Abstract
We have examined the male-specific phylogeography of the Levant and its surroundings by analyzing Y-chromosomal haplogroup distributions using 5,874 samples (885 new) from 23 countries. The diversity within some of these haplogroups was also examined. The Levantine populations showed clustering in SNP and STR analyses when considered against a broad Middle-East and North African background. However, we also found a coastal-inland, east-west pattern of diversity and frequency distribution in several haplogroups within the small region of the Levant. Since estimates of effective population size are similar in the two regions, this strong pattern is likely to have arisen mainly from differential migrations, with different lineages introduced from the east and west.
Keywords: Y chromosome, Y-SNP, Y-STR, Levant
INTRODUCTION
The Levant lies in the eastern Mediterranean region, south of the mountains of Cilicia (South Turkey) and north of the Sinai Peninsula. Throughout human prehistory and history, this territory has been a key area, due to its geographical location linking three continents: Europe, Asia and Africa. It was on one of the early out-of-Africa migration routes (Bar-Yosef, 1992, Tchernov, 1994, Luis et al., 2004, Stringer et al., 1989, Lahr & Foley, 1998), is believed to the be the first recipient of migration waves from East Africa seeking milder climatic conditions after the Last Glacial Maximum (LGM) and was the corridor for Neolithic migrations from the Fertile Crescent to Europe and North Africa (Cavalli-Sforza, 1997).
Within recent millennia, the Levant has been a frequent focus of conquest by a variety of states and imperial powers (Issawi, 1988). It was home to some of the oldest cities (Jericho, Byblos and Damascus), a path of some of the oldest and later most important trade routes, and a cradle to three of the World’s Great Religions (Hitti, 1957). Signatures of the genetic legacy of the Phoenician expansion and the Diaspora (Zalloua et al., 2008a, Aubet, 1993), and traces of the Muslim expansion and the Crusaders (Zalloua et al., 2008b, Lamb, 1930) have been identified. However, there remain a number of known historical events, ranging from the Byzantine expansion, the Ottoman expansion, periods of Egyptian, Babylonian, Persian, Hittite (Hitti, 1957) and other subjugations, as well as the likelihood of many unknown prehistoric and historic events, whose genetic impact is yet to be revealed. During the Bronze Age, the land south-west of the Fertile Crescent was called the land of Canaan, part of which became Phoenician territory after 1200 BCE (Issawi, 1988). This land included most of the coastal territory of the eastern Mediterranean countries. These countries shared a common culture and history (Hourani, 1946) but some population expansions and migrations have affected Lebanon and Syria in different ways (Harris, 2003). Such movements include the Aramean rule in Syria (Harris, 2003), the Sea Peoples disruption to some coastal cities north and south of modern day Lebanon (in the Phoenician era) (Harden, 1971) and the Ottoman occupation (Akarli, 1993).
Modern Jewish populations have a special phylogeographical status within the Levant for several reasons. Even by the Roman era, there was a significant Diaspora, as attested to by Strabo, Philo, Josephus, and Cicero (Stern, 2007). The Diaspora fragmented into branches with distinct and well-studied genetic signatures, such as those of Sephardim (Adams et al., 2008) and Ashkenazi (Behar et al., 2003, Nebel et al., 2005) and incorporated significant levels of male admixture (Adams et al., 2008) (Adams et al., 2008). Current Jewish populations in the Levant derive largely from a complex pattern of resettlement from multiple sources within the last ~50 years and may not represent the pre-Diaspora distribution (Baron, 2007). Jewish genetics have been studied more than those of many other groups (Carmeli, 2004). The current study therefore seeks to focus on a geographical genetic profile through the Levant and surrounding regions among the relatively less well characterized populations, highlighting the product of other migratory and population processes that influenced the Levantine region.
Previous studies of Levantine Y-chromosomal diversity have either focused on a specific population (Zalloua et al., 2008b) or been limited in their sampling or genotyping (Cadenas et al., 2008). A general survey of the Levantine genetic landscape could provide significant guidance to construct and test hypotheses concerning known and unknown expansion events in the region. We have therefore assembled a comprehensive set of 5,874 samples representing 23 countries and 35 populations, to present a survey of the Y-lineage distributions throughout the Levant defined by 58 binary markers and, for some, 19 Y-STRs, and place these in the context of the surrounding Y-chromosomal landscape.
In this study we analyzed the geographical distribution of Y-chromosomal haplogroups and STR haplotypes to identify recent and old migration and expansion patterns involving the Levant. This work should shed light on the geographic gradient or structuring resulting from migrations, geographical expansions, and size fluctuations affecting the populations over time. The haplogroup variation and the geographic clustering of some of the Y-STR haplotypes observed in the Levant is suggestive of several succeeding waves of migrations and or expansions into the region that may have taken place post LGM.
MATERIALS AND METHODS
Subjects and Comparative Datasets
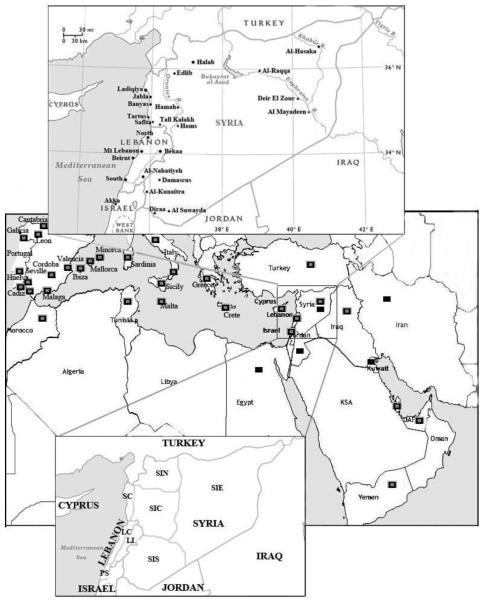
A total of 885 new samples from five populations (Syria, Jordan, Iran, Egypt and Kuwait) were collected and analyzed for this study. In addition, samples and genotyping data from 951 Lebanese, 200 Syrian and 101 Palestinian men (Akka) were already available (Zalloua et al., 2008b, Zalloua et al., 2008a). All the participants had three generations of paternal ancestry in their country of birth. Each provided detailed information on their geographical origin (Table S1) and gave informed consent for this study, which was approved by the IRB committee of the Lebanese American University. The 1,879 Levantine samples (Lebanese, Syrians, Jordanians and Palestinians) were classified into eight regions. These regions mainly reflect geographical subdivisions within modern day Lebanon and Syria (Table S3a and Figure 1). The Lebanese samples, collected from 20 different cities (Table S3b), were classified into the Lebanese coast (LC) which contains coastal Lebanese cities including Beirut, Tyre, Sidon, Jounieh and Tripoli, and Lebanese inland (LI) which contains samples from Bekaa and Nabatiyeh. The Palestinian samples (PS), were collected from men currently residing in Lebanon but who originated from Akka. Finally, Syrian samples were from 17 different Syrian cities (Table S3b) and were divided into coast (SC) and four inland regions: inland north (SIN), center (SIC), south (SIS) and east (SIE) (Table S3a and Figure 1).
Figure 1. Locations of samples analyzed.
