619—which are found in large numbers on introns and exons of both lncRNAs and protein-coding RNAs (Table 3). The miR-566 and miR-619 families have been previously reported as repeat-element-derived miRNAs positioned within AluSg repeats in the case of miR-566 and at the junction of nested transposition events between LIMC4 and AluSz6 for miR-619 (Borchert et al. 2006; Piriyapongsa et al. 2007). miR-1273 is the most abundant family of miRNAs found on the exonic and intronic sequences of both groups of transcripts (Table 3), and since its mature miRNA has been detected in a large-scale miRNA sequencing study (Morin et al. 2008), it is likely to be a bona fide miRNA. Our analysis indicated that similar to miR-566 and miR-619, miR-1273 is also derived from a repeat element (Table 3). Available data indicate that repeat elements contribute to miRNA genes in both plants and human (Smalheiser and Torvik 2005; Piriyapongsa et al. 2007); however, the functional significance of the presence of repeat-derived miRNAs at such a high copy number in the transcriptome is not known.
In addition to repeat-derived miRNAs, our analysis identified four exonic and four intronic full-length miRNA precursors in lncRNAs, including seven miRNA precursors that had been previously reported (Table 3; Rodriguez et al. 2004; Kim 2005; Kim et al. 2009). An miRNA precursor, hsa-mir-1247, is located within the exonic sequences of DIO3AS lncRNA, which to our knowledge has not been previously annotated as an miRNA host. Of the eight lncRNAs that host an miRNA, DIO3AS and MEG3 have isoforms that result from alternative splicing and/or the use of alternative promoters (Fig. 5C; Hernandez et al. 2004; Zhang et al. 2010). While in the case of MEG3 all isoforms contain the miRNA within their introns, a number of DIO3AS isoforms that originate from an alternative distal promoter (Hernandez et al. 2004) do not contain mir-1247 (Fig. 5C). In the DIO3AS isoforms that do contain the miRNA, it is positioned at the very 5′ end of the transcript, which is close to the promoter of the DIO3 gene. It is thought that transcription from this proximal promoter interferes with DIO3 transcription (Hernandez et al. 2004). Thus, regulation of transcription from the proximal versus distal promoter not only controls the expression of mir-1247, but it also determines the rate of transcription of the DIO3 gene at the same time, providing an example of the complex lncRNA-mediated regulatory networks.
In summary, other than a few lncRNAs that host characterized miRNAs, the functional lncRNAs do not seem to host well-defined intronic or exonic miRNAs. An important question raised by these observations is whether the lncRNAs that do host miRNAs have an independent cellular function. In the case of intronic miRNAs, it is likely that the spliced message independently performs its function, as has been observed with MEG3 lncRNA (Zhang et al. 2010) and protein-coding RNAs hosting intronic miRNAs. In the case of exonically encoded miRNA precursors, it is similarly possible that after the processing and removal of the miRNA sequences, the remainder of the RNA assumes an independent cellular role, as has been previously observed in the case of MALAT1 and Menɛ/β/Neat1 RNAs, which harbor a tRNA-like element (Wilusz et al. 2008; Sunwoo et al. 2009; Bernard et al. 2010; Tripathi et al. 2010). Alternatively, the processing of the miRNA precursor can be used as a means of regulation of the abundance of the lncRNA that harbors it. In addition to hosting miRNAs, several lncRNAs host snoRNAs in their introns, including the GAS5 and MEG8 lncRNAs, imparting an additional functional dimension to these transcripts (Smith and Steitz 1998; Koerner et al. 2009).
