Membrane proteins account for over one-fifth of the encoded proteins but their crystallization is challenging, particularly for multi-protein complexes such as the polysaccharide export system of Escherichia coli.[1, 2] One major component of this system is the translocation channel Wza, an octameric outer membrane protein of 320 kDa whose closed-state crystal structure has been recently determined.[3] Wza is thought to interact with different other proteins which, in part, reside in the inner membrane and cytosol, to form a periplasm-spanning molecular machine.[1, 2] The open-state structure of Wza is presumably stabilized during interaction with other proteins and X-ray crystallography alone may not give a view of this dynamic complex. Pulsed electron-electron double resonance (PELDOR) is a powerful tool for measuring distances up to 80 Å.[4-6] Recently the approach has been applied to study the maltose ATP binding cassette membrane protein complex and quantified the internal motions during the catalytic cycle. [7] However, the approach has yet to be applied to large highly symmetric integral membrane proteins such as Wza. This is an important gap as many cellular processes involve highly symmetric membrane proteins. We show that PELDOR is well suited for the study of such a system and a comparison of the PELDOR data with the crystal structure demonstrates the accuracy of the distance fingerprint. This fingerprint provides a convenient ruler by which to assess conformational changes.
Two spin-labelled Wza species were made by expressing the single mutants G58C and Q335C of Wza and then reacting these mutants with the nitroxide MTSSL. Mass spectrometry was used to confirm the labelling. Since Wza is an eight fold symmetric multimer, the eight labels in each biological unit give rise to four principle distances (Figure 1).
Figure 1.
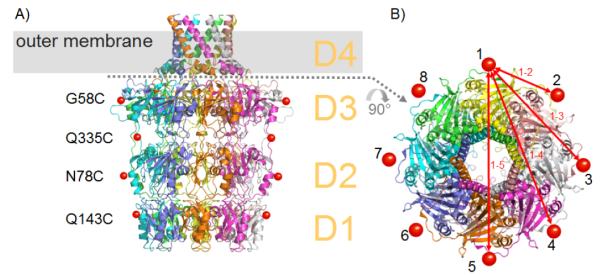
Structure of Wza. A) Each Wza monomer is coloured differently to indicate the octameric structure. Domains D1-D4 are labelled and the position of the outer membrane is indicated by the grey bar. Red spheres indicate where nitroxide spin labels were attached. B) View from the extracellular space, the colours correspond to A).
PELDOR experiments were performed on these proteins solubilised in n-dodecyl-β-D-maltopyranoside (DDM) micelles, yielding modulated time traces and distance distributions (Figure 2A,B). For both mutants, two distinct distance peaks could be measured, for WzaQ335C at 28.6 Å and 51 Å and for WzaG58C at 36.7 Å and 66 Å (Table 1). Taking the additional length of ~9 Å of each label into account, these distances agree well with the Cβ-Cβ distances 1-2 and 1-3 (Figure 1B) inferred from the crystal structure (Table. 1).
Figure 2.
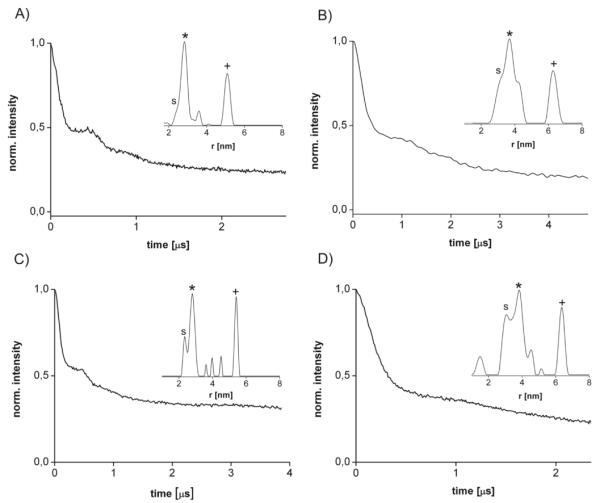
PELDOR time traces and distance distributions (insets) for A) WzaQ335C, B) WzaG58C, C) sWzaQ335C and D) sWzaG58C. The labels (*) and (s) indicate the two conformers for the principle distance 1-2 and the label (+) marks principle distance 1-3.
Table 1.
Distances obtained for Wza and sWza by PELDOR and X-ray crystallography.
| Label | Spin-Spin Distances [Å] | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 1 – 2 | 1 – 3 | ||||||
|
|
|||||||
| PELDORa | Cβ-Cβb | X-rayc | PELDORa | Cβ-Cβb | X-rayc | ||
| Wza | Q335C | 23.6, 28.6 | 24.9 | --- | 51 | 45.9 | --- |
| G58C | 31.8, 36.7 | 33.5b | --- | 66 | 62.3b | --- | |
|
| |||||||
| sWza | Q335C | 23.7, 28.1 | 24.9 | 23.4, 29.0 | 54 | 45.9 | 53.1 |
| G58C | 32.1, 36.6 | 33.5d | --- | 64 | 62.3b | --- | |
| N78C | 33.8, 37.5 | 33.0 | --- | 59 | 61.0 | --- | |
| Q143C | 29.6, 32.1 | 27.5 | --- | 61 | 50.5 | --- | |
The error in the PELDOR distances is 0.4 Å and 4 Å for principle distances 1-2 and 1-3, respectively.