The map in the center shows the Middle Eastern and North African countries sampled in the study. The black boxes designate the populations with newly generated data and the gray boxes populations with data obtained from the literature. The Levantine region under study is highlighted in the open box. The upper inset shows the Levantine cities that were sampled and analyzed in this study. The lower inset shows the Levantine area (1,613 samples), as classified here, with Lebanon and Syria subdivided geographically, as detailed in the methods and in Table S3a. LC = Lebanese coast, LI = Lebanese inland, PS = Palestinian samples, SC = Syrian coast, SIN = Syrian inland North, SIC = Syrian inland center, SIS = Syrian inland South, and SIE = Syrian inland East.
For comparative data on haplogroup frequencies, we used published sources (Table S2). In addition to the previously typed Levantine samples (1,252) and the samples typed here (885), 3,737 additional samples were assembled from 28 additional sites (16 countries) in the Middle East and North African region as well as some Mediterranean countries (Table1). These included countries neighboring the Levant, and Arabic countries in the region: Cyprus (Zalloua et al., 2008a, Capelli et al., 2006), Turkey (Cinnioglu et al., 2004), Iraq (Al-Zahery et al., 2003), and Qatar, UAE and Yemen (Cadenas et al., 2008). In addition, Mediterranean North African and European countries, including Morocco (Zalloua et al., 2008a), Tunisia (Zalloua et al., 2008a, Capelli et al., 2006), Spain (Flores et al., 2004, Zalloua et al., 2008a), Italy (Capelli et al., 2007), Portugal (Goncalves et al., 2005), Greece and Crete (Di Giacomo et al., 2003) as well as Malta, Sicily and Sardinia (Capelli et al., 2006) were represented.
Table 1. Percentage of haplogroups and sa mple sizes in the following countries.
Population frequencies of the main haplogroups (J1, J2, R1b, E1b1b1 and I) obtained from the current study or derived from the literature (Table S1).
| Country/Region | J1 | J2 | R1(xR1a1) | E1b1b1 | I | Other | Sample size | Reference |
|---|---|---|---|---|---|---|---|---|
| Lebanon | 18.9 | 29.4 | 7.9 | 16.2 | 2.9 | 27.6 | 951 | Zalloua et.al, 2008b(*) |
| Syria | 33.6 | 20.8 | 4.5 | 12 | 2.3 | 29.1 | 554 | This study and Zalloua et.al, 2008a(*) |
| Akka | 39.2 | 18.6 | 0 | 26.4 | 0 | 15.8 | 101 | Zalloua et.al, 2008a(*) |
| Kuwait | 33.3 | 9.5 | 9.5 | 14.3 | 0 | 33.4 | 42 | This study(*) |
| Jordan | 35.5 | 14.6 | 9 | 23 | 1.1 | 17.9 | 273 | This study(*) |
| Cyprus | 9.6 | 12.9 | 10.2 | 27.4 | 0.5 | 39.9 | 164 | Zalloua et.al, 2008a(*) |
| Morocco | 1 | 0.2 | 3.4 | 52.5 | 0 | 42.9 | 316 | Zalloua et.al, 2008a(*) |
| Tunisia | 0 | 8 | 0 | 82.3 | 0 | 9.7 | 62 | Zalloua et.al, 2008a(*) |
| Turkey | 9.1 | 24.2 | 15.8 | 10.7 | 5.3 | 40.2 | 523 | Cinnioglu et al. 2004(*) |
| Egypt | 19.8 | 7.6 | 6.8 | 42.7 | 0 | 23.1 | 124 | This study(*) |
| Iraq | 33.1 | 25.1 | 10.8 | 10.8 | 0.7 | 20.2 | 117 | Al-Zahery et.al, 2003(*) |
| Qatar | 58.3 | 8.3 | 1.4 | 5.5 | 0 | 26.5 | 72 | Cadenas et.al, 2008(*) |
| UAE | 34.7 | 10.3 | 4.3 | 11.5 | 0 | 39.2 | 164 | Cadenas et.al, 2008(*) |
| Yemen | 72.5 | 9.6 | 0 | 12.9 | 0 | 5 | 62 | Cadenas et.al, 2008(*) |
| Iran | 3.2 | 25 | 6.5 | 8.7 | 1.1 | 56.6 | 92 | This study(*) |
| Malta | 7.8 | 21.1 | 32.22 | 8.9 | 12.2 | 62.2 | 90 | Capelli et al. 2005 |
| Sicily | 5.2 | 22.6 | 24.05 | 23.1 | 8.5 | 49.1 | 212 | Capelli et al. 2005 |
| Sardinia | 4.9 | 9.9 | 21 | 9.9 | 30.8 | 75.3 | 81 | Capelli et al. 2005 |
| Crete | 3.5 | 35 | - | - | 14 | 61.5 | 143 | Di Giacomo et al. 2003 |
| Cadiz | 3.6 | 14.3 | 53.5 | 3.6 | 14.3 | 67.8 | 28 | Flores et al. 2004(**) |
| Malaga | 0 | 15.4 | 42.5 | 23 | 0 | 50.1 | 26 | Flores et al.2004(**) |
| Mallorca | 1.61 | 8.07 | 66.13 | 6.45 | 8.07 | 17.74 | 62 | Zalloua et.al, 2008a |
| Ibiza | 0 | 3.7 | 57.41 | 7.41 | 1.85 | 31.48 | 54 | Zalloua et.al, 2008a |
| Sevilla | 3.2 | 7.8 | 60 | 6.4 | 12.3 | 80 | 155 | Flores et al.2004(**) |
| Valencia | 2.74 | 5.48 | 64.38 | 10.96 | 9.59 | 16.44 | 31 | Zalloua et.al, 2008a |
| Huelva | 0 | 13.7 | 59.1 | 9 | 9.2 | 72.8 | 22 | Flores et al.2004(**) |
| Cordoba | 0 | 14.7 | 55.8 | 11.1 | 14.7 | 74.2 | 27 | Flores et al.2004(**) |
| Italy | 2 | 20 | 40.1 | 12.7 | 7.4 | 17.8 | 699 | Capelli et al. 2007 |
| Greece | 1.9 | 18.1 | - | - | 18.2 | 80 | 154 | Di Giacomo et al. 2003 |
| Portugal | 4.3 | 6.9 | 55.4 | 14.5 | 6.2 | 18.9 | 303 | Gonçalves et al. 2005 |
| Leon | 1.7 | 5 | 61.5 | 10 | 3.4 | 80 | 60 | Flores et al.2004(**) |
| Galicia | 5.3 | 0 | 63.1 | 26.3 | 0 | 68.4 | 19 | Flores et al.2004(**) |
| Cantabria | 2.9 | 2.9 | 58.4 | 8.6 | 5.7 | 75.6 | 70 | Flores et al.2004(**) |
| Castille | 0 | 9.5 | 52.4 | 4.8 | 33.3 | 71.4 | 21 | Flores et al.2004(**) |
R1-M173(xR1a-M17) is R1b
R1(xR1a1) = R1*(xR1a1xR1b3dxR1b3f) + R1b3d + R1b3f
Genotyping
DNA samples were extracted from blood or buccal swabs by standard methods (Wells et al., 2001, Behar et al., 2007). Samples were genotyped with a set of 58 Y-chromosomal binary markers on the non-recombining portion of the Y chromosome (Table S1 and Figure S1). The markers were genotyped by TaqMan RealTime PCR assays. Previously typed samples with a derived allele for each biallelic polymorphism were used as positive controls. These markers define 53 haplogroups (including paragroups), 21 of which were present in the typed samples. The phylogenetic relationships of the relevant Y-chromosomal haplogroups are illustrated in Figure S1 and follow the 2008 YCC convention (Table S1) (Karafet et al., 2008). Published data used in this study were converted to the 2008 YCC nomenclature (Table S2).
