The 5′ end of eukaryotic mRNA carries a N7-methylguanosine residue linked by a 5′-5′ triphosphate bond. This cap moiety (7mGpppN) is an essential RNA structural modification allowing its efficient translation, limiting its degradation by cellular 5′ exonucleases and avoiding its recognition as “nonself” by the innate immunity machinery. In vitro synthesis of capped RNA is an important bottleneck for many biological studies. In the present work, the authors combined a chemical synthesis method and an enzymatic methylation assay to produce large amounts of RNA oligonucleotides carrying a cap-0 or cap-1. The RNAs carrying different cap structures (cap, cap-0, or cap-1) act as bona fide substrates mimicking cellular capped RNAs and can be used for biochemical and structural studies.
Keywords: 5′-capped mRNA, RNA chemical synthesis, enzymatic methylation, guanine-N7-methyl transferase, solid-phase synthesis
Abstract
The 5′ end of eukaryotic mRNA carries a N7-methylguanosine residue linked by a 5′-5′ triphosphate bond. This cap moiety (7mGpppN) is an essential RNA structural modification allowing its efficient translation, limiting its degradation by cellular 5′ exonucleases and avoiding its recognition as “nonself” by the innate immunity machinery. In vitro synthesis of capped RNA is an important bottleneck for many biological studies. Moreover, the lack of methods allowing the synthesis of large amounts of RNA starting with a specific 5′-end sequence have hampered biological and structural studies of proteins recognizing the cap structure or involved in the capping pathway. Due to the chemical nature of N7-methylguanosine, the synthesis of RNAs possessing a cap structure at the 5′ end is still a significant challenge. In the present work, we combined a chemical synthesis method and an enzymatic methylation assay in order to produce large amounts of RNA oligonucleotides carrying a cap-0 or cap-1. Short RNAs were synthesized on solid support by the phosphoramidite 2′-O-pivaloyloxymethyl chemistry. The cap structure was then coupled by the addition of GDP after phosphorylation of the terminal 5′-OH and activation by imidazole. After deprotection and release from the support, GpppN-RNAs or GpppN2′-Om-RNAs were purified before the N7-methyl group was added by enzymatic means using the human (guanine-N7)-methyl transferase to yield 7mGpppN-RNAs (cap-0) or 7mGpppN2′-Om-RNAs (cap-1). The RNAs carrying different cap structures (cap, cap-0 or, cap-1) act as bona fide substrates mimicking cellular capped RNAs and can be used for biochemical and structural studies.
INTRODUCTION
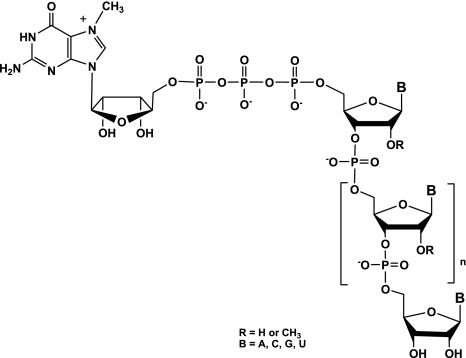
The 5′-cap structure found at the 5′ end of eukaryotic messenger RNAs (mRNAs) and many viral RNAs consists of a N7-methylguanosine nucleoside linked to the 5′-terminal nucleoside of the pre-mRNA via a 5′-5′ triphosphate linkage (Fig. 1; Shatkin 1976; Shuman 2001; Decroly et al. 2011). This modification is critical for recognition by the translation factor eIF4E protein and mRNA translation into proteins by ribosomes (Hodel et al. 1998), as well as for protection from degradation by 5′ exonucleases. RNA capping is also important for other processes, such as RNA splicing and export from the nucleus and to avoid recognition of mRNA by the cellular innate immunity machinery (Daffis et al. 2010; Zust et al. 2011). Indeed, most RNA viruses have evolved mechanisms to protect their RNA 5′-end with a cap moiety indistinguishable from cellular cap structure (Decroly et al. 2012). Viral RNA cap structures are essential for virus replication. Typically, four enzymatic activities are involved in eukaryotic RNA capping. The capping reaction begins with hydrolysis of the phosphate present at the 5′ end of pppRNA by an RNA triphosphatase (RTPase). A guanylyltransferase (GTase) next transfers a GMP molecule onto the mRNA, leading to the formation of GpppN-RNA (cap structure). The GpppN-RNAs are then methylated at their N7 position by an S-adenosyl-1-methionine (AdoMet)-dependent (guanine-N7)-methyltransferase (N7-MTase). This step renders the GTase reaction irreversible and allows the formation of 7mGpppN-RNA (cap-0 structure). Further methylation at the 2′-O position of the ribose of the first nucleotide by an AdoMet-dependent (nucleoside 2′-O)-methyltransferase (2′-O-MTase) leads to 7mGpppN2′-Om-RNA (cap-1 structure). Initially, RNA viruses were believed to synthesize their RNA cap structure using viral enzymes and following the same steps in the same sequence order as that of eukaryotes. In fact, several recent studies reveal that viral RNA capping pathways are highly diverse in enzymes, reaction sequences, and mechanisms (Decroly et al. 2012) and constitute, therefore, novel targets for antiviral drug design.
FIGURE 1.
mRNA cap structure. The cap consists of an N7-methylguanosine linked to the 5′ nucleoside of the messenger RNA chain through a 5′-5′ triphosphate bridge. The methyl group of the guanosine at its N7-position is surrounded, and the 2′-O position of the first nucleotide residue can be linked to an H atom or a CH3 group forming the cap and cap-1 structures, respectively.
However, structural and mechanistic studies, as well as structural studies on guanylyltransferase and MTase complexes with products, have been greatly limited by the lack of appropriate amounts of pure RNA substrates required for biochemical characterization of mRNA cap MTases. Consequently, very few crystal structures of RNA virus RNA capping enzymes exist in complex with RNA or capped RNA because the availability of 5′-capped RNA substrates of defined and appropriate length has remained an important bottleneck.
In vitro enzymatic synthesis of capped mRNAs also remains challenging since it is difficult to produce 5′-capped RNAs of defined sequences in great amounts. In particular, while the cellular machinery is able to cap any 5′ RNA sequence, it is known that viral enzymes cap defined and specific RNA sequences present at the RNA 5′ end (i.e., for flaviviruses, pppAGUUGU [Bollati et al. 2010] and coronaviruses, pppAUAUUA). These sequences are very difficult to synthesize using T3, T7, or SP6 DNA-dependent RNA polymerases (DdRp), because the bacteriophagic promoter prefers to initiate polymerization with GTP and allows mainly the synthesis of RNA beginning with pppGG (T3 and T7) or GA (sp6). The bacteriophage T7 φ 2.5 promoter has also been reported to initiate the RNA synthesis with pppAG (Coleman et al. 2004). Nevertheless, such substitutions in the initiation start site reduced the efficiency of RNA synthesis and favored a slippage mechanism resulting in alternative initiation products (Imburgio et al. 2000). After RNA in vitro transcription, a cap structure can be added to the 5′ triphosphate end of RNA using vaccinia virus capping enzyme that contains RNA triphosphatase, guanylyltransferase, and N7-MTase activities (Brownlee et al. 1995). Another possibility to synthesize very short-capped RNAs consists of using a DNA primase that recognizes a specific codon on a given DNA template and initiates RNA synthesis containing two nucleotides complementary to the initiation codon. Accordingly, primase such as that of the bacteriophage T7 (gene 4) was reported to produce 7mGpppACn and GpppACn efficiently (1 ≤ n ≤ 7) in the presence of 7mGpppA and GpppA, respectively (Peyrane et al. 2007). An obvious limitation of this method is an obligate pppAC RNA starting sequence.