The Cβ-Cβ distances of course do not allow for the length of the label and side chain.
The X-ray distances are measured as the nitroxides O-to-O distances.
Cα-Cα distance.
The purification of Wza is time-consuming, so we sought to investigate whether a soluble version of the protein could be produced that retained the same structural properties as full length Wza. We expressed a truncated Wza 24-345 mutant (sWza) (see supplementary information), which lacks the signal sequence and the C-terminal transmembrane domain D4. The structure of the sWza was essentially identical to the corresponding domains from the full-length protein (see Supplementary Information). As the C-terminal transmembrane helices are unlikely to be involved in protein protein complex formation, sWza is a suitable system for further study. A set of four mutant constructs was made, sWzaQ335C, sWzaG58C, sWzaN78C and sWzaQ143C, spanning the full length and width of octameric sWza. PELDOR experiments on sWzaQ335C and sWzaG58C give the same peaks identified for the corresponding full-length construct, but the spectra appear sharper. PELDOR measurements on sWzaN78C and sWzaQ143C yielded principle distances 1-2 and 1-3 (Table 1 and supplementary information) matching the predictions from the crystal structure. As with WzaG58C and WzaQ335C, the peak for the 1-2 distance appears to be split and can be resolved into two distances for all sWza mutants.
We had three concerns that needed to be addressed. First, we have failed to observe the longer 1-4 and 1-5 distances. These distances of about 75 Å would have calculated modulation periods longer than 8 μs. Within the time window accessible such long modulations are difficult to resolve from the superimposed decay caused by intermolecular spin-spin interactions.[4, 5] This appears to be an experimental limitation of the approach.
Second, the PELDOR time traces reveal a modulation depth λ of only 0.4 to 0.6 whereas theory predicts a λ of 0.98 for eight interacting spin-labels.[8, 9] Biochemical studies indicated that in solution spin-labelled Wza and sWza are octameric and the measured 1-2 and 1-3 distances are consistent with the octameric arrangement. The only variable that we can identify is incomplete labelling leading to a mixture of octamers some with less 8 labels. Experiments on organic model systems have shown that in mixtures of spin oligomers the refocused echo of the detection sequence for long time windows (as in this case) is dominated by the slower relaxing spin-systems.[10] Thus systems with fewer interactions spins would be predicted to contribute more to the echo intensity than the faster relaxing fully occupied system. If this is correct, then for Wza, λ is dominated by incompletely labelled Wza molecules within our timing window. Hence, shorter time windows should lead to an increase in λ. Shortening the time window for sWzaQ335C, does indeed increase λ significantly, and extrapolating the data back to t = 0 yields a λt=0 of 0.94 matching the theoretical value of 0.94 with a labelling efficiency of 85% (see supplementary information). It is hard to confirm independently labelling efficiency, since mass spectrometry and chemical analysis are not accurate enough to quantify small amounts of unlabelled protein. The time window analysis however identifies an experimental approach to detecting and accounting for incomplete labelling. Rapid relaxation could also be a limitation on signal quality in systems where the label itself was inserted within the lipid bi-layer.
Third, the presence of two peaks, a shoulder or a broad peak at the principle distance 1-2 was puzzling. We were concerned there was some structural artifact, or an anomaly in the data acquisition or analysis. The former issue was eliminated as sWza and labelled sWzaQ335C are in essence identical to the full length Wza for superimposable atoms (see supplementary information). Eight-fold ncs-averaged simulated-annealing 2.9 Å omit-maps of sWzaQ335C reveal strong evidence for two different conformations of the MTSSL side chain (Figure 3B). One of the conformers shows stronger electron density (I in Figure 3B), suggesting a higher occupancy compared to the second conformer (II in Figure 3B). Conformer I is involved in several weak van-der-Waals interactions, mostly with hydrophobic parts of neighboring residues, whereas conformer II sticks out into the solvent. Conformations I and II were modeled with an occupancy of 0.7 and 0.3, respectively, and refined independently. These conformers are compatible with a library of MTSSL rotamers derived form MTSSL-labelled T4-lysozyme structures.[11, 12] Using the crystallographic locations for the spin labels, an occupancy-weighted distance histogram was calculated, which shows an excellent fit to the PELDOR data reproducing the splitting of the 1-2 peak (Figure 3C).
Figure 3.