All samples were additionally amplified at 19 Y-chromosomal STR loci in two multiplexes. Multiplex I contained the standard 17 loci of the Applied Biosystems Y-filer™ PCR Amplification kit (ww.appliedbiosystems.com). The remaining two loci, DYS388 and DYS426, were genotyped in a separate multiplex (multiplex II), for which we developed an allelic ladder by amplifying and mixing previously typed samples with different number of repeats at the desired locus. The Y-STR data generated in this study are shown in Table S1. STR alleles were named according to current recommendations (Gusmao et al., 2006).
Contour maps
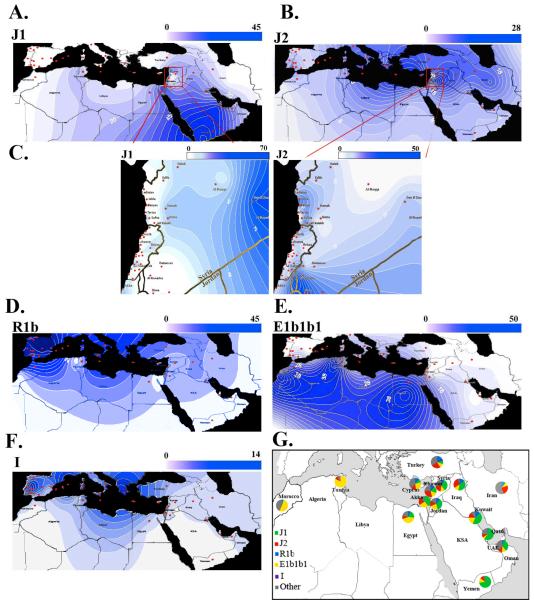
The geographical coordinates of the sample sites are shown in Table S3b (http://itouchmap.com/latlong.html). These include coordinates for the different cities in Lebanon and Syria that were used to make the high resolution maps shown in Figure 2C, as well as the coordinates for the different countries (including Lebanon and Syria as a whole) used to make the maps in Figure 2 A-B and D-F. Haplogroup-frequency surfaces were inferred using the Surfer System version 8.09 (Golden Software, Inc.) as previously described (Semino et al., 2004). The haplogroup data used to construct Figure 2 A-F are shown in Table 1 and the J1 and J2 frequency distribution across the cities of the Levant is shown in Table S3c.
Figure 2. Y-chromosomal variation in the Levant and the Mediterranean region. A-F) Haplogroup-frequency contour maps of the Mediterranean region.
The haplogroup percentages used for the contour map are shown in Table 1. Each panel shows a different haplogroup: J1 (A), J2 (B), R1b (or R1(xR1a1)-as indicated in Table 1) (D), E1b1b1 (E) and I (F). C) High resolution J1 and J2 frequency contour maps of the Levant. The locations of the cities used the haplogroup frequencies are shown in Table S3c. The scales reflect the percentages of haplogroups, where dark blue is the highest value and white is zero. The filled red circles indicate the sample sites. G) Haplogroup frequency analyses. The data used to make the pie charts are shown in Table 1.
Statistical analyses
Population pairwise comparisons and group comparisons were performed using the chi-square test of independence for qualitative variables. Spatial analysis of molecular variance (SAMOVA) (Dupanloup et al., 2002) was performed with the package SAMOVA 1.0 (http://web.unife.it/progetti/genetica/Isabelle/samova.html). SAMOVA is based on AMOVA (Excoffier et al., 1992) which provides a measure of variation between groups of populations, accounting for variations due to drift within populations by means of a nested two-way analysis of variance. Autocorrelation indices for DNA analysis (AIDA) was calculated as described: http://web.unife.it/progetti/genetica/Giorgio/giorgio_soft.html. AIDA is a spatial autocorrelation analysis that tests the dependence of the values of a variable on the values of the same variable at another geographical location, in order to reveal geographical patterns of gene variations (Bertorelle & Barbujani, 1995). RST values based on Y-STR data were calculated using Arlequin 3.11(Excoffier et al., 2005) and displayed as a multidimensional scaling (MDS) plot with SPSS 14.0. The plot shown in Figure S2 was a good fit to the data with a stress value of 0.08 and RSQ of 0.98.
Networks
The phylogenetic relationships between the microsatellite haplotypes were elucidated through reduced-median networks (Bandelt et al., 1995) using the program Network 4.5.0.1 (2008 version) (Fluxus-Engineering). Based on their consistent representation in the dataset, the following 10 loci were retained for analysis: DYS19, DYS388, DYS389I, DYS389b, DYS390, DYS391, DYS392, DYS393, DYS437 and DYS439. Weights applied to each locus were selected to be inversely proportional to the variance of that STR locus (specifically, the weight was 10 times the average variance divided by the locus variance). A SNP weighted at 99 marking the distinction between J1 and J2 was introduced to join those networks. The reduction coefficient was set to 1.0.
Principal Component Analysis
Principal Component Analysis (PCA) (Jolliffe, 1986) was performed on haplogroup frequencies of the Levantine samples (Table S1, Table S2) as well as Middle-Eastern and North African samples (Table S1, Table S2 and Table 1). The data were displaced about the means, and not normalized by standard deviation (Novembre & Stephens, 2008) resulting in a diagonalization of the covariance matrix. Principal Component selection followed the method of Cattell (Cattell, 1966).
Nei’s diversity
Nei’s genetic diversity measures the probability that any two chromosomes drawn from the population will not share the same type (Nei, 1973, Nei, 1978). Here, we use “types” defined in two ways. First, we computed diversity by haplogroup (displayed in Table 2A). Second, we considered STRs. Since a number of STR haplotypes are shared by multiple haplogroups (see Table S6), we used a combination of STRs and SNPs to define haplotypes. Since diversities are so close to 1 with such large numbers of types (Xue et al., 2006), we report diversities in terms of deviations from 1, measuring of the probability two chromosomes share the same type. Further, we also report the expected standard error for the diversity estimator to ensure the deviations between such narrowly deviating measures of diversity were meaningful. The reciprocal of the probability that two chromosomes share the same type can be interpreted as a characteristic number of dominating types in the region.
Table 2. Percentage of haplogroups in the North/South and coast/inland axes in the Levant.