Parallels between lncRNAs and 3′ UTRs
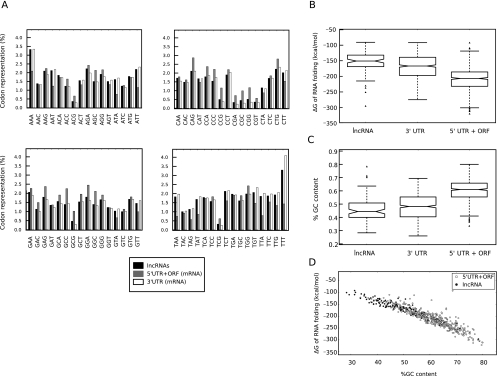
Several major characteristics of the lncRNAs described above are reminiscent of the 3′ untranslated region (3′ UTR) of protein-coding RNAs, another class of RNA regulatory sequences that through interactions with various cellular factors regulate several aspects of the metabolism of protein-coding RNAs. Similar to lncRNAs, the 3′ UTRs differ from the rest of the mRNA sequences in lacking protein-coding capacity and show a codon composition that was markedly different from that in 5′ UTRs and ORFs and similar to what is observed in lncRNAs (Fig. 6A). In another parallel to lncRNAs, the 3′ UTRs are highly intron-poor with ∼95% being intron-less (Figs. 4C, 1B,C). Analysis of the 3′ UTRs indicated a low GC content (42.9%), similar to what was observed in lncRNAs (43.6%), which is significantly below that of the protein-coding RNAs (51.8%) (Table 1). The lower GC content of lncRNAs and 3′ UTRs could potentially mean that these sequences contain fewer stably base-paired structures, and thus, their primary sequence may be more accessible for interaction with the rest of the cellular factors. However, the structural stability of RNAs strongly depends on their sequence, and even among RNAs with identical length and GC content, differences in sequence composition (despite the identical nucleobase composition) can lead to dramatic differences in structural stability. To compare the structural stability of the lncRNAs and 3′ UTRs to that of protein-coding sequences, we selected 500-nt-long fragments within lncRNAs and 3′ UTRs or the region in protein-coding RNAs comprising the 5′ UTR and the annotated ORF (5′UTR+ORF) and determined the folding energy of their most stable predicted folded structure (Fig. 6B). Analysis of 95 such fragments from lncRNAs and 593 fragments chosen from mRNAs indicated that, indeed, the 5′UTR+ORFs had significantly more stable secondary structures compared with both 3′ UTRs and lncRNAs (P-values of 9 × 10−46 and 2 × 10−30, respectively, as determined by the KS test) (Fig. 6B). Thus, the lncRNAs and 3′ UTRs have unexpected similarities in their genomic organization and structural flexibility. We also attempted to determine whether this lower structural stability was a direct result of the lower GC content of these sequences, or if additional selection for less stable structural elements was involved. As a first step in this direction, we first ensured that the 500-nt-long fragments used in the above studies had GC contents representative of the transcript groups they had been derived from, which was indeed the case (Figs. 6C, 2E). We next compared the free energy of global folding of the lncRNAs and the 5′UTR+ORF fragment of mRNAs versus their GC content (Fig. 6D), which suggested that the GC content was the main determinant of the free energy of global folding. However, further studies focusing on local structural features are required to address the possibility of additional selection for less stable secondary structure elements in lncRNAs.
FIGURE 6.
lncRNAs and 3′ UTRs share structural and gene organization features. (A) The codon composition of the 5′ UTRs and ORFs in protein-coding RNAs differs from those of 3′ UTRs and lncRNAs. The identity of each triplet is shown at the bottom. (mRNAs) Protein-coding RNAs. To address the possibility that lncRNAs contain ORFs with noncanonical start codons, the codon compositions of all three reading frames were calculated, and the sum of representation of each triplet in the three reading frames is graphed. Similar analyses were performed on the 5′ UTR + protein-coding ORF of mRNAs and on their 3′ UTRs. The codon composition of the protein-coding ORFs in mRNAs in the translated reading frame is shown in Figure 4A. (B) The predicted folding energy of 3′ UTRs and lncRNAs is consistent with a less rigid secondary structure compared with protein-coding RNAs. (C) The 500-nt fragments used in the folding analysis have GC contents representative of the RNA sequences from which they are derived. (D) GC content is the main determinant of the free energy of global folding in the RNA sequences studied.