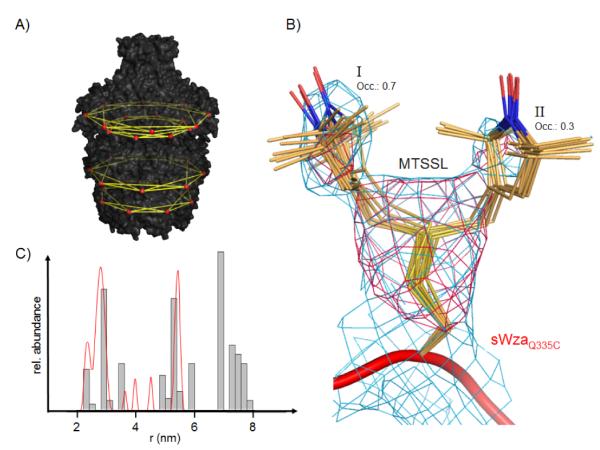
PELDOR distance fingerprint of Wza. A) Distances measured by PELDOR are indicated by yellow lines, illustrating the extended distance web over Wza (black surface). The position of spin labels is indicated by red spheres. B) Close-up of the MTSSL label in crystallized sWzaQ335C. All of the individually refined MTSSL residues in the octameric structure have been superposed and rendered as sticks. Conformation I has been refined at an occupancy of 0.7 and II of 0.3. The pink mesh represents an 8-fold NCS-averaged simulated-annealing omit map contoured at 4.5 σ. The blue mesh shows the 8-fold NCS-averaged 2Fo-Fc map contoured at 1.0 σ. C) Comparison between distances from PELDOR (red line) and X-ray structure of sWzaQ335C (grey histogram). The histogram is corrected for the different occupancies of MTSSL conformations I and II.
That we are capable of experimentally resolving such fine detail, points to the power and potential of PELDOR in even complex systems such as Wza. The data are another example of discrete multiple locations of the label [11-13] rather than static or so called smooth cone distribution. [14]
PELDOR has now been applied to a large highly symmetrical membrane protein oligomer expanding the demonstrated utility of the technique. We have carefully analysed this system to quantify not only the accuracy but also the limitations of the approach. We have created a series of site-specifically labelled mutants providing us with a molecular fingerprint that can monitor changes in distances associated with molecular re-arrangements of Wza. Thus, when combined with other structural techniques as e.g. X-ray, cryo-electron microscopy, fluorescence or molecular dynamics simulations, PELDOR can contribute compelling molecular insights into complex biological phenomena.
Supplementary Material
[**] Acknowledgement
Funding from The BBSRC, the EPSRC “Hyper” project and The Wellcome Trust program grant (JHN and CW). CW is supported by a Canada Research Chair and funding from CIHR and OS by a fellowship from the Research Councils of the UK.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Experimental Section See supplementary material for a full experimental details.
References
- [1].Whitfield C, Naismith JH. Curr.Opin.Struct.Biol. 2008;18:466. doi: 10.1016/j.sbi.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Whitfield C. Annu.Rev.Biochem. 2006;75:39. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- [3].Dong C, Beis K, Nesper J, Brunkan-Lamontagne AL, Clarke BR, Whitfield C, Naismith JH. Nature. 2006;444:226. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jeschke G. Macromolecular Rapid Communications. 2002;23:227. [Google Scholar]
- [5].Milov AD, Salikohov KM, Shirov MD. Fizika Tverdogo Tela. 1981;23:975. [Google Scholar]
- [6].Schiemann O, Prisner TF. Q.Rev.Biophys. 2007;40:1. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- [7].Grote M, Bordignon E, Polyhach Y, Jeschke G, Steinhoff HJ, Schneider E. Biophysical Journal. 2008;95:2924. doi: 10.1529/biophysj.108.132456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Milov AD, Maryasov AG, Tsvetkov YD. Applied Magnetic Resonance. 1998;15:107. [Google Scholar]
- [9].Milov AD, Ponomarev AB, Tsvetkov YD. Chemical Physics Letters. 1984;110:67. [Google Scholar]
- [10].Bode BE, Margraf D, Plackmeyer J, Durner G, Prisner TF, Schiemann O. J.Am.Chem.Soc. 2007;129:6736. doi: 10.1021/ja065787t. [DOI] [PubMed] [Google Scholar]
- [11].Guo Z, Cascio D, Hideg K, Hubbell WL. Protein Sci. 2008;17:228. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Langen R, Oh KJ, Cascio D, Hubbell WL. Biochemistry. 2000;39:8396. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]
- [13].Hilger D, Polyhach Y, Padan E, Jung H, Jeschke G. Biophysical Journal. 2007;93:3675. doi: 10.1529/biophysj.107.109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hustedt EJ, Stein RA, Sethaphong L, Brandon S, Zhou Z, DeSensi SC. Biophysical Journal. 2006;90:340. doi: 10.1529/biophysj.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Beis K, Whitfield C, Booth I, Naismith JH. Int.J.Biol.Macromol. 2006;39:10. doi: 10.1016/j.ijbiomac.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Drummelsmith J, Whitfield C. EMBO J. 2000;19:57. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jeschke G, Panek G, Godt A, Bender A, Paulsen H. Applied Magnetic Resonance. 2004;26:223. [Google Scholar]
- [18].Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. Applied Magnetic Resonance. 2006;30:473. [Google Scholar]
- [19].Otwinowski Z, Minor W. Macromolecular Crystallography, Pt A. 1997;276:307. [Google Scholar]
- [20].Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Journal of Applied Crystallography. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bailey S. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760. doi: 10.1107/S0907444993011898. [DOI] [PubMed] [Google Scholar]
- [22].Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1948. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