A. Haplogroups percentages and Nei diversities of the North/South and Coast/Inland axes in the Levant, used for Figure 3B. The p-values were obtained using the chi-square test. B. Coastal and inland Levantine Nei diversities by groups determined by SNPs and STRs
| Region | J1 | J2 | R1b | E1b1b1 | I | R1a | G | K2 | L | Other | Nei Diversitye |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SC+LC+PS | 19.8 | 26.8 | 6.9 | 17.4 | 3.9 | 2.3 | 6.1 | 4.2 | 4.8 | 7.3 | 0.792 |
| SIN+SIC+LI+SIS | 28.8 | 27.2 | 5.0 | 14.4 | 1.5 | 5.4 | 6.0 | 1.5 | 5.0 | 4.6 | 0.809 |
| SIE | 48.2 | 3.4 | 0.8 | 4.3 | 0.8 | 4.3 | 2.5 | 0 | 31.0 | 4.3 | 0.669 |
| SC+SIC+SIN+SIE | 35.3 | 21.0 | 3.8 | 10.2 | 1.2 | 4.8 | 4.8 | 1.0 | 13.5 | 3.8 | 0.796 |
| LC+LI+PS+SIS | 21.4 | 26.9 | 6.5 | 17.2 | 3.4 | 3.0 | 6.1 | 3.7 | 4.6 | 7.0 | 0.835 |
| Region | Across regions | In each region |
||||
|---|---|---|---|---|---|---|
| All haplogroups | J1 | J2 | p-value (J1 vs. J2) |
|||
| Chi-Square | df | p-value | p-value | |||
| SC+LC+PS SIN+SIC+LI+SIS SIE |
224.0 | 29 | <0.001 | <0.001 | <0.001 | <0.001 |
| <0.001 | ||||||
| 0.002 | ||||||
| SC+SIC+SIN+SIE LC+LI+PS+SIS |
94.1 | 19 | <0.001 | 0.001 | 0.016 | <0.001 |
| <0.001 | ||||||
The Nei correction adjusting for sampling (Nei, 1978) follows from an assumption of sampling from a multinomial distribution (Nei & Roychoudhury, 1974), which leads to the variance of the Nei estimator to be . The derivation of this estimator is provided in the supplementary materials, and is consistent with Nei’s estimate of the variance (Nei & Roychoudhury, 1974). The results of the estimator for haplogroup data are displayed in Table 2A, while the estimator applied to coastal vs. inland groups identified by SNP+STR loci are presented in Table 2B. Further detailed description of the method used is included in supplemental methods.
Effective Population sizes
Effective population sizes for coastal and inland populations were estimated using BATWING (Wilson et al., 2003), applied to the 564 coastal Levantine samples and 255 inland Levantine samples for which all of the 17 SNPs (M96, M35, M78, M123, M89, M201, M170, 12f2.1, M172, M12, M9, M70, M20, M45, M173, M269, M17) and 11 STR loci (DYS19, DYS388, DYS389I, DYS389b, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, and DYS439) were typed. We use setting and priors described earlier (Xue et al., 2006) with a constant-sized then expanding population and ran the program for 250,000 Monte-Carlo cycles for the inland set, and 300,000 for the coastal set. Satisfactory convergence was demonstrated by plotting the trajectory of Na (Figure S3). Cycles 100,000-300,000 were used to determine posterior estimates of parameters for the coastal sample, and cycles 100,000-250,000 for the inland sample.
RESULTS
The region under study herein is shown in Figure 1. The map shows the Levantine countries and regions (modern borders), including Lebanon, Syria, Akka and Jordan.
The Y-chromosomal hg distribution in the Levantine population was compared to the surrounding Middle Eastern and North African countries (Figure 1) using 884 newly collected samples in addition to our existing Middle Eastern population database. (Table S1 and Table S2). As previously reported (Zalloua et al., 2008b), the most frequent haplogroups present in the Levant are J1, J2, R1b, E1b1b1 and I (Figure S1), and these are shown Figures 2, 4 and 5, although the statistical analyses, such as Nei diversity and AIDA, were performed on all the haplogroups found in each sample, as shown in Table 2A. We first consider these distributions individually.
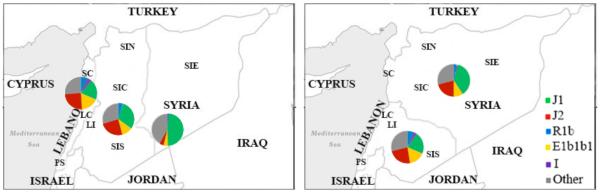
Figure 4. Coastal/inland and north/south classifications of haplogroup frequencies in the Levant.
The Levantine regions (Table S3a) were grouped in two ways and their haplogroup frequencies presented in pie charts. The right hand panel shows the North/South grouping. The left hand panel shows cost/inland grouping, where the regions were split into coastal cities, center regions and deep inland regions. The data used to make the pie charts are shown in Table 2A. The legend explains the color coding for the different haplogroups used in the pie charts.
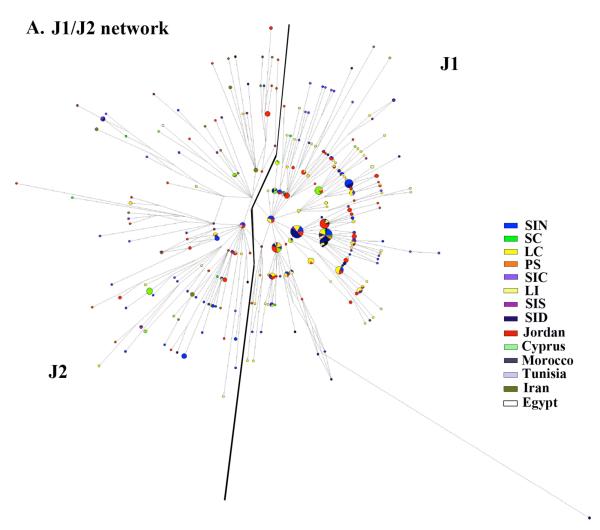
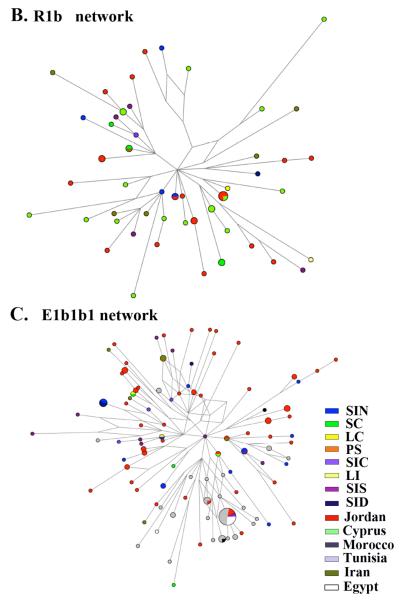
Figure 5. Network analysis.
Reduced-median networks were constructed for the following haplogroups, J1 and J2 (A), R1b (B), and E1b1b1 (C). Networks were constructed from 10 STRs (Table S1 and S2) as described in materials and methods. For each network, the smallest circles represent a count of one individual. Branch lengths are proportional to the number of mutational steps separating two haplotypes.