To overcome this bottleneck, we have developed a straightforward strategy for the synthesis of 5′-capped RNAs with high yields and without any limitation concerning the nucleotides present at 5′ of the substrate RNA. While the automated large-scale chemical synthesis of RNA on solid support was established more than 20 years ago, the synthesis of the 5′-capped RNA is still a challenge, even at the milligram scale. Although a few attempts for their synthesis in solution were reported (Sekine et al. 1985; Iwase et al. 1988; Sekine et al. 1989, 1996; Sawai et al. 1999; Zubereck et al. 2003; Koukhareva and Lebedev 2004; Mikkola et al. 2005), the associated yields were always rather low, with long reaction times (6–10 d), and associated purification difficulties to obtain material in large quantity. Regarding the synthesis of 5′-capped RNAs on solid support, the instability of N7-methylguanosine under acidic and basic conditions has limited the efforts in this area. Because of the positive charge on the N7-methylguanosine, the nucleoside is hydrolytically less stable than other purine nucleosides. Under basic conditions used for standard RNA deprotection, the opening of the imidazole ring of the 7-methylguanine would occur. For this reason, the synthesis of 7mGpppRNAs completely achieved on solid support excludes the ammonia treatment to deprotect and to release 5′-capped RNAs. In fact, two examples of a successful synthesis of the same short 5′-capped oligonucleotide GpppApUpA on solid support have been described in the literature, but in both cases, the linker between the RNA chain and the support was not base-labile (Kadokura et al. 2001; Jemielity et al. 2005). The first solid-phase synthesis of the analog 2,2,7-trimethylguanosine (TMG)-capped trinucleotide was carried out in 2001 with an acid-labile phosphoramidate linker (Kadokura et al. 2001). The successive steps were the conversion of the 5′-OH end of RNA into its 5′-diphosphate by two successive phosphorylations, followed by the nucleobases’ deprotection in basic conditions, then addition of the cap structure using the 5′-phosphoroimidazolide N7-methylguanosine, and finally, release of the expected 5′-capped RNA with acetic acid. The desired compound was isolated in an overall 20% yield. More recently, an alternative synthesis of the same 5′-capped short oligomer was proposed using a disulfide linker between the solid support and the 5′-phosphate RNA and using the unprotected N7-methylguanosine 5′-diphosphoroimidazolide as a capping agent (Jemielity et al. 2005). The total efficacy of capped RNA production was not evaluated, but the reported yield of the capping step was <50%. Two other examples of synthetic capped RNA of much longer sequence (42-mer and 130-mer) have been reported. The 42-mer was synthesized in three fragments that were enzymatically ligated (Iwase et al. 1992). The 130-mer sequence was chemically assembled by using 2-cyanoethoxymethyl as the 2′-OH protecting group, then the 5′ end was pyrophosphorylated to add the cap structure with a capping enzyme (Nagata et al. 2010). The need of a universal and efficient method to obtain synthetic 5′-capped RNA in significant quantities and high purity has been growing, in particular with increasing availability of crystal structures of apo-enzymes involved in viral RNA capping (Egloff et al. 2007; Bouvet et al. 2010; Decroly et al. 2011).
Here, we report the successful production of good yields of 5′-7mGppp-RNAs of various lengths (from 4 to 18 nt) and different sequences corresponding to the 5′ end of Dengue virus and SARS coronavirus genomes. The originality of this strategy over the existing reported methods is based upon the automated RNA assembly and the capping reaction entirely performed on solid support with commercially available materials, which makes it easy to achieve by other researchers without laborious and time-consuming preparation of reagents. Chemical synthesis of RNAs was performed by the phosphoramidite solid-phase method involving the base-labile 2′-O-pivaloyloxymethyl (PivOM) groups and developed by our group (Lavergne et al. 2008). The capping reaction with GDP was derived from the synthesis of 5′-triphosphate DNA, RNA on solid support that allows the production of oligonucleotides in good yields and fair purity (Zlatev et al. 2010). After solid-phase elongation, RNA is converted into its 5′-H-phosphonate derivative which is activated as its phosphoroimidazolide by amidative oxidation. The next step is the coupling of the commercial guanosine diphosphate (GDP) on RNA bound to the solid support, followed by deprotection and release by ammonia, affording the expected Gppp-RNA in solution. All these chemical steps are performed while RNA is attached to the support, enabling excess reagents to be removed by simple washing under argon which makes the synthesis simple and convenient. Finally, the N7-methyl group is enzymatically added by means of purified recombinant human (guanine N7)-methyl transferase (N7-hMTase), an enzyme involved in the capping of cellular mRNA in human cells.
RESULTS AND DISCUSSION
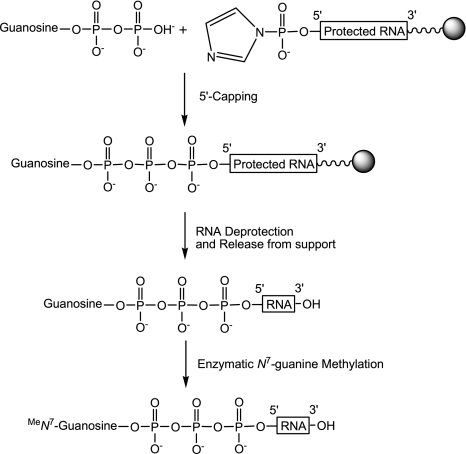
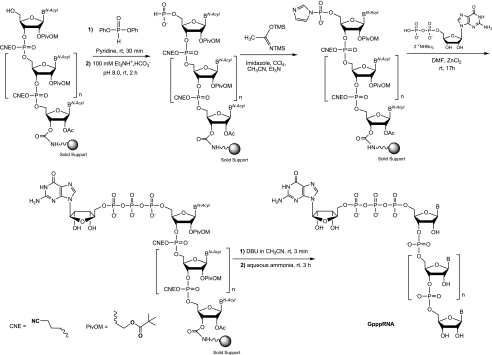
The chemical synthesis of small-capped RNAs is a bottleneck for biological and structural studies enzymes involved in the RNA capping pathway. Moreover, the lack of methods allowing the production of large amount capped RNAs has probably impeded the crystal structure determination of enzymes in complex with such capped RNAs. Although there are several possible methods for chemical synthesis of capped oligoribonucleotides, most of those methods produce a very limited amount of capped RNA (Kadokura et al. 2001; Jemielity et al. 2005). Therefore our aim was to synthesize RNA carrying various cap structures (cap, cap-0 and cap-1) in great quantities using the combination of chemical synthesis of Gppp-RNAs followed by enzymatic methylation. The general course of the synthesis of 7mGppp-RNAs is outlined in Scheme 1.
SCHEME 1.
General course of the synthesis of 5′-7mGppp-RNAs.
