Abstract
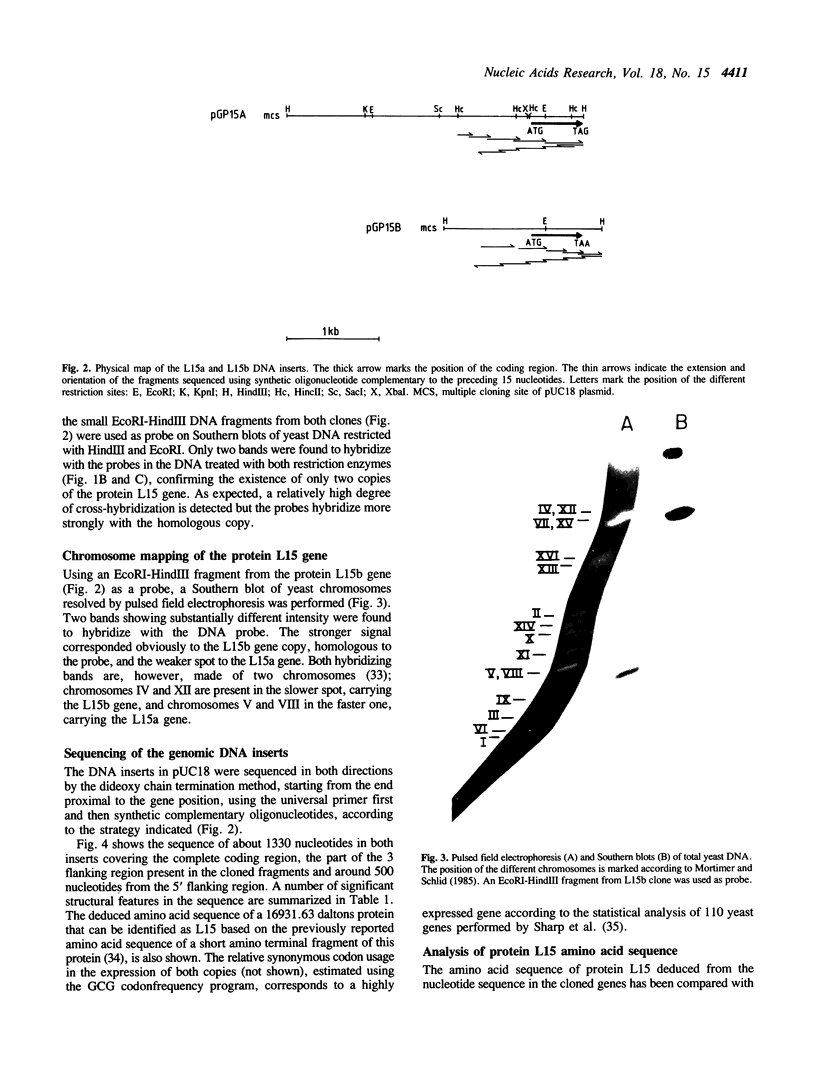
Transformant phages expressing L15, a yeast ribosomal protein which binds to 26S rRNA and interacts with the acidic ribosomal proteins, were isolated by screening a yeast cDNA expression library in lambda gt11 with specific monoclonal antibodies. Using yeast DNA HindIII fragments that hybridize with the cDNA insert from the L15-expressing clones, minilibraries were prepared in pUC18, which were afterward screened with the same cDNA probe. In this way, plasmids carrying two different types of genomic DNA inserts were obtained. The inserts were subcloned and sequenced and we found a similar coding sequence in both cases flanked by 5' and 3' regions with very low homology. Sequences homologous to the consensus TUF-binding UAS boxes are present in the 5' flanking regions of both genes. Southern analysis revealed the presence of two copies of the L15 gene in the Saccharomyces cerevisiae genome, which are located in different chromosomes. The encoded amino acid sequence corresponds, as expected, to protein L15 and shows a high similarity to bacterial ribosomal protein L11.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballesta J. P., Vazquez D. Activities of ribosomal cores deprived of proteins L7, L10, L11 and L12. FEBS Lett. 1974 Nov 15;48(2):266–270. doi: 10.1016/0014-5793(74)80483-7. [DOI] [PubMed] [Google Scholar]
- Beauclerk A. A., Cundliffe E., Dijk J. The binding site for ribosomal protein complex L8 within 23 s ribosomal RNA of Escherichia coli. J Biol Chem. 1984 May 25;259(10):6559–6563. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijk J., Garrett R. A., Müller R. Studies on the binding of the ribosomal protein complex L7/12-L10 and protein L11 to the 5'-one third of 23S RNA: a functional centre of the 50S subunit. Nucleic Acids Res. 1979 Jun 25;6(8):2717–2729. doi: 10.1093/nar/6.8.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman J. C., Doorenbosch M. M., Maurer C. T., de Winde J. H., Mager W. H., Planta R. J., Grivell L. A. An ARS/silencer binding factor also activates two ribosomal protein genes in yeast. Nucleic Acids Res. 1989 Jul 11;17(13):4917–4923. doi: 10.1093/nar/17.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon K. B., Parnassa A. P., Foster C. L. Lupus autoantibodies target ribosomal P proteins. J Exp Med. 1985 Aug 1;162(2):459–471. doi: 10.1084/jem.162.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamil K. G., Nam H. G., Fried H. M. Constitutive transcription of yeast ribosomal protein gene TCM1 is promoted by uncommon cis- and trans-acting elements. Mol Cell Biol. 1988 Oct;8(10):4328–4341. doi: 10.1128/mcb.8.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Juan-Vidales F., Sánchez Madrid F., Saenz-Robles M. T., Ballesta J. P. Purification and characterization of two ribosomal proteins of Saccharomyces cerevisiae. Homologies with proteins from eukaryotic species and with bacterial protein EC L11. Eur J Biochem. 1983 Nov 2;136(2):275–281. doi: 10.1111/j.1432-1033.1983.tb07738.x. [DOI] [PubMed] [Google Scholar]
- Kearney J. F., Radbruch A., Liesegang B., Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979 Oct;123(4):1548–1550. [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Langridge J., Langridge P., Bergquist P. L. Extraction of nucleic acids from agarose gels. Anal Biochem. 1980 Apr;103(2):264–271. doi: 10.1016/0003-2697(80)90266-3. [DOI] [PubMed] [Google Scholar]