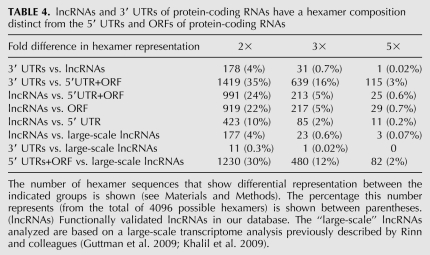
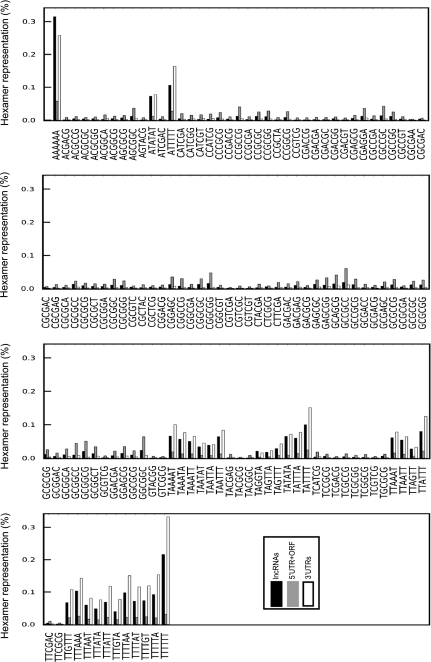
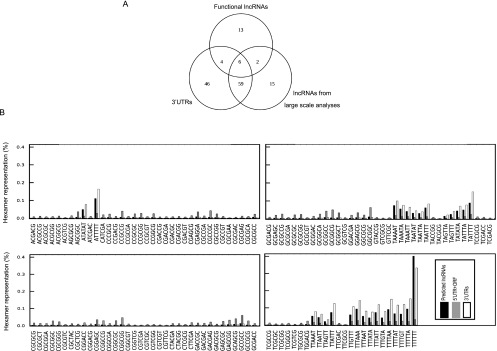
We next analyzed the sequence composition of the lncRNAs, 3′ UTRs, and the 5′UTR+ORF region of protein-coding RNAs using hexamer analysis by a sliding window advanced at single-nucleotide steps. By inclusion of all possible hexamers within a sequence, this approach provides insight into the functional features of a transcript including its potential interactions with other cellular elements. Interestingly, the 3′ UTRs and lncRNAs had a highly similar hexamer profile. Out of 4096 possible hexamers, <5% showed more than twofold differential representation in 3′ UTRs and lncRNAs (Table 4). In contrast, over 24% and 34% of hexamers showed a representation difference of twofold or more when the hexamer composition of 5′UTR+ORF of protein-coding RNAs was compared with lncRNAs and 3′ UTRs, respectively. The difference in hexamer composition between ORFs and lncRNAs was at least twofold larger than that of 5′ UTRs versus lncRNAs, suggesting that the protein-coding sequences indeed have a sequence composition distinct from that of noncoding sequences (Table 4). A large fraction of the hexamers that showed an altered representation in lncRNAs compared with 5′UTR+ORFs in protein-coding RNAs were also differentially represented when the 3′ UTRs were compared with 5′UTR+ORFs (Figs. 7, 9A). Only a single hexamer showed more than fivefold differential representation between 3′ UTRs and lncRNAs, compared with 25 and 115 hexamers between 5′UTR+ORFs of protein-coding RNAs and lncRNAs and 3′ UTRs, respectively (Table 4).
TABLE 4.
lncRNAs and 3′ UTRs of protein-coding RNAs have a hexamer composition distinct from the 5′ UTRs and ORFs of protein-coding RNAs
FIGURE 7.
The hexamer composition of 3′ UTRs and lncRNAs shows a pronounced difference from the 5′ UTRs and ORFs of protein-coding RNAs. The hexamer content of the 5′ UTRs and ORFs of mRNAs (protein-coding RNAs), lncRNAs, and 3′ UTRs of mRNAs has been analyzed using a sliding window advanced at single-nucleotide steps. The hexamers that show more than fivefold under- or over-representation between any two of the three groups of the studied sequences is included in this figure. The identity of each hexamer is shown below each group of three columns.
FIGURE 9.
The hexamer composition of predicted lncRNAs described in large-scale transcriptome analyses resembles that of 3′ UTRs and is markedly different from 5′ UTRs and ORFs of protein-coding genes. (A) The overlap between the three sets of hexamers that show a fivefold over- or under-representation in each of the three groups of noncoding sequences compared with the 5′UTR+ORF of protein-coding RNAs. The low number of hexamers in the functional lncRNAs class is due to the smaller number of the RNAs in this group. (B) The hexamer composition of predicted lncRNAs, 3′ UTRs, and 5′ UTRs and ORFs. The analysis is done in an identical fashion to that in Figure 7, but instead of functionally characterized lncRNAs, the lncRNAs described in a large-scale transcriptome study have been analyzed (Guttman et al. 2009; Khalil et al. 2009). The hexamers that showed more than fivefold difference in representation between any two of the three groups of sequences are shown in the figure.
