Abstract
A recently-published study has used next-gen sequencing technology to resequence two Y chromosomes separated by 13 generations and discovered four single-base differences in ~10 Mb DNA, suggesting that the Y chromosome euchromatin accumulates around one mutation per generation. Y-SNPs therefore now offer the best resolution of Y haplotypes and promise to distinguish almost every Y chromosome. This work illustrates the promise of current sequencing technology for forensically-relevant applications.
Keywords: Next-gen sequencing, Y-SNP, Y-STR, Haplotype resolution, forensic applications
Would you bet on the hare or the tortoise winning the race? The hare can run faster and rushes away at the beginning, but if you remember Aesop’s fable, you will recall that the slow plodding tortoise wins in the end. For almost a quarter of a century, forensic genetics has depended on highly-variable minisatellites and STRs to characterize DNA from humans and other species, generating ‘DNA fingerprints’ or ‘DNA profiles’ that are, apart from in identical twins, individual-specific [1]. But now the tortoise of the field, SNPs, has received a boost from developments in sequencing technology [2] and is challenging the hare. Our money is on the tortoise.
We have been exploring the potential of one of these technologies to resequence Y chromosomes from related males and measure the base substitution mutation rate on this chromosome [3]. Such an esoteric study is far from the realities of casework that confront forensic geneticists every day, but nevertheless has far-reaching implications that stimulate us to write this commentary.
Forensic and other geneticists often need to discriminate between Y chromosomes. In a forensic context, an example would be the need to decide whether or not a Y-chromosomal DNA sample from a crime scene matches that from a suspect, or a profile in a database of population samples. To address such questions, Y-STRs have generally been used. Each STR locus has multiple alleles and a combination of 9-17 loci, as commonly used, defines a haplotype whose frequency can be estimated from a suitable database [4]. But Y-STRs have limitations: because of the lack of recombination on the Y chromosome and finite Y-STR mutation rate, a son will generally carry the same haplotype as his father, and male-line relatives less than 20 generations apart are more likely than not to carry an identical haplotypes even with 17 Y-STRs. While this association between genetic similarity and family relationship is central to some applications, such as testing for paternity or more distant male-line relationships [5], it means that close male-line relatives may not be distinguished on the basis of a Y-STR profile. In such cases, higher haplotype resolution would be advantageous, and additional Y-STRs have been discovered [6] and datasets of 67-Y-STR haplotypes produced [7]. But it was notable that in a set of 590 males from worldwide populations, 18 high-resolution haplotypes of 67 Y-STRs were still shared by more than one (and up to five) individuals [7]. While the number of Y-STRs could in principle be increased a few-fold more, these findings point to the value of exploring alternative ways of increasing phylogenetic resolution.
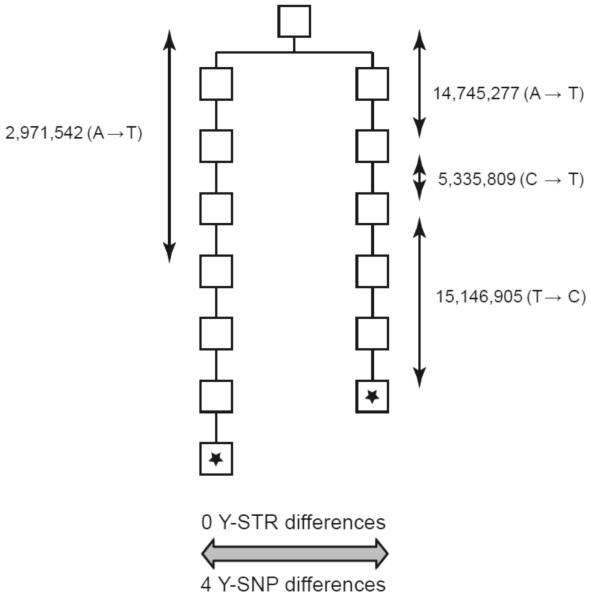
Y-SNPs are generally thought of as providing low phylogenetic resolution: they are currently used mainly to define haplogroups, the major branches in the Y tree. But this property reflects the ascertainment of the commonly-used Y-SNPs more than the intrinsic properties of Y-SNPs. Investigators have laboriously sought Y-SNPs shared by many individuals and have generally paid little attention to the more numerous rare or individual-specific SNPs. Since there are around 24 million nucleotides in the euchromatic Y-specific section of the chromosome, there are plenty of opportunities for SNP variation to occur. Next-gen sequencing technology allows entire Y chromosomes to be sequenced, so this vast potential resource can be accessed. Our study compared two Y chromosomes from the same family separated by 13 generations [3]. These chromosomes were genotyped with the 67 Y-STRs mentioned above, and showed no Y-STR differences. But sequencing them revealed four Y-SNP differences (Figure 1). Detecting this small number of differences presented formidable technical challenges and more than 30,000 false positives had to be eliminated, including eight in vitro mutations that had arisen in the cell lines that formed the source of the DNA that was sequenced rather than in vivo within the individuals. The four true mutations were confirmed by standard capillary sequencing of blood DNA from the same individuals, and three of the four by their presence in other members of the family. For technical reasons, this study focused on ~10 Mb of single-copy DNA, so if the entire 24 Mb of Y-specific euchromatin had been analysed, then around 10 mutations would be expected (with wide confidence limits), close to one Y-SNP mutation per generation and in excellent agreement with expectations from comparisons of human and chimpanzee Y chromosomes.