The haplogroup frequency for J1 peaked in the Arabian Peninsula (Yemen, UAE, and Kuwait) and decreased beyond the Middle-East and North Africa (Figure 2A). J1 frequencies in Syria, Akka and Jordan were more comparable to Lebanon than to the remaining Arabic countries (58.3% in Qatar and 72.5% in Yemen; Figure 2G). Hg J2, in contrast, was present at its highest frequency in the Lebanese population (29.4%) and was significantly more frequent there, than in the remaining Levantine regions (p < 0.05) (Table 1). As previously reported (Zalloua et al., 2008a), it decreases towards the west in North African countries and towards the east in the Arabian Peninsula (29.4% in Lebanon compared to 7.6% in Egypt and 8.3% in Kuwait; Figure 2G and Table 1).
The frequencies of the R1b and I haplogroups (Rootsi et al., 2004) peaked in Europe and the gradient faded beyond the Levant (Figure 2D and 2F). R1b showed some variability in the Levant (4.5% - 9%), had minimal presence in Qatar (1.4%) and was absent from the Yemen sample. Iraq and Kuwait showed significantly higher frequencies of R1b (10.8% and 9.5% respectively; Figure 2G) which may be explained by the strong historical Ottoman influence (Al-Zahery et al., 2003) .
Finally, E1b1b1 (previously E3b), showed the highest concentrations in North African and Berber-speaking populations (Egypt, Morocco and Tunisia; Figure 2E) (Cruciani et al., 2002, Bosch et al., 2001). It showed significant variability in frequency among the Levantine regions (16.2% in Lebanon, 12% in Syria, 26.4% in Akka and 23% in Jordan) (pairwise comparisons: p-value Lebanon vs. Syria = 0.015, Lebanon vs. Akka = 0.009 and Lebanon vs. Jordan = 0.007); however, E1b1b1 frequencies in the Levant as a whole were significantly lower than those in North African countries (42.7% in Egypt, 51.3% in Tunisia and 52.5% in Morocco; Figure 2G) (p-values for pairwise comparisons with Lebanon all < 0.001).
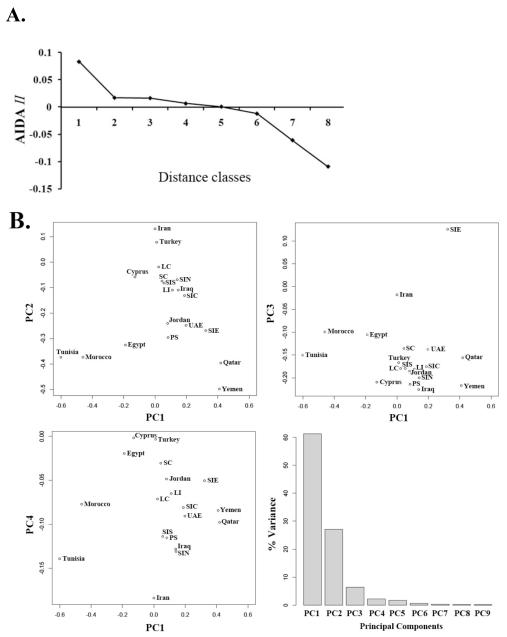
We next investigated the overall Y-chromosomal genetic structure of the region. In an autocorrelation (AIDA) analysis, the autocorrelation index II decreased from positive to negative values with increasing geographical distance (Figure 3A), demonstrating an underlying clinal pattern: nearby populations tend to be similar (positively correlated), while distant populations tend to be dissimilar (negatively correlated). SAMOVA, however, invariably distinguished additional single samples as the number of groups specified was increased, revealing a lack of distinct clusters of geographically contiguous samples, perhaps reflecting the sampling strategy which provided multiple Levantine samples and diverse set of more distant ones (Table S4).
Figure 3. Analysis of binary marker data in Middle-Eastern and North African populations.
A) Correlogram of the AIDA II indices and B) Principal component analysis. The data are the haplogroup frequencies in the Middle-Eastern and North-African countries (Table 1) and in the Levantine regions as defined in Table S3a (Table S1 and S2). The principal component variances are shown in the bar graph in the last panel
In order to examine the haplogroup distribution further, we performed a PCA analysis on the frequencies of the nine haplogroups listed in Table 2, with any additional rare haplogroups combined into a single ‘others’ category. We included the Levantine regions plus samples from Egypt, Morocco, Tunisia, Cyprus, Turkey, Iran, Iraq, Jordan, Qatar, UAE, and Yemen. The percentages of variance associated with each principal component are shown in Figure 3B (lower right panel). PC1 captures 61.3% of the variation, followed by a substantially smaller 27.1% for PC2, with PC3 and PC4 explaining 6.4% and 2.2% respectively. Following this, the remaining PCs carry 3.0% of the variation (Table S5).
PC1 increases with decreasing E1b1b and increasing J1 (Table S5). It showed the largest variation across North Africa, reflecting the high frequency of E1b1b across this region, together with the more localized distribution of J1 in the East. The PC1 scores place the Levantine sites close to each other and close to Cyprus and Egypt in the span from Morocco to the West to Qatar and Yemen to the East. Within this group, SC, LC, and SIS show a reduced J1 score relative to inland Levantine regions. PC2 increases with increasing J2, and decreasing J1 and E1b1b (Table S5). Its distribution identified a gradient in the south to north direction through the Levant, placing Africa in the southern portion of the Levant along with UAE, SIE, PS and Qatar. However, the localized J1 and J2 gradients show increasing values for more coastal sites LC, SC, SIN, LI, and SIC compared to PS and SIE. PC3 increases with decreasing J2 and increasing L (Table S5). Almost all of the regions appeared similar to each other, including all the North African samples, except for Iran and SIE. PC4 increases with increasing R1b and G, decreasing R1a and J2 (Table S5). This principal component shows the largest spread among Levantine sites. In this case, SC, LC, LI, and SIE show larger values, while SIC, SES, PS, and SIN show smaller ones.
The principal components capturing the largest variations primarily establish the Levant within the context of the larger-scale Neolithic signal across North Africa, as well as variations between Iran and Iraq, and the Arabian Peninsula. PC4 shows the strongest signal differentiating among Levantine sites.
An MDS analysis of Y-STR-based genetic distances RST (2D MDS showed Stress = 0.08178 and RSQ = 0.98172), showed similar general features as the two leading principal components. Levantine populations were mostly clustered, while North African populations were progressively more distinct as the distance west increased. Two exceptions were SIE, which however is seen to be divergent in PC3, and the similarity between Jordan and Cyprus, not observed among any of the PCs.
A higher resolution contour map of haplogroup frequency distribution among Levantine cities (Table S3b and S3c) revealed coast/inland opposing gradients for J1 and J2 (Figure 2C). We then regrouped the Levant into northern and southern regions (as shown in Figure 4) and into three regions going from west to east (coast, inland and further inland regions). J1 frequencies but not J2 were significantly different along the South to North axis of the Levant. More strikingly, going from coastal to inland regions in the Levant, there was a significant increase of J1 frequencies (19.8 to 48.2%, p<0.001) compared to a significant decrease of J2 frequencies (26.8% to 3.4%, p<0.001) (Table 2A). This steep difference between the coast and inland regions was particularly remarkable considering the small geographical area under consideration.