- Mager W. H. Control of ribosomal protein gene expression. Biochim Biophys Acta. 1988 Jan 25;949(1):1–15. doi: 10.1016/0167-4781(88)90048-6. [DOI] [PubMed] [Google Scholar]
- Mitra G., Warner J. R. A yeast ribosomal protein gene whose intron is in the 5' leader. J Biol Chem. 1984 Jul 25;259(14):9218–9224. [PubMed] [Google Scholar]
- Mitsui K., Motizuki M., Endo Y., Yokota S., Tsurugi K. Characterization of a novel acidic protein of 38 kDa, A0, in yeast ribosomes which immunologically cross-reacts with the 13 kDa acidic ribosomal proteins, A1/A2. J Biochem. 1987 Dec;102(6):1565–1570. doi: 10.1093/oxfordjournals.jbchem.a122205. [DOI] [PubMed] [Google Scholar]
- Mitsui K., Nakagawa T., Tsurugi K. On the size and the role of a free cytosolic pool of acidic ribosomal proteins in yeast Saccharomyces cerevisiae. J Biochem. 1988 Dec;104(6):908–911. doi: 10.1093/oxfordjournals.jbchem.a122581. [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tsurugi K. Identification of A1 protein as the fourth member of 13 kDa-type acidic ribosomal protein family in yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1989 Jun 30;161(3):1001–1006. doi: 10.1016/0006-291x(89)91342-9. [DOI] [PubMed] [Google Scholar]
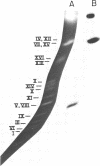
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton C. H., Shimmin L. C., Yee J., Dennis P. P. A family of genes encode the multiple forms of the Saccharomyces cerevisiae ribosomal proteins equivalent to the Escherichia coli L12 protein and a single form of the L10-equivalent ribosomal protein. J Bacteriol. 1990 Feb;172(2):579–588. doi: 10.1128/jb.172.2.579-588.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. J., McMaster G. K., Trachsel H. Cloning of eukaryotic protein synthesis initiation factor genes: isolation and characterization of cDNA clones encoding factor eIF-4A. Nucleic Acids Res. 1985 Oct 11;13(19):6867–6880. doi: 10.1093/nar/13.19.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwint R. T., Mager W. H., Maurer K. C., Planta R. J. Mutational analysis of the upstream activation site of yeast ribosomal protein genes. Curr Genet. 1989 Apr;15(4):247–251. doi: 10.1007/BF00447039. [DOI] [PubMed] [Google Scholar]
- Nieuwint R. T., Molenaar C. M., van Bommel J. H., van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. The gene for yeast ribosomal protein S31 contains an intron in the leader sequence. Curr Genet. 1985;10(1):1–5. doi: 10.1007/BF00418486. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Remacha M., Santos C., Ballesta J. P. Disruption of single-copy genes encoding acidic ribosomal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2182–2190. doi: 10.1128/mcb.10.5.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacha M., Sáenz-Robles M. T., Vilella M. D., Ballesta J. P. Independent genes coding for three acidic proteins of the large ribosomal subunit from Saccharomyces cerevisiae. J Biol Chem. 1988 Jul 5;263(19):9094–9101. [PubMed] [Google Scholar]