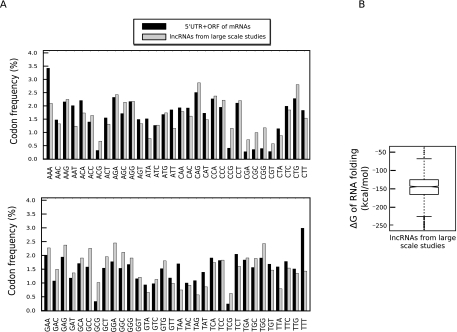
To further confirm that the distinct sequence composition of 3′ UTRs and lncRNAs compared with 5′UTR+ORFs is a common property of noncoding RNA regulatory sequences, we repeated this analysis on a database of long noncoding RNAs that have been discovered in large-scale transcriptome studies (Guttman et al. 2009; Khalil et al. 2009). Although these RNAs have not yet been functionally validated, they exhibit a high level of conservation across species and, thus, have a high likelihood of being functional RNAs. Interestingly, analysis of sequence features of these transcripts indicated that although they have a GC content distribution similar to the protein-coding messages (Fig. 1F), they resembled the lncRNAs in our database in codon composition pattern and the sequence context of their small predicted ORFs (Figs. 8A and 3F, respectively). This observation is consistent with previously published studies on this set of RNAs that indicated the short length and low level of phylogenetic conservation of the predicted ORFs in these transcripts and thus suggested a lack of protein-coding capacity (Guttman et al. 2009; Khalil et al. 2009). Furthermore, analysis of the structural stability of these RNAs indicated that despite having a GC content similar to that observed in protein-coding transcripts, the secondary structure of these RNAs was considerably more flexible and closely resembled that of lncRNAs in our database (cf. Fig. 8B with 6B). This observation strongly suggested that despite having a higher GC content, the lncRNAs from the large-scale study should have a sequence composition that results in lack of stable base-paired structures. To determine if this is indeed the case, we performed a hexamer analysis similar to the one performed above on mRNA sequences and lncRNAs in our database. Remarkably, the lncRNAs from the large-scale study had a hexamer composition highly similar to functionally characterized lncRNAs and 3′ UTRs and significantly differed from the 5′UTR+ORFs (Fig. 9A,B; Table 4). These results also indicate that the observed differences in sequence composition and structural stability when lncRNAs and 3′ UTRs were compared with 5′UTR+ORFs cannot be simply ascribed to the variance in GC content between these groups, since the lncRNAs for the large-scale study, which have an overall GC content similar to that in protein-coding genes (Fig. 1F), still exhibited the same difference in sequence composition.
FIGURE 8.
lncRNAs discovered in large-scale studies resemble the functionally validated lncRNAs in sequence composition and structural stability. (A) The codon composition of the lncRNAs described by Khalil et al. (2009) compared with that of 5′ UTRs and ORFs in protein-coding RNAs in the three reading frames. (B) The lncRNAs from large-scale studies resemble the functionally validated lncRNAs in terms of structural stability.
Taken together, analysis of the functionally characterized lncRNAs elucidated several specific features of these RNAs that can potentially contribute to different aspects of their biogenesis and function. Importantly, a detailed analysis of their coding capacity indicates that it is highly unlikely that they perform their function via coding for short functional peptides. Furthermore, these studies have revealed the presence of remarkable similarities between the lncRNAs and 3′ UTRs of protein-coding RNAs. Interestingly, recent global transcriptome analyses suggest that many 3′ UTRs give rise to independent lncRNA transcripts, and there is at least one example of a viral lncRNA that is generated from a 3′ UTR after the rest of the original protein-coding message has been degraded (Iwakawa et al. 2008; Mercer et al. 2011). These findings, together with the results discussed above, suggest that the different classes of cellular RNA regulatory sequences share features that may contribute to their RNA-mediated mode of regulation. Whether they are part of a protein-coding RNA or an independent transcript, the regulatory sequences seem to be part of a ubiquitous “RNA regulome” that has evolved under evolutionary pressures that are distinct from those acting on protein-coding sequences and likely reflect fundamental features required for RNA-mediated regulation and intrinsic to their mode of interaction with the cellular factors.