Chemical synthesis of SARS coronavirus and Dengue virus 5′-RNA sequences
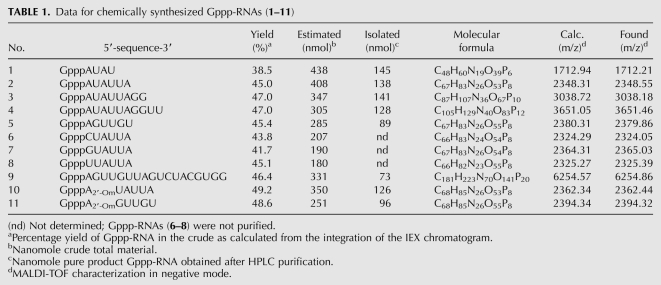
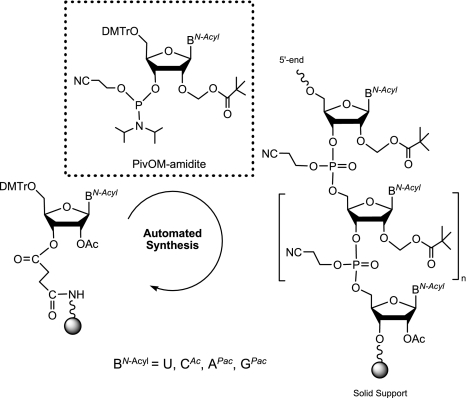
We first synthesized several RNA sequences of various lengths (Table 1) corresponding to the 5′-end of Dengue virus and SARS coronavirus genomes, two important human pathogenic viruses, encoding their own capping enzymes (Bollati et al. 2010; Bouvet et al. 2010). The RNA assembly was performed with the pivaloyloxymethyl (PivOM) technology recently introduced by our group for RNA synthesis on solid support (Lavergne et al. 2008). The major feature of this technology is to use base-labile protecting groups exclusively: pivaloyloxymethyl group (PivOM) for 2′-OH protection, cyanoethyl for phosphates protection, and acyl groups for nucleobase protection. They are removed under basic conditions without RNA damage (i.e., isomerization or strand breaking). Moreover the PivOM technology takes advantage of the standard TBDMS chemistry for RNA synthesis because the deprotection process is a mild ammonia treatment that prevents degradation of the triphosphate moiety of the cap structure. Indeed, in our previous work, we showed that a treatment with Et3N-3HF/N-methylpyrrolidone at 65°C, used to remove TBDMS groups, resulted in a total hydrolysis of the TP moiety; hence, we should perform a treatment with 1 M TBAF at room temperature for 24 h, leading to low degradation (Zlatev et al. 2010). Furthermore, the ammonia deprotection is more convenient for RNA recovery that is made by simple evaporation and avoids tedious desalting procedures necessary after TBAF treatment. RNA oligomers were prepared on a 1-μmol scale from the assembly of 2′-O-PivOM phosphoramidites on CPG solid support through the succinyl linker as previously described (Scheme 2; Lavergne et al. 2008). Amidites used at 0.1-M-concentration in CH3CN and activated by 5-benzylmercaptotetrazole (BMT, 0.3 M) were reacted with 5′-OH supported nucleoside for a 180 sec coupling step. The capping reaction was performed for 160 sec with phenoxyacetic anhydride instead of the regular acetic anhydride to avoid the replacement of the Pac group of G by the acetyl group which would not be removed during mild ammonia deprotection. The terminal adenosine in the SARS sequence AUAUUA was also replaced during the automated synthesis by a 2′-O-methyl adenosine to give A2′-OmUAUUA. The chemical introduction of commercially available 2′-O-methyl ribonucleoside amidites should allow the synthesis of RNA carrying a cap-1, after chemical guanylyltransfer followed by an enzymatic methylation of the N7-guanine. The coupling of 2′-O-Me adenosine amidite was performed under the same conditions as the assembly of the 2′-O-PivOM amidites and with comparable efficiency.
TABLE 1.
Data for chemically synthesized Gppp-RNAs (1–11)
SCHEME 2.
Automated solid-phase synthesis of RNA by the PivOM method.
Synthesis of solid-supported 5′-phosphoroimidazolidate RNA
Our present approach involves the triphosphate motif formation by the reaction of commercially available GDP with an activated phosphate group at the 5′ terminus of the solid-supported and protected RNA. Among the different existing activation methods, we used 5′-phosphoroimidazolidate RNA, prone to give triphosphate derivatives in good yield. Another way would have been to activate GDP with imidazolide (Im) and to react with 5′-phosphorylated supported RNA (Lewdorowicz et al. 2004). This capping strategy requires more steps for the preparation of Im (GDP) and 5′-monophosphorylated RNA instead of just modifying the 5′-OH supported RNA. Imidazole activation in organic solvent for the synthesis of 5′-capped oligoribonucleotides was initially used by Sekine group (Sekine et al. 1989) and seems to be the most powerful activation method for synthesis of a phosphate chain (Jemielity et al. 2010). In fact, phosphoroimidazolide derivatives are known to react with numerous nucleophiles as pyrophosphate or nucleoside mono-, di-, or triphosphates. Usually they are formed using coupling reagents such as carbonyldiimidazole (Iwase et al. 1988; Sekine et al. 1989; Yanachkov et al. 2011) or triphenylphosphine/2,2′-dithiopyridine starting from a 5′-phosphate monoester (Sawai et al. 1999). To our knowledge, these activation methods have not been applied to the 5′-phosphate of a solid-supported oligonucleotide. In our original work (Zlatev et al. 2010), we found that the fastest and easiest way to prepare an activated phosphate at the 5′ end of a supported RNA was through the H-phosphonate intermediate which was readily obtained using commercially available reagents and simply activated by amidative oxidation.
Thus, after RNA assembly, the 5′-OH of the fully protected oligomers still on the solid support were converted to their H-phosphonate monoester with >95% yield by reaction with diphenyl H-phosphonate (Sobkowski et al. 1998) to obtain the unstable H-phosphonate diesters which were subsequently hydrolyzed with triethylammonium bicarbonate buffer to the stable 5′-H-phosphonate supported RNAs (Scheme 3). These entities were then oxidized in strict anhydrous conditions with carbon tetrachloride in the presence of imidazole and N,O-bis-trimethylsilylacetamide (BSA) to give the solid-supported activated 5′-phosphoroimidazolide RNAs (Im-pRNAs) with quantitative yield ready for the next step.
SCHEME 3.
Solid-phase synthesis of 5′-Gppp-RNAs.
Synthesis, deprotection, and release of 5′-Gppp-RNAs (1–11) from solid support
As for the synthesis of 5′-triphosphate RNAs (Zlatev et al. 2010) in different reaction conditions, the imidazolide-activated monophosphate at the 5′ end of RNA was then substituted by guanosine 5′-diphosphate in anhydrous DMF. The poor solubility of GDP in organic solvents and, consequently, its lack of reactivity required the presence of ZnCl2 as a catalyst. Actually, the addition of metal chlorides was reported to generate a clear homogenous solution and to accelerate the coupling reaction (Kadokura et al. 1997). Among several divalent metal salts tested, ZnCl2 was demonstrated to be the best catalyst in anhydrous organic solvent because of the high affinity of Zn2+ ions for the phosphate oxygens and for the nitrogen atom of the imidazolidate. In this work, the triphosphate bond formation between GDP bis (tri-n-butylammonium) salt and 5′-phosphoroimidazolidate supported RNAs was performed at 30°C overnight in the presence of 400 mM ZnCl2 in dry DMF. After removal of the capping solution from the synthesis columns and several washes using CH3CN, solid-supported cap-RNAs were treated first with 1 M DBU solution in dry CH3CN for 3 min to remove cyanoethyl protecting groups from the phosphates, then with 30% aqueous ammonia solution to deprotect nucleobases, 2′-OH, and to release the capped Gppp-RNAs (1–11) from the solid support (Table 1). The conversion of RNAs into the desired capped Gppp-RNAs (1–11) reached a satisfactory average yield between 40% and 50%. The yields were calculated by integration of the major peaks corresponding to the capped RNAs in the HPLC chromatograms of the crude materials (Fig. 2). The other noticeable peaks correspond to the 5′-phosphoroimidazolidate RNA resulting from the incomplete capping reaction and leading to the 5′-phosphate RNA after hydrolysis. It is noteworthy that the conversion yield would not be dependent either on the length or on the 5′-terminal nucleotide (A, C, G, or U) of the RNA sequence, since the yields were quite similar. Furthermore, the capping reaction was very efficient with RNA sequences (10–11) incorporating a 2′-O-methyl ribonucleotide at the 5′ end, the yields reaching 50%. After purification by anion-exchange HPLC, the capped GpppN-RNAs and GpppN2′-Om-RNAs were isolated, with satisfactory yields (ranging from 33% to 42%) and with high purity (>97%) (Fig. 2).