Application of the Nei diversity estimator to haplogroup frequency data from the individual Levantine populations showed a minimum value 0.669 for SIE. This would suggest that that population is dominated by roughly 3 haplogroups. Haplogroups J1 and L are the most frequent, with a number of other rarer haplogroups. The rest of the entries in Table 2A show diversities in the 0.7 to 0.8 range, suggesting a rough average number of dominating haplogroups in the range of 3.3 to 5. Table 2A shows that these populations are dominated by two or three haplogroups, but with slightly lower relative frequencies than in the SIE population, and with higher frequencies among the remaining haplogroups. The coastal and near coastal groups SC+LC+PS and SIN+SIC+LI+SIS show similar Nei diversity values, while the far-inland SIE shows the greatest reduction in Nei diversity.
Diversity values were further investigated by including both haplogroup and STR haplotype, and varying the number of STR loci used. The number of SNP+STR types ranged from 257 for 2 STR loci through to 1286 for 10 STR loci (Table 2B). For such large numbers of STR+SNP types, the probability that any two chromosomes drawn from the population may be expected to share the same type is small. For all samples, the estimated probability that two chromosomes would share the same STR+SNP type was roughly twice as high for inland samples than for coastal samples, indicating consistently lower inland diversity. Further, the differences between inland and coastal diversities were much larger than the corresponding estimated standard errors (Table 2B). Estimation of the effective population size using BATWING, however, led to similar numbers for the two regions: ~1,200 (95% CI 840 - 1550) near the coast and ~1,230 (95% CI 934 - 1534) inland.
We sought, through STR network analysis, to assess whether or not the observed geographic distribution of each haplogroup was reflected in geographic variations of STR haplotype distributions. The J1 and J2 (Figure 5A) sister clades depicted a clear non-uniform geographic distribution of STR haplotypes and few instances of haplotype sharing across geographic regions. Consistent with previous analyses, coastal Levantine regions were well represented in the J2 network. Some evidence of sharing with Jordan was also apparent. The J1 network was dominated by inland Levantine samples (mainly Jordan and inland Lebanon and Syria). The R1b network showed much less geographic correlation, possibly because most of the R1b chromosomes have entered the region recently (Figure 5B). In fact, without extra-Levantine representation of R1b to establish context, it is difficult to identify where these R1bs originated. Finally, E1b1b showed a clear demarcation between the Levantine STR haplotypes and North African STR haplotypes, with a lower diversity among North African STR haplotypes than among Levantine STR haplotypes (Figure 5C). 91 STR haplotypes belonged to the E1b1b1 haplogroup within the Levantine population compared to 60 STR haplotypes within the N. African population (data not shown).
DISCUSSION
In this study, we have used a combination of novel and published data to explore the Y-chromosomal landscape of the Levant and its surrounding regions. On a large geographical scale, the Levantine samples clustered together and were readily distinguished from North African or Arabian Peninsula samples (Figures 2 and 3). This pattern of correlation between genetics and geography is expected from many previous studies of human variation (Cavalli-Sforza, 1997) and is particularly marked for the Y chromosome (Jobling & Tyler-Smith, 2003). But within the Levant, there was significant heterogeneity, with a predominantly east-west, coastal-inland structure (Figure 4). Such a high level of differentiation within a small geographical region is striking, and we consider here the historical and other factors that might have contributed to it.
The Levantine genetic structure consists largely of decreasing frequencies of the major haplogroups J2 and E1b1b1 inland and a corresponding increasing frequency of J1, as well as similar patterns in several lower-frequency haplogroups such as L, R1b and I. Diversity is higher on the coast. Such a pattern could be interpreted in two ways: as arising from genetic drift within a geographically stable population that differs in effective population size between the coast and interior (Kimura & Crow, 1964), or alternatively as arising from differential migration from distinct sources into populations of similar size. These interpretations are not mutually exclusive, and we next explore the possible contributions of each.
The coastal region lies within the western section of the Fertile Crescent and has been densely inhabited for all of recorded history and much of prehistory, as illustrated by the location of several of the earliest known cities here. In contrast, most of the interior is dry and now experiences desert or semi-desert conditions, and consequently supports a lower population density. These conditions are likely to have prevailed for much of the Holocene. A lower level of haplogroup diversity inland may therefore be expected from the demographic history, although the specific haplogroups that increased or decreased in frequency would be a matter of chance.
Recent migrations impacting the region under consideration included impacts from Hittites, Babylonians, Persians, Phoenicians (Zalloua et al., 2008a) and subsequent trade, Romans, Sassanids, the Muslim expansion (Zalloua et al., 2008b), the movement of the Omayyad capital from Baghdad to Damascus, the Crusades (Zalloua et al., 2008b), The Ottoman Empire, and European Colonialism as well as significant caravan trade through the region throughout this entire time period. Against this background, estimation of an effective population size is problematical. BATWING reports very similar population sizes for inland and coastal populations, arguing against a detectable gradient of effective population size driving differential drift, although the BATWING results must be interpreted with caution because of the complex demography. Nevertheless, the differences in diversity suggest that migration has most likely been the determining factor in distinguishing coastal diversity from nearby inland regions.
Hg J is believed to have split into J1 and J2 about 18 Kya (Semino et al., 2004). These two sister clades showed distinct histories and geographical localizations with a coastal range that is predominantly J2 and an inland range that is predominantly J1. Chiaroni, King and Underhill, report the same inland vs. coastal divergence pattern of J1 and J2, and correlate the expansion during the rise of agriculture in the Fertile Crescent to the patterns of rainfall distribution (Chiaroni, 2008). They suggest that J2 haplogroup marked agricultural populations that followed the coasts, whereas the J1 haplogroup appears to have fixed in herdsmen populations that remained inland (Chiaroni, 2008).
The diversified J2 reduced-median network and high coastal frequency suggest a sustained and non-interrupted presence of this Hg along the Eastern coast of the Mediterranean. While our network analysis excluded the Arabian Peninsula and mainly focused on the Levantine regions, some haplotypes appear to have originated in the Peninsula, and to have been recently carried into the Levant with the Islamic expansion. Tofanelli et al. (Tofanelli et al., 2009) argue that the initial origin of spread of J1 into the Arabian Peninsula from North Africa is evident when correlated with Arabic nations’ haplotypes. Further, they date the expansion to before the Islamic expansion (Tofanelli et al., 2009). While we observe the geographical gradients consistent with the expansions from the earlier refugia, it is also clear that the more recent Muslim expansion has had a significant impact on the elevated J1 frequency distribution in the Levant.
Migration and back-migration between the Levant and North Africa were evident from the haplogeography of hg E1b1b1 and its haplotype network structure. E1b1b1 shows the highest frequency in North Africa (Egypt, Morocco and Tunisia) and drops gradually as it spills out of North Africa through the horn of Africa and the Levantine corridor into the Arabian Peninsula and central Asia, and through the Strait of Gibraltar into Spain and South Europe. A historical event that spread of E1b1b1 to Iberia most likely occurred through the Strait of Gibraltar culminating with the Islamic conquests of Iberia. Their armies bore large numbers of Berber recruits, whose presence started with the Umayyad Caliphates (661-75CE) and continued uninterrupted for several hundred years. This distinction between Arabic and Berber pools of E1b1b1 in North Africa was highlighted by a study conducted on populations from Morocco, Tunisia and Algeria (Gerard et al., 2006).