- Reyes R., Vázquez D., Ballesta J. P. Peptidyl transferase center of rat-liver ribosome cores. Eur J Biochem. 1977 Feb 15;73(1):25–31. doi: 10.1111/j.1432-1033.1977.tb11288.x. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmin L. C., Ramirez C., Matheson A. T., Dennis P. P. Sequence alignment and evolutionary comparison of the L10 equivalent and L12 equivalent ribosomal proteins from archaebacteria, eubacteria, and eucaryotes. J Mol Evol. 1989 Nov;29(5):448–462. doi: 10.1007/BF02602915. [DOI] [PubMed] [Google Scholar]
- Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- Strycharz W. A., Nomura M., Lake J. A. Ribosomal proteins L7/L12 localized at a single region of the large subunit by immune electron microscopy. J Mol Biol. 1978 Dec 5;126(2):123–140. doi: 10.1016/0022-2836(78)90355-8. [DOI] [PubMed] [Google Scholar]
- Sáenz-Robles M. T., Vilella M. D., Pucciarelli G., Polo F., Remacha M., Ortíz B. L., Vidales F. J., Ballesta J. P. Ribosomal protein interactions in yeast. Protein L15 forms a complex with the acidic proteins. Eur J Biochem. 1988 Nov 15;177(3):531–537. doi: 10.1111/j.1432-1033.1988.tb14405.x. [DOI] [PubMed] [Google Scholar]
- Sánchez-Madrid F., Reyes R., Conde P., Ballesta J. P. Acidic ribosomal proteins from eukaryotic cells. Effect on ribosomal functions. Eur J Biochem. 1979 Aug 1;98(2):409–416. doi: 10.1111/j.1432-1033.1979.tb13200.x. [DOI] [PubMed] [Google Scholar]
- Towbin H., Ramjoué H. P., Kuster H., Liverani D., Gordon J. Monoclonal antibodies against eucaryotic ribosomes. Use to characterize a ribosomal protein not previously identified and antigenically related to the acidic phosphoproteins P1/P2. J Biol Chem. 1982 Nov 10;257(21):12709–12715. [PubMed] [Google Scholar]
- Vidales F. J., Robles M. T., Ballesta J. P. Acidic proteins of the large ribosomal subunit in Saccharomyces cerevisiae. Effect of phosphorylation. Biochemistry. 1984 Jan 17;23(2):390–396. doi: 10.1021/bi00297a032. [DOI] [PubMed] [Google Scholar]
- Vidales F. J., Sanchez-Madrid F., Ballesta J. P. The acidic proteins of eukaryotic ribosomes. A comparative study. Biochim Biophys Acta. 1981 Nov 27;656(1):28–35. doi: 10.1016/0005-2787(81)90022-8. [DOI] [PubMed] [Google Scholar]
- Vignais M. L., Woudt L. P., Wassenaar G. M., Mager W. H., Sentenac A., Planta R. J. Specific binding of TUF factor to upstream activation sites of yeast ribosomal protein genes. EMBO J. 1987 May;6(5):1451–1457. doi: 10.1002/j.1460-2075.1987.tb02386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989 Jun;53(2):256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt L. P., Mager W. H., Nieuwint R. T., Wassenaar G. M., van der Kuyl A. C., Murre J. J., Hoekman M. F., Brockhoff P. G., Planta R. J. Analysis of upstream activation sites of yeast ribosomal protein genes. Nucleic Acids Res. 1987 Aug 11;15(15):6037–6048. doi: 10.1093/nar/15.15.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Baradi T. T., de Regt V. C., Einerhand S. W., Teixido J., Planta R. J., Ballesta J. P., Raué H. A. Ribosomal proteins EL11 from Escherichia coli and L15 from Saccharomyces cerevisiae bind to the same site in both yeast 26 S and mouse 28 S rRNA. J Mol Biol. 1987 Jun 20;195(4):909–917. doi: 10.1016/0022-2836(87)90494-3. [DOI] [PubMed] [Google Scholar]
- van Agthoven A., Kriek J., Amons R., Möller W. Isolation and characterization of the acidic phosphoproteins of 60-S ribosomes from Artemia salina and rat liver. Eur J Biochem. 1978 Nov 15;91(2):553–565. doi: 10.1111/j.1432-1033.1978.tb12709.x. [DOI] [PubMed] [Google Scholar]