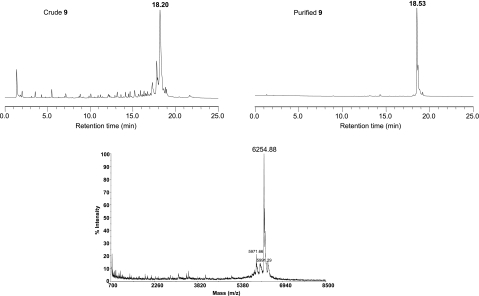
FIGURE 2.
Anion-exchange HPLC profiles of the crude mixture (left panel) and purified (right panel) GpppAGUUGUUAGUCUACGUGG (9). MALDI-TOF mass spectrum of purified RNA (9).
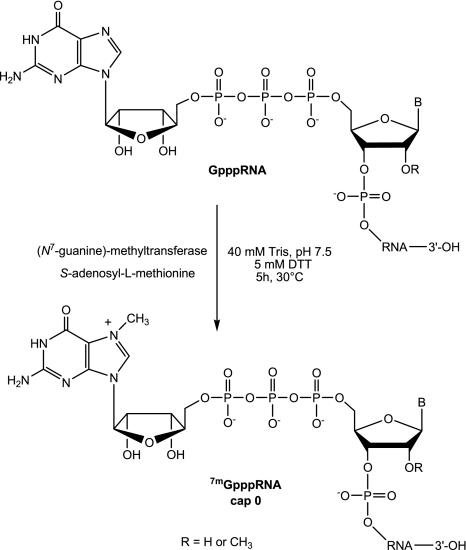
Enzymatic methylation of cap structure by human N7-MTase
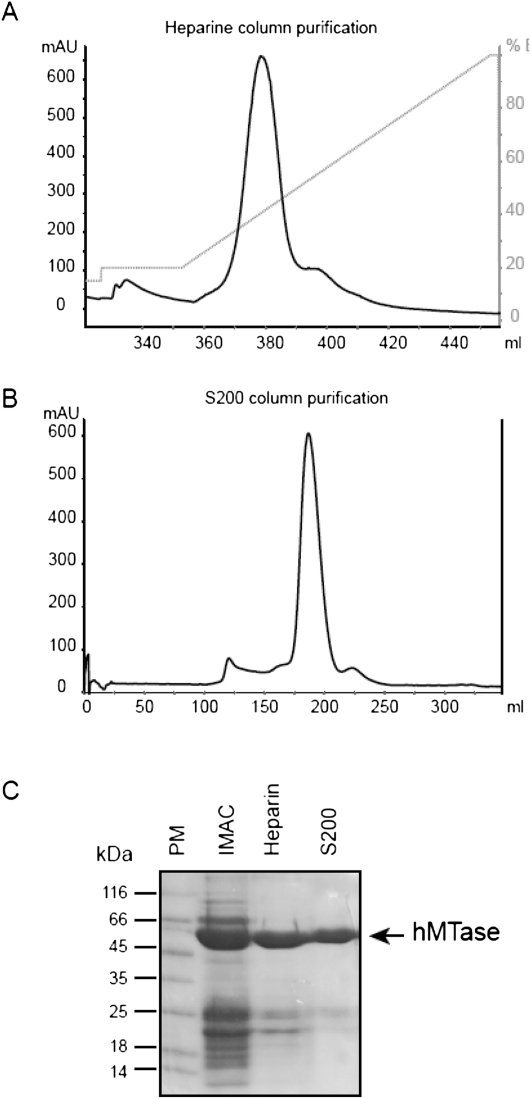
The RNA cap structure obtained by solid-phase chemistry corresponds to the product of the GTase reaction (GpppN-RNA) and is not methylated in its N7 position. Those RNAs may be used as substrate for N7- MTase in crystallographic or biochemical studies. In contrast, cellular mRNAs used as a translation template by the ribosomal complex are methylated at their N7 position by cellular N7-MTase. Since to our knowledge no N7-MTases are commercially available (or only VV N7-MTase coupled the GTase activity), we decided to clone and purify the human N7-hMTase in order to convert GpppN-RNAs (2–4), (9) into 7mGpppN-RNAs (12–15) and GpppN2′-Om-RNA (10) into 7mGpppN2′-Om-RNA (16) (Table 2; Scheme 4). The use of human N7-hMTase would additionally allow us to confirm that cellular MTase recognizes the chemically prepared compounds. We, therefore, cloned the N7-hMTase carrying a six histidine tag into a bacterial expression plasmid. After bacterial expression, the protein was purified in two steps on affinity columns (Ni chelating sepharose and heparin). The N7-hMTase was subsequently purified by gel filtration and highly purified N7-hMTase was eluted as a dimer from the column (Fig. 3). The N7-hMTase stored at −20° in 50% glycerol is stable and keeps its MTase activity for >2 yr.
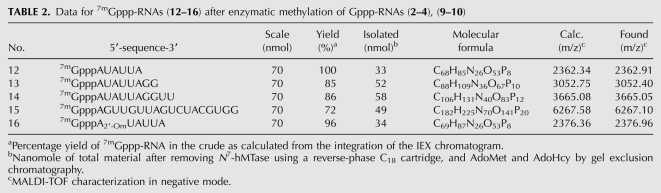
TABLE 2.
Data for 7mGppp-RNAs (12–16) after enzymatic methylation of Gppp-RNAs (2–4), (9–10)
SCHEME 4.
(Guanine-N7)-methyltransfer reaction on 5′-Gppp-RNA by human methyltransferase.
FIGURE 3.
Human N7-methyl transferase purification: The N7-hMTase was expressed in E. coli and purified by two affinity chromatography steps (IMAC and heparin) followed by gel filtration. (A) The elution profile of N7-hMTase followed by monitoring absorption at 280 nm from the heparin column. The percentage of B elution buffer is indicated in gray on the right of the graph. (B) The elution profile of N7-hMTase on a S200 gel filtration column equally followed by monitoring absorption at 280 nm. The main peak eluting after 190 mL corresponds to a dimer of N7-hMTase (110 kDa). (C) The N7-hMTase recovered after each purification step was analyzed by 12% SDS-PAGE stained with Coomassie blue. Lane 1 corresponds to the molecular weight marker, lanes 2, 3, and 4 to the protein eluted from IMAC, heparin, and S200 column, respectively.
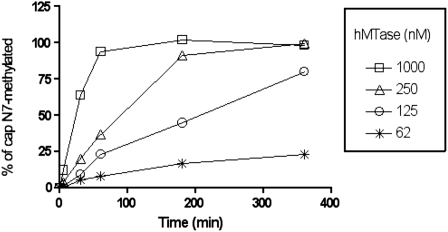
We first incubated Gppp-RNA (5) corresponding to the 5′ end of Dengue virus genome with increasing concentrations of purified hMTase (62, 125, 250, and 1000 nM) in the presence of the methyl donor (AdoMet) containing a tritiated methyl group. The reaction products were then separated from free AdoMet by filter binding assays using DEAE filtermats. As shown in Figure 4, the N7-hMTase allowed the methylation of Gppp-RNA (5) in a time-dependent manner. At the lower enzyme concentrations used (62 and 125 nM), the conversion of Gppp-RNA (5) into 7mGppp-RNA was incomplete. In contrast, at higher N7-hMTase concentrations (250 and 1000 nM), we observed that a plateau was reached after 3–4 h of the incubation period at 30°C. Quantitation of the [3H]-methyl incorporated into the capped RNA indicated that the plateau corresponded to the conversion of 100% of Gppp-RNA (5) into 7mGppp-RNA. The reaction products were also digested by P1 nuclease, and TLC analysis confirmed that the cap structure was methylated at its N7 position as expected (data not shown). Therefore, these results demonstrate that the capped RNA produced by solid-phase chemistry is efficiently recognized by the recombinant cellular N7-hMTase and that it is possible to perform a quantitative N7-methylation of the synthesized RNAs.