The E1b1b1 frequency gradient, considered in the light of the haplotype diversity, suggests an early migration (Neolithic) from the Levant into North Africa that is consistent with a limited gene flow into Africa followed by a rapid expansion and later punctuated by some back migrations as a result of migratory events in the Mediterranean (Arredi et al., 2004). According to this model, the E1b1b1 frequency gradient between coastal Levant and inland Levant would reflect an origin and expansion near the coast, and limited migration inland. The STR network for the E1b1b1 haplotype lineages (Figure 5C) dissects these migration events. The network shows geographical separation of the lineages marked by STRs between North African countries (Morocco and Tunisia) and the Levantine countries. Another independent measure is the number of mutations along a lineage. In Arredi et al., the authors estimated the time to the most recent common ancestor for E1b1b1 lineages in North Africa (E1b1b1b-M81) and this was found to coincide with a possible Neolithic origin (Arredi et al., 2004).
In conclusion, the current Levantine Y-chromosomal landscape is dominated by the coastal-inland contrast in haplogroup frequencies and diversity that reflects largely the influence of successive migrations, which have tended to reinforce one another by introducing different Y lineages from the east and west.
Supplementary Material
Acknowledgements
We thank the sample donors for taking part in this study, Ms. Janet Ziegel and Mr. Pandikumar Swamikrishnan for their help with genotyping and data organization. YX and CTS are supported by The Wellcome Trust. The Genographic Project is supported by funding from the National Geographic Society, IBM and the Waitt Family Foundation.
References
- Adams SM, Bosch E, Balaresque PL, Ballereau SJ, Lee AC, Arroyo E, Lopez-Parra AM, Aler M, Grifo MS, Brion M, Carracedo A, Lavinha J, Martinez-Jarreta B, Quintana-Murci L, Picornell A, Ramon M, Skorecki K, Behar DM, Calafell F, Jobling MA. The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. Am J Hum Genet. 2008;83:725–36. doi: 10.1016/j.ajhg.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akarli ED. The Long Peace: Ottoman Lebanon, 1861-1920. I.B. Tauris; London: 1993. [Google Scholar]
- Al-Zahery N, Semino O, Benuzzi G, Magri C, Passarino G, Torroni A, Santachiara-Benerecetti AS. Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol. 2003;28:458–72. doi: 10.1016/s1055-7903(03)00039-3. [DOI] [PubMed] [Google Scholar]
- Arredi B, Poloni ES, Paracchini S, Zerjal T, Fathallah DM, Makrelouf M, Pascali VL, Novelletto A, Tyler-Smith C. A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet. 2004;75:338–45. doi: 10.1086/423147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubet M-E. The Phoenicians and the West: Politics, Colonies and Trade. Cambridge University Press; Cambridge: 1993. [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–53. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yosef O. The role of western Asia in modern human origins. Philos Trans R Soc Lond B Biol Sci. 1992;337:193–200. doi: 10.1098/rstb.1992.0097. [DOI] [PubMed] [Google Scholar]
- Baron SW. Population. In: Berenbaum M, Skolnik F, editors. Encyclopaedia Judaica. 2nd ed. Macmillan; Detroit, MI: 2007. Encyclopaedia Judaica. [Google Scholar]
- Behar DM, Rosset S, Blue-Smith J, Balanovsky O, Tzur S, Comas D, Mitchell RJ, Quintana-Murci L, Tyler-Smith C, Wells RS. The Genographic Project public participation mitochondrial DNA database. PLoS Genet. 2007;3:e104. doi: 10.1371/journal.pgen.0030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Thomas MG, Skorecki K, Hammer MF, Bulygina E, Rosengarten D, Jones AL, Held K, Moses V, Goldstein D, Bradman N, Weale ME. Multiple origins of Ashkenazi Levites: Y chromosome evidence for both Near Eastern and European ancestries. Am J Hum Genet. 2003;73:768–79. doi: 10.1086/378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle G, Barbujani G. Analysis of DNA diversity by spatial autocorrelation. Genetics. 1995;140:811–9. doi: 10.1093/genetics/140.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J. High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet. 2001;68:1019–29. doi: 10.1086/319521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas AM, Zhivotovsky LA, Cavalli-Sforza LL, Underhill PA, Herrera RJ. Y-chromosome diversity characterizes the Gulf of Oman. Eur J Hum Genet. 2008;16:374–86. doi: 10.1038/sj.ejhg.5201934. [DOI] [PubMed] [Google Scholar]
- Capelli C, Brisighelli F, Scarnicci F, Arredi B, Caglia A, Vetrugno G, Tofanelli S, Onofri V, Tagliabracci A, Paoli G, Pascali VL. Y chromosome genetic variation in the Italian peninsula is clinal and supports an admixture model for the Mesolithic-Neolithic encounter. Mol Phylogenet Evol. 2007;44:228–39. doi: 10.1016/j.ympev.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Capelli C, Redhead N, Romano V, Cali F, Lefranc G, Delague V, Megarbane A, Felice AE, Pascali VL, Neophytou PI, Poulli Z, Novelletto A, Malaspina P, Terrenato L, Berebbi A, Fellous M, Thomas MG, Goldstein DB. Population structure in the Mediterranean basin: a Y chromosome perspective. Ann Hum Genet. 2006;70:207–25. doi: 10.1111/j.1529-8817.2005.00224.x. [DOI] [PubMed] [Google Scholar]
- Carmeli DB. Prevalence of Jews as subjects in genetic research: figures, explanation, and potential implications. Am J Med Genet A. 2004;130A:76–83. doi: 10.1002/ajmg.a.20291. [DOI] [PubMed] [Google Scholar]
- Cattell R. The scree test for the number of factors. Multiv. Behav. Res. 1966;1:245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL. Genes, peoples, and languages. Proc Natl Acad Sci U S A. 1997;94:7719–24. doi: 10.1073/pnas.94.15.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaroni KAU. Correlation of annual precipitation with human Y-chromosome diversity and the emergence of Neolithic agricultural and pastoral economies in the Fertile Crescent. Antiquity. 2008;82:281–289. [Google Scholar]
- Cinnioglu C, King R, Kivisild T, Kalfoglu E, Atasoy S, Cavalleri GL, Lillie AS, Roseman CC, Lin AA, Prince K, Oefner PJ, Shen P, Semino O, Cavalli-Sforza LL, Underhill PA. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–48. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, Wallace DC, Oefner PJ, Torroni A, Cavalli-Sforza LL, Scozzari R, Underhill PA. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet. 2002;70:1197–214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacomo F, Luca F, Anagnou N, Ciavarella G, Corbo RM, Cresta M, Cucci F, Di Stasi L, Agostiano V, Giparaki M, Loutradis A, Mammi C, Michalodimitrakis EN, Papola F, Pedicini G, Plata E, Terrenato L, Tofanelli S, Malaspina P, Novelletto A. Clinal patterns of human Y chromosomal diversity in continental Italy and Greece are dominated by drift and founder effects. Mol Phylogenet Evol. 2003;28:387–95. doi: 10.1016/s1055-7903(03)00016-2. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol Ecol. 2002;11:2571–81. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–91. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores C, Maca-Meyer N, Gonzalez AM, Oefner PJ, Shen P, Perez JA, Rojas A, Larruga JM, Underhill PA. Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography. Eur J Hum Genet. 2004;12:855–63. doi: 10.1038/sj.ejhg.5201225. [DOI] [PubMed] [Google Scholar]