MATERIALS AND METHODS
The functional lncRNA database
The lncRNAs included in the database (http://www.valadkhanlab.org/database) were manually culled from the literature and are restricted to transcripts that have been individually studied and been ascribed a cellular function. The recent availability of another lncRNA database (Amaral et al. 2011) allowed us to add another 12 studied lncRNAs to our database. As control, we created a parallel database containing all annotated human protein-coding RNAs including all of their annotated protein-coding isoforms. The sequence of the mature transcripts, coordinates of introns and exons, the annotated protein-coding ORFs, and the assigned name for the protein product of protein-coding transcripts were obtained from the “knownGene” table of the hg19 genome database downloaded from the UCSC Genome Bioinformatics website (Fujita et al. 2011). We omitted transcripts that were not explicitly annotated as protein-coding transcripts and lacked a name assigned to their protein product. Any poly(A) sequences at the 3′ ends of the annotated transcript sequences were removed to increase the accuracy of sequence composition analyses.
Using the sequence of the mature transcripts of lncRNAs, we obtained the major structural data including the number and coordinates of introns and exons by a locally installed BLAT program (downloaded from http://www.soe.ucsc.edu/∼kent along with the human and mouse genomes in 2-bit format) (Fujita et al. 2011). These results, along with the results of all subsequent analyses, were stored as Structured Query Language (SQL) enabled relational tables in a MySQL RDBMS (Relational Database Management System) database.
To determine the open reading frames (ORFs) on each RNA, a locally installed “getorf” application from the EMBOSS project repository of applications (emboss.sourceforge.net) (Rice et al. 2000) was used. The minimal length for the predicted ORFs was set to 100 nt. To ensure the accuracy of the ORF predictions, we compared the predicted ORFs with the annotated ORFs in the protein-coding RNAs, which confirmed the robustness of our ORF predictions and also indicated that in >95% of cases, the longest predicted ORF in protein-coding RNAs corresponds to the one annotated as the coding ORF in the public databases.
To find possible miRNA precursors or miRNA-like sequences in our transcripts, a database containing all annotated miRNA precursor hairpins was downloaded from miRBase (ftp://mirbase.org/pub/mirbase/-CURRENT/hairpin.fa.gz) (Griffiths-Jones et al. 2008). The sequence matches were determined using the BLAST executables package (http://blast.ncbi.nlm.nih.gov) from NCBI. This analysis was performed both on primary unspliced transcripts and on mature spliced ones. To eliminate the weak or fortuitous matches, we filtered matches that had a bitscore below 60 (roughly corresponding to an E-value above 10−10) from our subsequent analyses. For the miRNA precursor matches in lncRNAs that did not originate from a repeat element and had an E-value <10−10, the degree of sequence similarity was manually examined, and the matched sequences that were unlikely to form a hairpin structure were eliminated.
Data analysis
The scripts used for data analysis were written in Python programming language (http://www.python.org) with the use of SciPy and NumPy packages as the implementations for processing logic and matplotlib as the main plotting and graphing package on Linux operating systems. To demonstrate the population distribution in our analyses, we used box plots that offer nonparametric data visualization without any assumption about underlying statistical distribution. The box plots were drawn using matplotlib, with the top and bottom of the box corresponding to the 75th and 25th percentiles among the data points and the line in the middle of the box indicating the 50th percentile (the median). The whiskers indicate the boundary of outlier data points assuming a normal distribution. Specifically, the lower and upper whiskers cover data in a distance equal to 1.5 times of IQR (interquartile range) from the lower and upper quartile, respectively. IQR is a robust statistical parameter equal to the difference between the third and first quartiles (25th and 75th percentile) as shown by the top and bottom of the box.