FIGURE 4.
Time course analysis of guanine-N7-methylation of GpppAGUUGU by N7-hMTase: 62, 125, 250, and 1000 nM of N7-hMTase was incubated with GpppAGUUGU (5) in the presence of methyl donor (AdoMet) containing a tritiated methyl group. The reaction products, collected after increasing time periods, were separated from free AdoMet by filter binding onto DEAE filtermats. The percentage of methylated Gppp-RNA (5) was quantitated by measurement of the radioactivity transfer onto the substrate RNA.
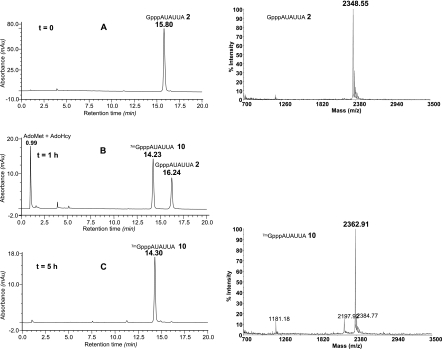
We next set up large-scale N7-methylation assays in order to methylate large amounts (∼70 nmol) of GpppN-RNAs (2–4), (9) or GpppN2′Om-RNA (10) using N7-hMTase (Table 2). For this purpose, the synthetic capped RNAs (2–4), (9–10) were incubated in the presence of 250 nM N7-hMTase and 0.4 mM of the methyl donor (AdoMet) for 5 h at 30°C. The progress of the methylation reaction was followed by HPLC analysis after 1 h and 5 h of incubation (Fig. 5). In the case of GpppN-RNA (2) (Fig. 5A), after 1 h of incubation, the peak corresponding to unmethylated capped GpppN-RNA (2) (RT 16.2) had gradually disappeared and was converted into a new peak corresponding to 7mGpppN-RNA (12) (RT 14.2) (Fig. 5B). After 5 h of incubation, the conversion of RNA (2) in N7-methylated cap-RNA (12) was complete since no trace of the starting material (2) was observed (Fig. 5C). Moreover, no product resulting from RNA degradation was detected; only the presence of the remaining AdoMet and the formed AdoHcy as by-products was noticed at RT 0.99. The 7mGppp-RNAs (12–16) were next separated from the N7-hMTase using a Sep Pak C18 cartridge, and AdoMet and AdoHcy were eliminated by gel exclusion chromatography on a Sephadex G-25 column. 7mGppp-RNAs (12–16) were eluted using TEA buffer, and fraction purity was analyzed by anion-exchange chromatography before lyophilization. The methylation of the cap structure was again confirmed by MALDI-TOF mass spectrometry analysis (Fig. 5). As attested by the yields (Table 2), the methylation reaction of the cap structure was very efficient since the starting capped Gppp-RNAs (2–4), (9–10) were completely converted into N7-methylated Gppp-RNAs (12–16). The lower yields of 85% and 86% obtained for RNA (13) and (14), respectively, may be explained by a slight RNA degradation which was considered acceptable for short sequences. We believe that the higher recovery of RNA (13) and (14) (52 and 58 nmol) compared to RNA (12) and (16) (33 and 34 nmol) originates from a decreased retention of 7mGppp-RNAs (13–14) on the Sephadex column due to the use of a smaller amount of Sephadex gel (5 g instead of 12 g of Sephadex G-25).
FIGURE 5.
Guanine N7-methylation reaction of 5′-GpppAUAUUA (2). Left panel: (A) Anion-exchange HPLC profile of (2) before methylation. (B) Anion-exchange HPLC profile of reaction medium at t = 1 h of incubation with hMTase. (C) Anion-exchange HPLC profile of the crude 5′-7mGpppAUAUUA (12) after 5 h of incubation with hMTase, then desalting through a Sep-pak cartridge to remove proteic material and gel exclusion chromatography to get rid of AdoMet and AdoHcy. Right panel: MALDI-TOF mass spectra of 5′-GpppAUAUUA (2) and of 5′-7mGpppAUAUUA (12).
It is noteworthy that hMTase-mediated N7-methylation was equally efficient irrespective of the substitution at the 2′ position of the 5′ end ribonucleotide (2′’-OH or 2′-O-methyl). This observation is interesting because it means that N7-methylation of the cap structure can occur even when the 5′-terminal ribonucleotide is already 2′-O-methylated.
In conclusion, we succeeded in the synthesis of several capped RNAs through a straightforward and efficient method based on solid-phase RNA assembly followed by the 5′-functionalization with the cap structure Gppp. Both steps were performed on solid support with commercially available materials, and GpppN-RNAs (cap) as well as GpppN2′-Om-RNAs could be obtained with high yields whatever the nature of nucleotides present in 5′ of the RNAs. Using this method, we were able to synthesize large amounts of several RNAs (from 4 to 18 nt) carrying unmethylated cap structure at its N7-position with similar capping yield. It is noteworthy that our chemical synthesis, in contrast to enzymatic methods, allows the production of great amounts of RNA with a defined length and without any limitation regarding 5′ RNA end sequences. Moreover, our procedure is advantageous over existing strategies to synthesize Gppp-RNAs. Indeed, the synthesis of Gppp-RNAs using vaccinia virus guanylyltranferase is rather inefficient because of the reversibility of the capping reaction. We also set up the enzymatic assay allowing methylation of N7-guanine in order to produce cap-0 and cap-1 structure. Since no commercially N7-Mtase is available, this methylation was performed using human N7-Mtase with quantitative yield, and 7mGpppN-RNAs or 7mGpppN2′-Om-RNAs were obtained as pure materials after a simple purification process (filtration through cartridges). The easier access to capped RNAs and their availability are very attractive and promising for structural and mechanistic studies of their complexes with RNA capping enzymes.
MATERIALS AND METHODS
General methods
Pyridine, acetonitrile, triethylamine, and tri-n-butylamine were distilled under calcium hydride. Anhydrous dimethylformamide, carbon tetrachloride, and most of the reagents purchased from Aldrich were used without further purification. Analytical and semipreparative high performance liquid chromatographies were performed on a Dionex DX 600 HPLC system equipped with anion-exchange DNAPac PA100 columns (4 × 250 mm or 9 × 250 mm, Dionex). The following HPLC solvent systems were used: 5% CH3CN in 25 mM Tris-HCl buffer, pH 8 (buffer A) and 5% CH3CN containing 400 mM NaClO4 in 25 mM Tris-HCl buffer, pH 8 (buffer B).
Flow rates were 1.5 mL/min and 5 mL/min for analysis and semipreparative purposes, respectively. MALDI-TOF mass spectra were recorded on a Voyager-DE spectrometer (PerSeptive Biosystems) using a 10:1 (m/m) mixture of 2,4,6-trihydroxyacetophenone/ammonium citrate as a saturated solution in acetonitrile/water (1:1, v/v) for the matrix. Analytical samples were mixed with the matrix in a 1:5 (v/v) ratio, crystallized on a 100-well stainless steel plate and analyzed. UV quantitation of RNAs was performed on a Varian Cary 300 Bio UV/Visible spectrometer by measuring absorbance at 260 nm.