Figure 1.
Two Y chromosomes (from the starred individuals) from a deep-rooting pedigree were genotyped and resequenced. They showed zero Y-STR differences after typing 67 Y-STRs, but four base substitutions after comparing ~10 Mb DNA sequence. Typing additional members of the pedigree allowed these four mutations to be placed on the pedigree in the locations indicated.
The implication of this study is thus that it should be possible to find a SNP specific for almost any Y chromosome, distinguishing even between fathers and sons. But when those faced with trace stains from casework and urgent deadlines hear that the study required the establishment of cell lines (taking months) and flow-sorting of the Y chromosome (taking weeks), as well as the funding and sequencing resources of a genome centre, they may be sceptical of its relevance to their day-to-day work. However, sequencing technology is improving rapidly. Numerous personal genome sequences have been published, e.g. [8, 9] and the 1000 Genomes Project has made ~180 whole-genome sequences available (http://www.1000genomes.org/page.php). Sequencing costs are falling rapidly. At some point, the first step in a forensic genetic will be to sequence DNA from the stain: degraded fragments, contaminants and all, and then sort out the information in silico. The human sequence will be present, along with other potentially-informative sequences from the environment. Then it will be natural for analyses of the Y chromosome to concentrate on the SNPs, and benefit from their increased resolution: the full sequence provides the maximum information we can extract. This time may arrive sooner than we expect.
Acknowledgements
Our work is supported by The Wellcome Trust.
Footnotes
Commentary for Forensic Science International: Genetics
References
- 1.Jobling MA, Gill P. Encoded evidence: DNA in forensic analysis. Nat. Rev. Genet. 2004;5:739–751. doi: 10.1038/nrg1455. [DOI] [PubMed] [Google Scholar]
- 2.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Xue Y, Wang Q, Long Q, Ng BL, Swerdlow H, Burton J, Skuce C, Taylor R, Abdellah Z, Zhao Y, Asan DG, MacArthur MA, Quail NP, Carter H, Yang, Tyler-Smith C. Human Y chromosome base substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr. Biol. 2009 doi: 10.1016/j.cub.2009.07.032. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willuweit S, Roewer L. Y chromosome haplotype reference database (YHRD): update. Forensic Science International. 2007;1:83–87. doi: 10.1016/j.fsigen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 5.Foster EA, Jobling MA, Taylor PG, Donnelly P, de Knijff P, Mieremet R, Zerjal T, Tyler-Smith C. Jefferson fathered slave’s last child. Nature. 1998;396:27–28. doi: 10.1038/23835. [DOI] [PubMed] [Google Scholar]
- 6.Kayser M, Kittler R, Erler A, Hedman M, Lee AC, Mohyuddin A, Mehdi SQ, Rosser Z, Stoneking M, Jobling MA, Sajantila A, Tyler-Smith C. A comprehensive survey of human Y-chromosomal microsatellites. Am. J. Hum. Genet. 2004;74:1183–1197. doi: 10.1086/421531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vermeulen M, Wollstein A, van der Gaag K, Lao O, Xue Y, Wang Q, Roewer L, Knoblauch H, Tyler-Smith C, de Knijff P, Kayser M. Improving global and regional resolution of male lineage differentiation by simple single-copy Y-chromosomal short tandem repeat polymorphisms. Forensic Science International. 2009 doi: 10.1016/j.fsigen.2009.01.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O’Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, Guo Y, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z, Zheng H, Hellmann I, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, Qin J, Ma L, Li G, Yang Z, Zhang G, Yang B, Yu C, Liang F, Li W, Li S, Li D, Ni P, Ruan J, Li Q, Zhu H, Liu D, Lu Z, Li N, Guo G, Zhang J, Ye J, Fang L, Hao Q, Chen Q, Liang Y, Su Y, San A, Ping C, Yang S, Chen F, Li L, Zhou K, Zheng H, Ren Y, Yang L, Gao Y, Yang G, Li Z, Feng X, Kristiansen K, Wong GK, Nielsen R, Durbin R, Bolund L, Zhang X, Li S, Yang H, Wang J. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]