To determine the statistical significance of our observations, we analyzed our data by both a Student's t-test and the Kolmogorov-Smirnov test (KS test). Since the two-sample KS test, which is a nonparametric method for comparing two samples, is sensitive to differences in both location and shape of the empirical cumulative distribution functions of the two samples, it was particularly suitable for defining the statistical significance of our analyses. When the P-values obtained from both the Student's t-test and KS-test were <0.001, the differences were considered statistically significant.
For analysis of the 5′-UTR elements and drawing the diagrams for defining positional nucleotide abundance, the protein-coding transcripts in which the 5′ UTR was not included in the annotated transcript sequence were discarded, and except when noted, the translation-related sequence motifs in all three reading frames were analyzed. A local installation of the Weblogo application (weblogo.berkeley.edu) (Crooks et al. 2004) was used for making the pictograms. Since a small number of lncRNAs had several alternatively processed isoforms, in order to prevent a biased result, when appropriate all but one of the isoforms were eliminated from the analysis.
To determine the folding energy of the sequences in each group, all lncRNAs that were longer than 500 nt were analyzed. A 500-nt-long fragment was selected from each RNA such that the midpoint of the RNA fell on the midpoint of the 500-nt fragment. For the protein-coding RNAs, we randomly selected 593 transcripts in which the 3′ UTRs and 5′UTR/ORF regions were longer than 500 nt. Since the median length of 3′ UTRs in protein-coding RNAs is 700 nt (Fig. 4F), this approach did not bias our analysis toward a small subset of transcripts. The first and last 500-nt fragments of these RNAs, which corresponded to a fragment of 5′UTR/ORFs and 3′ UTRs, respectively, were selected for analysis. Since the median length of 5′ UTRs is 200 nt and >75% of protein-coding RNAs contained 5′ UTRs that were ∼350 nt long or shorter (Fig. 4D), the selected fragments contained sequences representing both 5′ UTRs and ORFs of mRNAs. The fragments were analyzed using the program EnsembleEnergy from the RNAstructure software suite (RNA.urmc.rochester.edu/RNAstructure.html) (Reuter and Mathews 2010).
ACKNOWLEDGMENTS
We thank Dr. Tim Nilsen, Dr. Ahmad Khalil and Dr. Mark Adams for critical review of the manuscript, suggestions, and providing the coordinates for the database of noncoding RNAs derived from large-scale analyses; Justin Pruttivarasin and Faiza Khimji for help in making the database; and Cen Guo and Jing Li for assistance with the initial phase of construction of the database. We also thank Youngmin Park for help with the database website. This work is supported by startup funds from Case Western Reserve University to S.V.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029520.111.
REFERENCES
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS 2011. lncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res 39: D146–D151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. 2010. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29: 3082–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL 2006. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13: 1097–1101 [DOI] [PubMed] [Google Scholar]
- Cai X, Cullen BR 2007. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13: 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci 106: 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-L, Carmichael GG 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-L, Carmichael GG 2010. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol 22: 357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES 2007. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci 104: 19428–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS 2008. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput Biol 4: e1000176 doi: 10.1371/journal.pcbi.1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS 2009. NRED: A database of long noncoding RNA expression. Nucleic Acids Res 37: D122–D126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA 2010a. Emerging roles for natural microRNA sponges. Curr Biol 20: R858–R861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA 2010b. MicroRNA sponges: Progress and possibilities. RNA 16: 2043–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Bailey TL, Kasukawa T, Mignone F, Kummerfeld SK, Madera M, Sunkara S, Furuno M, Bult CJ, Quackenbush J, et al. 2006a. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol 3: 40–48 [DOI] [PubMed] [Google Scholar]
- Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, Grimmond SM 2006b. The abundance of short proteins in the mammalian proteome. PLoS Genet 2: e52 doi: 10.1371/journal.pgen.0020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. 2011. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res 39: D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, O'Neill BL, Stewart CL, Kozlov SV, Croce CM 2009. At least ten genes define the imprinted Dlk1–Dio3 cluster on mouse chromosome 12qF1. PLoS ONE 4: e4352 doi: 10.1371/journal.pone.0004352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Rivas FV, Murchison EP, Steitz JA 2006. The expanding universe of noncoding RNAs. Cold Spring Harb Symp Quant Biol 71: 551–564 [DOI] [PubMed] [Google Scholar]
- Hernandez A, Martinez ME, Croteau W, St Germain DL 2004. Complex organization and structure of sense and antisense transcripts expressed from the DIO3 gene imprinted locus. Genomics 83: 413–424 [DOI] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA 2008. Selection on codon bias. Annu Rev Genet 42: 287–299 [DOI] [PubMed] [Google Scholar]