Synthesis of RNA on solid support
RNA synthesis was performed on an ABI 394 synthesizer (Applied Biosystems) from commercially available (Link Technologies) long chain alkylamine controlled-pore glass (LCAA-CPG) solid support with a pore size of 1000 Å derivatized through the succinyl linker with 5′-O-dimethoxytrityl-2′-O-acetyl-[uridine, N4-acetyl cytidine, N6-phenoxyacetyl adenosine, or N2-dimethylformamide guanosine]. RNA sequences were assembled on a 1-μmol scale in Twist oligonucleotide synthesis columns (Glen research) using the PivOM amidites (5′-O-DMTr-2′-O-PivOM-[U, CAc, APac or GPac]-3′-O-(O-cyanoethyl-N,N-diisopropyl-phosphoramidite) prepared according to the reported procedure (Lavergne et al. 2010). The PivOM amidites are also commercially available at Chemgenes. The 5′-O-DMTr-2′-O-Me-ABz-3′-O-(O-cyanoethyl-N,N-diisopropyl-phosphoramidite) was purchased from Chemgenes. Phosphoramidites were vacuum dried prior to their dissolution in extra dry acetonitrile (Glen Research) at 0.1 M. For the coupling reaction, the activator was 5-benzylmercaptotetrazole (BMT, Chemgenes) used at 0.3 M concentration. Dichloroacetic acid (3% in CH2Cl2) (Glen Research) was the detritylation reagent. The capping step was performed with a mixture of 5% phenoxyacetic anhydride (Pac2O) in THF and 10% N-methylimidazole in THF (Link Technologies). The oxidizing solution was 0.1 M iodine in THF/pyridine/H2O (78:20:2; v/v/v) (Link Technologies). After RNA assembly completion, the column was removed from the synthesizer and dried under a stream of argon.
5′-H-phosphitylation of solid-supported RNA
A solution (2 mL) of 0.1 M diphenyl phosphite in dry pyridine was manually passed with a glass syringe through a column containing RNA still attached to the solid support. Using another syringe, the solution was pushed back and forth for 5 min and left to stand for 30 min at room temperature. The support was washed with CH3CN (4 × 2 mL), then a 0.1 M solution of TEAB (2 mL) was pushed back and forth for 5 min and left to react for 120 min at 30°C. The solution was removed from the column, the support was washed with dry CH3CN (4 × 2 mL) and then dried under a stream of argon.
Solid-supported 5′-phosphoroimidazolide RNA by amidative oxidation
The oxidation solution was prepared as follows: to a stirred anhydrous solution of imidazole (150 mg, 2 mmol) in N,O-bis-trimethylsilylacetamide (0.4 mL, 1.64 mmol), CH3CN (0.75 mL), CCl4 (0.75 mL), and triethylamine (0.1 mL) were added under argon. The column containing the 5′-H-phosphonate oligonucleotide was flushed with argon and activated 3-Å molecular sieves (five beads) were added to the glass syringes. The solution was pushed back and forth through the synthesis column for 5 min, then left to react for 5 h at 30°C. The solution was removed from the column, and the support was washed twice with anhydrous CH3CN (2 × 2 mL), followed by a 1 min flush with argon.
Capping reaction with GDP
Guanosine-5′-diphosphate, bis(tri-n-butylammonium) salt, suitable for phosphorylation reactions was prepared from the commercially available guanosine-5′-diphosphate, sodium salt purchased from Jena Bioscience. GDP sodium salt (0.5 g, 1.2 mmol) was dissolved in milliQ water (25 mL), and the solution was passed through a glass column filled with 20 mL of wet DOWEX-50W X 8 resin, H+ form, and then was collected in a 250-mL round flask containing absolute ethanol (12 mL) and tri-n-butylamine (0.72 mL), stirred at 0°C. The Dowex column was rinsed with water (80 mL) to reach pH 5–6. The solvents were evaporated from the collected solution, and the residue was coevaporated four times with absolute ethanol, then was lyophilized from water to afford a white hygroscopic powder (0.938 g, 1.15 mmol, 96%). The desired GDP, tri-n-butylammonium salt was characterized by two doublets at −10.55 and −11.20 ppm (31P-NMR, 121 MHz, D2O). It can be stored as a solid for several weeks at −20°C.
In a dry 4-mL screw-capped glass vial, bis (tri-n-butylammonium) guanosine diphosphate (103 mg, 0.14 mmol), zinc chloride (28 mg, 0.2 mmol, Alfa Aesar), and activated 4-Å molecular sieves (five beads) were mixed in anhydrous DMF (0.5 mL). The Twist column containing the 5′-phosphoroimidazolidate RNA was flushed with argon, and activated 4-Å molecular sieves (five beads) were added to the glass syringes. The GDP solution (0.28 M) was applied to the column and left to react for 18 h at 30°C. The solution was removed, and the support was washed with water (2 × 2 mL), a 0.1 M aqueous solution of EDTA (pH 7, 2 × 2 mL), and dry CH3CN (4 × 2 mL). Finally, the column was dried by blowing argon through it for 1 min.
Deprotection and release of GpppN-RNAs (1–9) and GpppN2′-Om-RNAs (10–11)
The solid-supported Gppp-RNAs (1–11) were deprotected and released from the support as follows: first, a 1 M solution of 1,8-diazadicyclo-[5,4,0]undec-7-ene (DBU) in anhydrous CH3CN was applied to the column for 3 min. Then the solution was removed and the solid support was washed with anhydrous CH3CN. The support was dried by a 1 min flush with argon. Second, a 30% aqueous ammonia solution was applied to the column in three batches (1.5 mL, 1mL, 0.5 mL) for 30 min each. The three ammonia fractions were collected in a 4-mL screw-capped glass vial and were left to react at room temperature for 1.5 h. The fully deprotected oligonucleotides (1–11) were transferred to 50-mL round-bottomed flasks, and isopropylamine (15% of total volume: 0.45 mL) was added to the solutions. Then, the mixtures were evaporated under reduced pressure with a bath at 30°C maximum until the volumes were reduced to 0.3 mL. The mixtures were coevaporated three times with 1 mL of water following the same protocol. The residues were redissolved in water (1.5 mL divided in three portions for flask rinse: 0.8 mL, 0.4 mL, 0.3 mL) and transferred to 2-mL Eppendorf-vials, then lyophilized from water.
Analysis and purification of Gppp-RNAs (1–11)
The crude Gppp-RNAs (1–8), (10–11) were analyzed by anion-exchange HPLC using a 0%–30% or 0%–40% linear gradient of buffer B in buffer A, and they were characterized by MALDI-TOF spectrometry. The crude mixtures (1–5), (10–11) were then purified by semipreparative IEX-HPLC with a 0%–30% or 0%–40% linear gradient of buffer B in buffer A. The crude (9) was analyzed and purified by IEX-HPLC using a 0%–75% linear gradient of buffer B in buffer A at 50°C. The pure fractions of the Gppp-RNAs (1–5), (9–11) were pooled in 100-mL round-bottomed flasks and were concentrated to 0.5 mL under reduced pressure with a bath at 30°C. The mixtures were dissolved in 3 mL of 50 mM TEAAc buffer and loaded on a Sep Pak C18 cartridge. Elution was performed with 10 mL of 50 mM TEAAc, pH 7, then with 10 mL of 50% CH3CN in 12.5 mM TEAAc, pH 7. The second fraction containing the desired RNA was collected in a 100-mL round-bottomed flask and was freeze-dried. The residue was dissolved in 1.5 mL water (divided in three portions of 0.8 mL, 0.4 mL, and 0.3 mL for flask rinse) and transferred to a 2-mL Eppendorf-vial and lyophilized from water. Lyophilized Gppp-RNAs (1–11) were stored at −20°C for several months without any degradation.