- Gerard N, Berriche S, Aouizerate A, Dieterlen F, Lucotte G. North African Berber and Arab influences in the western Mediterranean revealed by Y-chromosome DNA haplotypes. Hum Biol. 2006;78:307–16. doi: 10.1353/hub.2006.0045. [DOI] [PubMed] [Google Scholar]
- Goncalves R, Freitas A, Branco M, Rosa A, Fernandes AT, Zhivotovsky LA, Underhill PA, Kivisild T, Brehm A. Y-chromosome lineages from Portugal, Madeira and Acores record elements of Sephardim and Berber ancestry. Ann Hum Genet. 2005;69:443–54. doi: 10.1111/j.1529-8817.2005.00161.x. [DOI] [PubMed] [Google Scholar]
- Gusmao L, Butler JM, Carracedo A, Gill P, Kayser M, Mayr WR, Morling N, Prinz M, Roewer L, Tyler-Smith C, Schneider PM. DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Int J Legal Med. 2006;120:191–200. doi: 10.1007/s00414-005-0026-1. [DOI] [PubMed] [Google Scholar]
- Harden D. The Phoenicians. Penguin Books; London: 1971. [Google Scholar]
- Harris WW. (Princeton series on the Middle East).The Levant, a Fractured Mosaic. 2003 [Google Scholar]
- Hitti PK. Lebanon in History: From the Earliest Times to the Present. St. Martin’s Press; New York: 1957. [Google Scholar]
- Hourani AH. Syria and Lebanon. Oxford University Press; London: 1946. [Google Scholar]
- Issawi C. The Fertile Crescent, 1800-1914: A Documentary Economic History. Oxford University Press; New York: 1988. [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Jolliffe I. Principal Coponents Analysis. Second Edition Springer; New York, NY: 1986. [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–8. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Crow JF. The Number of Alleles That Can Be Maintained in a Finite Population. Genetics. 1964;49:725–38. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr MM, Foley RA. Towards a theory of modern human origins: geography, demography, and diversity in recent human evolution. Am J Phys Anthropol. 1998;(Suppl 27):137–76. doi: 10.1002/(sici)1096-8644(1998)107:27+<137::aid-ajpa6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Lamb H. The Crusades. Doubleday; New York: 1930. [Google Scholar]
- Luis JR, Rowold DJ, Regueiro M, Caeiro B, Cinnioglu C, Roseman C, Underhill PA, Cavalli-Sforza LL, Herrera RJ. The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet. 2004;74:532–44. doi: 10.1086/382286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebel A, Filon D, Faerman M, Soodyall H, Oppenheim A. Y chromosome evidence for a founder effect in Ashkenazi Jews. Eur J Hum Genet. 2005;13:388–91. doi: 10.1038/sj.ejhg.5201319. [DOI] [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A. 1973;70:3321–3. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Estimation of Average Heterozygosity and Genetic Distance from a Small Number of Individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Roychoudhury AK. Sampling Variances of Heterozygosity and Genetic Distance. Genetics. 1974;76:379–390. doi: 10.1093/genetics/76.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–9. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rootsi S, Magri C, Kivisild T, Benuzzi G, Help H, Bermisheva M, Kutuev I, Barac L, Pericic M, Balanovsky O, Pshenichnov A, Dion D, Grobei M, Zhivotovsky LA, Battaglia V, Achilli A, Al-Zahery N, Parik J, King R, Cinnioglu C, Khusnutdinova E, Rudan P, Balanovska E, Scheffrahn W, Simonescu M, Brehm A, Goncalves R, Rosa A, Moisan JP, Chaventre A, Ferak V, Furedi S, Oefner PJ, Shen P, Beckman L, Mikerezi I, Terzic R, Primorac D, Cambon-Thomsen A, Krumina A, Torroni A, Underhill PA, Santachiara-Benerecetti AS, Villems R, Semino O. Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in europe. Am J Hum Genet. 2004;75:128–37. doi: 10.1086/422196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Magri C, Benuzzi G, Lin AA, Al-Zahery N, Battaglia V, Maccioni L, Triantaphyllidis C, Shen P, Oefner PJ, Zhivotovsky LA, King R, Torroni A, Cavalli-Sforza LL, Underhill PA, Santachiara-Benerecetti AS. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet. 2004;74:1023–34. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M. Diaspora. In: Berenbaum M, Skolink F, editors. Encyclopaedia Judaica. 2nd ed. Gale Group, Macmillan Reference Books; Detroit, MI: 2007. Encyclopaedia Judaica. [Google Scholar]
- Stringer CB, Grun R, Schwarcz HP, Goldberg P. ESR dates for the hominid burial site of Es Skhul in Israel. Nature. 1989;338:756–8. doi: 10.1038/338756a0. [DOI] [PubMed] [Google Scholar]
- Tchernov E. New comments on the biostratigraphy of the Middle and Upper Pleistocene of the southern Levant. Late Quaternary Chronology and Paleoclimates of the Eastern Mediterranean. 1994:333–350. [Google Scholar]
- Tofanelli S, Ferri G, Bulayeva K, Caciagli L, Onofri V, Taglioli L, Bulayev O, Boschi I, Alu M, Berti A, Rapone C, Beduschi G, Luiselli D, Cadenas AM, Awadelkarim KD, Mariani-Costantini R, Elwali NE, Verginelli F, Pilli E, Herrera RJ, Gusmao L, Paoli G, Capelli C. J1-M267 Y lineage marks climate-driven pre-historical human displacements. Eur J Hum Genet. 2009 doi: 10.1038/ejhg.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Chariev A, Bodmer WF. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci U S A. 2001;98:10244–9. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I, Balding D, Weale M. Inferences from DNA Data: Population Histories, Evolutionary Processes and Forensic Probabilities. Journal of the Royal Statistical Society: Series A (Statistics in Society) 2003;166:155–188. [Google Scholar]
- Xue Y, Zerjal T, Bao W, Zhu S, Shu Q, Xu J, Du R, Fu S, Li P, Hurles ME, Yang H, Tyler-Smith C. Male demography in East Asia: a north-south contrast in human population expansion times. Genetics. 2006;172:2431–9. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalloua PA, Platt DE, El Sibai M, Khalife J, Makhoul N, Haber M, Xue Y, Izaabel H, Bosch E, Adams SM, Arroyo E, Lopez-Parra AM, Aler M, Picornell A, Ramon M, Jobling MA, Comas D, Bertranpetit J, Wells RS, Tyler-Smith C. Identifying genetic traces of historical expansions: Phoenician footprints in the Mediterranean. Am J Hum Genet. 2008a;83:633–42. doi: 10.1016/j.ajhg.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalloua PA, Xue Y, Khalife J, Makhoul N, Debiane L, Platt DE, Royyuru AK, Herrera RJ, Hernanz DF, Blue-Smith J, Wells RS, Comas D, Bertranpetit J, Tyler-Smith C. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet. 2008b;82:873–82. doi: 10.1016/j.ajhg.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.