- Hung T, Chang HY 2010. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol 7: 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2007. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Iwakawa H-O, Mizumoto H, Nagano H, Imoto Y, Takigawa K, Sarawaneeyaruk S, Kaido M, Mise K, Okuno T 2008. A viral noncoding RNA generated by cis-element-mediated protection against 5′ → 3′ RNA decay represses both cap-independent and cap-dependent translation. J Virol 82: 10162–10174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L 2010. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 16: 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Kondo T, Hashimoto Y 2011. Coding vs non-coding: Translatability of short ORFs found in putative non-coding transcripts. Biochimie 93: 1981–1986 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kelly SM, Corbett AH 2009. Messenger RNA export from the nucleus: A series of molecular wardrobe changes. Traffic 10: 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN 2005. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Koerner MV, Pauler FM, Huang R, Barlow DP 2009. The function of non-coding RNAs in genomic imprinting. Development 136: 1771–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y 2010. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329: 336–339 [DOI] [PubMed] [Google Scholar]
- Kozak M 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37 [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M 2006. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol 4: e180 doi: 10.1371/journal.pbio.0040180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, et al. 2009. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci 106: 2453–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Wilhelm D, Dinger ME, Soldà G, Korbie DJ, Glazov EA, Truong V, Schwenke M, Simons C, Matthaei KI, et al. 2011. Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res 39: 2393–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G, Yoshizawa A, Komori T, Asai K 2009. The functional RNA database 3.0: Databases to support mining and annotation of functional RNAs. Nucleic Acids Res 37: D89–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O 2010. Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega MA, Zhu Y, Plajzer-Frick I, Afzal V, Rubin EM 2004. Megabase deletions of gene deserts result in viable mice. Nature 431: 988–993 [DOI] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Stephen S, Engström PG, Tajul-Arifin K, Chen W, Wahlestedt C, Lenhard B, Hayashizaki Y, Mattick JS 2005. RNAdb—a comprehensive mammalian noncoding RNA database. Nucleic Acids Res 33: D125–D130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS 2006. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet 22: 1–5 [DOI] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF 2011. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheasant M, Mattick JS 2007. Raising the estimate of functional human sequences. Genome Res 17: 1245–1253 [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Mariño-Ramírez L, Jordan IK 2007. Origin and evolution of human microRNAs from transposable elements. Genetics 176: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. 2006a. Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2: e168 doi: 10.1371/journal.pgen.0020168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. 2006b. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443: 167–172 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Pääbo S, Rubin EM 2006. Accelerated evolution of conserved noncoding sequences in humans. Science 314: 786. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. 2006. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH 2010. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129 doi: 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis LA, Kastenmayer JP, Hüttenhofer AG, Green PJ 2008. Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci 13: 329–334 [DOI] [PubMed] [Google Scholar]
- Senapathy P 1986. Origin of eukaryotic introns: A hypothesis, based on codon distribution statistics in genes, and its implications. Proc Natl Acad Sci 83: 2133–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Spiridonov AN, Novichkov PS, Spiridonov NA, Koonin EV 2010. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol Biol Evol 27: 1745–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Emery LR, Zeng K 2010. Forces that influence the evolution of codon bias. Philos Trans R Soc Lond B Biol Sci 365: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI 2005. Mammalian microRNAs derived from genomic repeats. Trends Genet 21: 322–326 [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA 1998. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 18: 6897–6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS 2007. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays 29: 288–299 [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulveling D, Francastel C, Hubé F 2011. When one is better than two: RNA with dual functions. Biochimie 93: 633–644 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Nilsen TW 2010. Reprogramming of the non-coding transcriptome during brain development. J Biol 9: 5 doi: 10.1186/jbiol197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wethmar K, Smink JJ, Leutz A 2010. Upstream open reading frames: Molecular switches in (patho)physiology. BioEssays 32: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL 2009. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A 2010. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinology 151: 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]