Human (guanine N7)-methyltransferase (N7-hMTase) expression and purification
The cDNA coding for N7-hMTase was a kind gift from Aaron J. Shatkin (Center of Advanced Biotechnology and Medicine, Piscataway, NJ). It was cloned into expression vector pDest14 (Invitrogen) to yield pDest14/6His-hMTase, as described in Peyrane et al. (2007). E. coli Rosetta (DE3) cells were transformed with pDest14/6His-hMTase and grown in Luria-Bertani medium containing ampicillin and chloramphenicol. At an OD600 of 0.6, IPTG and ethanol were added to a final concentration of 100 mM and 2%, respectively, and expression was allowed to proceed for 18 h at 17°C. The cellular pellet was resuspended in 10 mL lysis buffer (50 mM Tris, pH 8.5, 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, and antiprotease cocktail [Complete, Roche]) supplemented with 10 mM imidazole, 100 μg/mL lysozyme, 1 μg/mL DNAse I, and 0.5% triton X100. After lysis by sonication and clarification, immobilized-metal-affinity chromatography (IMAC) was used for the first purification step (chelating sepharose fast-flow resin [GE Healthcare] loaded with Ni2+). The N7-hMTase was eluted with lysis buffer, pH 7.5, containing 250 mM imidazole. Fractions were then diluted fivefold in 50 mM Bis-Tris, pH 7.0, 25 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol and loaded onto a 5-mL heparin column (GE Healthcare). The N7-hMTase was then eluted with a gradient of NaCl (0–1 M) in the same buffer. The protein elutes between 150 and 350 mM NaCl. Fractions enriched in protein were concentrated using a centricon Plus 70 (10,000 MW cut-off, Millipore) device, loaded onto a gel HiLoad 16/60 Superdex 200 gel filtration column (GE Healthcare), and eluted using 50 mM Tris, pH 7.5, 300 mM NaCl, glycerol 10%, and 5 mM β-mercaptoethanol. Fractions containing N7-hMTase eluted as an apparent dimer in size. They were pooled, concentrated at 1 mg/mL, and dialyzed against 50 mM Tris, pH 7.5, 300 mM NaCl, 50% glycerol, and 5 mM β-mercaptoethanol and stored at −20°C.
Radioactive (guanine-N7)-methylation assay of GpppAGUUGU (5) by N7-hMTase
The radiocative methyltransferase assays were performed as described by Selisko et al. (2010), with the following modification. Briefly, the methylation reactions were set up in 80 μL of 40 mM Tris-HCl, pH 8, with 5 mM dithiothreitol, 10 μM GpppAGUUGU (5), and 0.4 mM S-adenosylmethionine (AdoMet, New England Biolabs) supplemented with 0.03 μCi (1.11 kBq) [3H]AdoMet per μL (GE Healthcare) in the presence of increasing concentrations of purified N7-hMTase (62, 125, 250, or 1000 nM). The reaction was started either with AdoMet after a pre-incubation of N7-hMTase with RNA. Reaction mixtures were incubated at 30°C for the given time periods and stopped by 10-fold dilution with chilled 100 μM AdoHcy. Samples were then transferred to glass-fiber filtermats (DEAE filtermats, Wallac) by a Filtermat Harvester (Packard Instruments). Filtermats were washed twice with 0.01 M ammonium formate (pH 8.0), twice with water, and once with ethanol, and then dried and transferred into sample bags. Betaplate Scint (Wallac) scintillation fluid was added, and the methylation of RNA substrates was measured in counts per minute (c.p.m.) using a Wallac 1450 MicroBeta TriLux Liquid Scintillation Counter.
Large-scale (guanine-N7)-methylation of GpppN-RNAs (2–4), (9) and GpppN2′-Om-RNA (10) by N7-hMtase
Methylation of purified Gppp-RNAs (2–4), (9–10) (∼70 nanomol) to give 7mGpppN-RNAs (12–15) and 7mGpppN2′-Om-RNA (16) was carried out using 0.25 μM N7-hMTase and 0.4 mM S-adenosylmethionine (New England Biolabs) in 40 mM Tris-HCl, pH 8, with 5 mM dithiothreitol in a 1725-μL reaction volume (Gppp-RNAs at a final concentration of 40 μM) at 30°C. N7-methylation was monitored using IEX-HPLC analysis of an aliquot from the reaction mixture and passed through ZipTip C18 (see procedure below) prior analysis to remove protein. Crude 7mGppp-RNAs (12–14), (16) were analyzed using IEX-HPLC and a step gradient of 0%–15% of solvent B for 5 min, then 15%–30% for 15 min (for 6-mers [12] and [16]) or 0%–20% for 5 min, then 20%–40% for 15 min (for 8-mer [13] and 10-mer [14]). Crude 18-mer (15) was analyzed with a step gradient of 0%–40% of eluent B for 5 min, then 40%–75% for 15 min at 50°C. After a 5-h incubation, the reaction was complete. The mixture was subsequently redissolved in 2 mL of 50 mM TEAAc, pH 7. Proteic material was removed using a Sep Pak C18 cartridge following the same desalting procedure as described above. Remaining AdoMet and the S-adenosylhomocysteine (AdoHcy) product were then removed as follows: the residue was dissolved with 1 mL of 12.5 mM TEAAc, pH 7, and loaded onto a Sephadex G-25 gel filtration column (12 g, h 120 × d 25 mm). Capped RNAs (12–16) were eluted using 30 mL of 12.5 mM TEAAc applied in batches of 5 mL each, and fractions were analyzed by IEX-HPLC. Then, batches containing the pure 7mGppp-RNA were pooled together in a 100-mL round-bottomed flask and lyophilized from water. The residue was dissolved in 0.8 mL, lyophilized twice with sequential dissolution in 0.4 mL, and finally 0.3 mL of water, and transferred to a 2-mL Eppendorf-vial and freeze-dried. Lyophilized 7mGppp-RNAs (12–16) were stored at −20°C for several months without any observable degradation.
ZipTip C18 procedure
ZipTip C18 (Millipore) was first conditioned with 40 μL of CH3CN (HPLC grade) followed by 80 μL of 50 mM TEAAc buffer, pH 7. An aliquot of the reaction mixture (40 μL) was mixed with buffer (40 μL) and loaded on ZipTip C18. Elution was performed with buffer (40 μL), then with 50% CH3CN in 12.5 mM TEAAc, pH 7 (40 μL). The second fraction containing the desired Gppp-RNA was collected in a 1-mL Eppendorf-vial and was concentrated to remove CH3CN under reduced pressure (speed-vac).
ACKNOWLEDGMENTS
We thank Dr. I. Zlatev for helpful discussions on purification and M.-C. Bergogne for assistance in the capped RNA bibliography search. This work was supported by CNRS funding through the Programme Interdisciplinaire: Maladies Infectieuses Emergentes, by the Ministère de l'Education Nationale de la Recherche et Technologie, by the project SILVER (Health-F3-2010-260644) of the European Union 7th Framework Programme, and by the Direction Générale de l'Armement (contract DGA 09co402). Furthermore, we thank M.C. Hugues Tolou for support and acknowledge support by the Fondation pour la Recherche Médicale (Programme Equipe FRM to B.C.). Y.T. thanks the MNRT for the award of a research studentship. F.M. is from INSERM.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030932.111.
REFERENCES
- Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, Cook S, Coutard B, Decroly E, de Lamballerie X, Gould EA, et al. 2010. Structure and functionality in flavivirus NS-proteins: Perspectives for drug design. Antiviral Res 87: 125–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, Decroly E 2010. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog 6: e1000863 doi: 10.1371/journal.ppat.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee GG, Fodor E, Pritlove DC, Gould KG, Dalluge JJ 1995. Solid-phase synthesis of 5′-diphosphorylated oligoribonucleotides and their conversion to capped m7Gppp-oligoribonucleotides for use as primers for influenza A virus RNA polymerase in vitro. Nucleic Acids Res 23: 2641–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman TM, Wang GC, Huang FQ 2004. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 φ2.5 promoter. Nucleic Acids Res 32: e14 doi: 10.1093/nar/gnh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li JQ, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong HP, et al. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468: 452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E, Debarnot C, Ferron F, Bouvet M, Coutard B, Imbert I, Gluais L, Papageorgiou N, Sharff A, Bricogne G, et al. 2011. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog 7: e1002059 doi: 10.1371/journal.ppat.1002059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E, Ferron F, Lescar J, Canard B 2012. Conventional and unconventional mechanisms for capping of virus mRNA. Nat Rev Microbiol 10: 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, Canard B 2007. Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol 372: 723–736 [DOI] [PubMed] [Google Scholar]
- Hodel AE, Gershon PD, Quiocho FA 1998. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol Cell 1: 443–447 [DOI] [PubMed] [Google Scholar]
- Imburgio D, Rong M, Ma K, McAllister W 2000. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry 39: 10419–10430 [DOI] [PubMed] [Google Scholar]
- Iwase R, Sekine M, Hata T, Miura K-I 1988. A new method for the synthesis of capped oligoribonucleotides by use of an appropriately protected 7-methylguanosine diphosphate derivative as a donor for the triphosphate bond formation. Tetrahedron Lett 29: 2969–2972 [Google Scholar]
- Iwase R, Maeda M, Fujiwara T, Sekine M, Hata T, Miura K 1992. Molecular design of a eukaryotic messenger RNA and its chemical synthesis. Nucleic Acids Res 20: 1643–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity J, Heinonen P, Lonnberg H, Darzynkiewicz E 2005. A novel approach to solid-phase chemical synthesis of oligonucleotide mRNA cap analogs. Nucleosides Nucleotides Nucleic Acids 24: 601–605 [DOI] [PubMed] [Google Scholar]
- Jemielity J, Kowalska J, Rydzik AM, Darzynkiewicz E 2010. Synthetic mRNA cap analogues with a modified triphosphate bridge—synthesis, applications, and prospects. New J Chem 34: 829–844 [Google Scholar]
- Kadokura M, Wada T, Urashima C, Sekine M 1997. Efficient synthesis of γ-methyl-capped guanosine 5′-triphosphate as a 5′-terminal unique structure of U6 RNA via a new triphosphate bond formation involving activation of methyl phosphoroimidazolidate using ZnCl2 as a catalyst in DMF under anhydrous conditions. Tetrahedron Lett 38: 8359–8362 [Google Scholar]
- Kadokura M, Wada T, Seio K, Moriguchi T, Huber J, Luhrmann R, Sekine M 2001. Solid-phase synthesis of a 5′-terminal TMG-capped trinucleotide block of U1 snRNA. Tetrahedron Lett 42: 8853–8856 [Google Scholar]
- Koukhareva II, Lebedev AV 2004. Chemical route to the capped RNAs. Nucleosides Nucleotides Nucleic Acids 23: 1667–1680 [DOI] [PubMed] [Google Scholar]
- Lavergne T, Bertrand JR, Vasseur JJ, Debart F 2008. A base-labile group for 2′-OH protection of ribonucleosides: A major challenge for RNA synthesis. Chemistry 14: 9135–9138 [DOI] [PubMed] [Google Scholar]
- Lavergne T, Janin M, Dupouy C, Vasseur J-J, Debart F 2010. Chemical synthesis of RNA with base-labile 2′-O-(pivaloyloxymethyl)-protected ribonucleoside phosphoramidites. In Current protocols in nucleic acid chemistry (ed. SL Beaucage et al.), pp. 3.19.11–13.19.27. Wiley, New York. [DOI] [PubMed] [Google Scholar]
- Lewdorowicz M, Yoffe Y, Zubereck J, Jemielity J, Stepinski J, Kierzek R, Stolarski R, Shapira M, Darzynkiewicz E 2004. Chemical synthesis and binding activity of the trypanosomatid cap-4 structure. RNA 10: 1469–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola S, Zhang Z, Mäki E, Lönnberg H 2005. Preparation and properties of mRNA 5′-cap structure. Curr Org Chem 9: 1–23 [Google Scholar]
- Nagata S, Hamasaki T, Uetake K, Masuda H, Takagaki K, Oka N, Wada T, Ohgi T, Yano J 2010. Synthesis and biological activity of artificial mRNA prepared with novel phosphorylating reagents. Nucleic Acids Res 38: 7845–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrane F, Selisko B, Decroly E, Vasseur JJ, Benarroch D, Canard B, Alvarez K 2007. High-yield production of short GpppA- and 7MeGpppA-capped RNAs and HPLC-monitoring of methyltransfer reactions at the guanine-N7 and adenosine-2′-O positions. Nucleic Acids Res 35: e26 doi: 10.1093/nar/gkl1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H, Wakai H, Nakamura-Ozaki A 1999. Synthesis and reactions of nucleoside 5′-diphosphate imidazolide. A nonenzymatic capping agent for 5′-monophosphorylated oligoribonucleotides in aqueous solution. J Org Chem 64: 5836–5840 [Google Scholar]
- Sekine M, Nishiyama S, Kamimura T, Osaki Y, Hata T 1985. Chemical synthesis of capped oligoribonucleotides, m7G5′ pppAUG, and m7G5′ pppAUGACC. Bull Chem Soc Jpn 58: 850–860 [Google Scholar]
- Sekine M, Iwase R, Hata T, Miura K 1989. Synthesis of capped oligoribonucleotides by use of protected 7-methylguanosine 5′-diphosphate derivatives. J Chem Soc, Perkin Trans 1 969–978 [Google Scholar]
- Sekine M, Kadokura M, Satoh T, Seio K, Wada T, Fischer U, Sumpter V, Luhrmann R 1996. Chemical synthesis of a 5′-terminal TMG-capped triribonucleotide m(3)(2,2,7)G(5′)pppAmpUmpA of U1 RNA. J Org Chem 61: 4412–4422 [DOI] [PubMed] [Google Scholar]
- Selisko B, Peyrane FF, Canard B, Alvarez K, Decroly E 2010. Biochemical characterization of the (nucleoside-2′-O)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n). J Gen Virol 91: 112–121 [DOI] [PubMed] [Google Scholar]
- Shatkin AJ 1976. Capping of eukaryotic messenger-RNAs. Cell 9: 645–653 [DOI] [PubMed] [Google Scholar]
- Shuman S 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol 66: 1–40 [DOI] [PubMed] [Google Scholar]
- Sobkowski M, Kraszewski A, Stawinski J 1998. The reactions of H-phosphonates with bifunctional reagents. Part V. Functionalization of support-bound oligonucleotides and synthesis of nonradioactive hybridization probes. Nucleosides Nucleotides Nucleic Acids 17: 253–267 [DOI] [PubMed] [Google Scholar]
- Yanachkov IB, Dix EJ, Yanachkova MI, Wright GE 2011. P1,P2-diimidazolyl derivatives of pyrophosphate and bis-phosphonates—synthesis, properties, and use in preparation of dinucleoside tetraphosphates and analogs. Org Biomol Chem 9: 730–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatev I, Lavergne T, Debart F, Vasseur J-J, Manoharan M, Morvan F 2010. Efficient solid-phase chemical synthesis of 5′-triphosphates of DNA, RNA, and their analogues. Org Lett 12: 2190–2193 [DOI] [PubMed] [Google Scholar]
- Zubereck J, Wyslouch-Cieszynska A, Niedzwiecka A, Dadlez M, Stepinski J, Augustyniak W, Gingras A-C, Zhang Z, Burley SK, Sonenberg N, et al. 2003. Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogs by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA 9: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, et al. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and nonself mRNA dependent on the RNA sensor Mda5. Nat Immunol 12: 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]