Abstract
Drug resistance during chemotherapy is the major obstacle to the successful treatment of many cancers. Here, we report that inhibition of NF-E2-related factor 2 (Nrf2) may be a promising strategy to combat chemoresistance. Nrf2 is a critical transcription factor regulating a cellular protective response that defends cells against toxic insults from a broad spectrum of chemicals. Under normal conditions, the low constitutive amount of Nrf2 protein is maintained by the Kelch-like ECH-associated protein1 (Keap1)-mediated ubiquitination and proteasomal degradation system. Upon activation, this Keap1-dependent Nrf2 degradation mechanism is quickly inactivated, resulting in accumulation and activation of the antioxidant response element (ARE)-dependent cytoprotective genes. Since its discovery, Nrf2 has been viewed as a ‘good’ transcription factor that protects us from many diseases. In this study, we demonstrate the dark side of Nrf2: stable overexpression of Nrf2 resulted in enhanced resistance of cancer cells to chemotherapeutic agents including cisplatin, doxorubicin and etoposide. Inversely, downregulation of the Nrf2-dependent response by overexpression of Keap1 or transient transfection of Nrf2–small interfering RNA (siRNA) rendered cancer cells more susceptible to these drugs. Upregulation of Nrf2 by the small chemical tert-butylhydroquinone (tBHQ) also enhanced the resistance of cancer cells, indicating the feasibility of using small chemical inhibitors of Nrf2 as adjuvants to chemotherapy to increase the efficacy of chemotherapeutic agents. Furthermore, we provide evidence that the strategy of using Nrf2 inhibitors to increase efficacy of chemotherapeutic agents is not limited to certain cancer types or anticancer drugs and thus can be applied during the course of chemotherapy to treat many cancer types.
Introduction
The transcription factor NF-E2-related factor 2 (Nrf2) was originally identified to be a critical regulator of intracellular antioxidants and phase II detoxification enzymes by the transcriptional upregulation of many ARE-containing genes (1,2). Nrf2 is negatively regulated by Kelch-like ECH-associated protein1 (Keap1), a substrate adaptor for the Cul3-dependent E3 ubiquitin ligase complex (3–5). Normally, Keap1 constantly targets Nrf2 for ubiquitin-dependent degradation and thus to maintain low constitutive expression of Nrf2-downstream target genes. Both stress and chemopreventive compounds are able to transcriptionally activate the Nrf2 target genes to trigger a cytoprotective response. Detailed mechanistic studies indicate that Keap1 is the molecular switch that controls activation and inactivation of the Nrf2 pathway. In response to oxidative stress or chemopreventive compounds, Keap1-dependent ubiquitin ligase activity is inhibited and the Nrf2 protein is accumulated. Subsequently, Nrf2 translocates to the nucleus to activate transcription of the ARE-containing genes (5,6). During the postinduction period, Keap1 travels into the nucleus to remove Nrf2 from the ARE regulatory sequences and to shuttle Nrf2 back into the cytoplasmic degradation machinery to turn off the Nrf2-dependent response (7).
Recent advance has been made in identifying the Nrf2-downstream target genes by comparing constitutive and inducible messenger RNA (mRNA) levels of genes using microarray approaches in the wild-type and Nrf2-knockout mice. Many genes, whose biological functions cannot be categorized as antioxidants or detoxification enzymes, have been demonstrated to be regulated by Nrf2. Thus, the Nrf2-downstream target genes have been expanded beyond antioxidant and detoxification genes to include stress response genes, xenobiotics-metabolizing genes, genes involving the ubiquitin-mediated proteasomal degradation system, intracellular redox-regulating genes, genes controlling cell growth and genes encoding transporters (8–12). Based on the function of these genes, it is clear that induction of the Nrf2-dependent response represents the cell’s attempt to defend itself from stressful conditions. Therefore, the Nrf2 signal pathway is currently regarded as a cell survival pathway. Indeed, accumulating in vivo evidence has demonstrated the importance of Nrf2 in protecting cells from toxic and carcinogenic effects of many environmental insults. The Nrf2-knockout mouse was prone to acute damages induced by acetaminophen, ovalbumin, cigarette smoke, pentachlorophenol and 4-vinylcyclohexene diepoxide (13–17). In addition, the Nrf2-knockout mouse had increased tumor formation when they were exposed to carcinogens such as benzo[a]pyrene, diesel exhaust and N-nitrosobutyl (4-hydroxybutyl) amine (18–20).
The role of Nrf2 has been underlined by the findings demonstrating the Nrf2-dependent protection against many human diseases or pathological states such as cancer, neurodegenerative diseases, aging, cardiovascular disease, inflammation, pulmonary fibrosis and acute pulmonary injury (18,21–25). Using dietary or synthetic compounds to boost the Nrf2-mediated cellular defense response for disease prevention has been under intensive study. Many Nrf2 activators have been identified and their efficacy in chemoprevention has been verified both in animal models and in human clinical trials (26,27).
Intriguingly, the Nrf2–Keap1 signal pathway has been reported to be impaired in many lung cancer tissues and lung carcinoma cell lines (28,29). There was a high frequency of loss of heterozygosity at 19p13.2 where Keap1 is located. Furthermore, mutations were detected in the linker and the Kelch domain of Keap1, which inactivated the repressor function of Keap1. As a consequence, the Nrf2-dependent defense response was fully activated in these cancer tissues and cell lines as demonstrated by the elevated Nrf2 protein level, nuclear localization of Nrf2 and increased mRNA expression of Nrf2 target genes. Therefore, it is highly possible that cancer cells acquire growth advantage during the course of transformation by eliminating the Keap1-mediated negative control of Nrf2, which leads to activation of the Nrf2-dependent defense response. In supporting of this notion, Nrf2 was found to be overexpressed in 91.5% of tumor tissues with head and neck squamous cell carcinoma (30).
Based on the established function of Nrf2 in conferring cell survival under detrimental conditions, it is conceivable that inhibition of the Nrf2-dependent protective response should render cancer cells more susceptible to chemotherapeutic agents. In this study, we have addressed the role of Nrf2 in determining drug response in lung carcinoma, breast adenocarcinoma and neuroblastoma. Our results show that upregulation of Nrf2 enhances cell resistance, whereas downregulation sensitizes cells to chemotherapeutic agents, demonstrating the feasibility of using Nrf2 inhibitors as adjuvants to chemotherapeutic agents to maximize cancer cell death. Furthermore, the Nrf2-dependent response to chemotherapeutic agents is not limited to a particular cell type or a specific class of anticancer drugs. Therefore, inhibiting Nrf2 during chemotherapy can be a general approach for treatment of tumors of different tissue origin.
Materials and methods
Real-time reverse transcription–polymerase chain reaction and the quinone oxidoreductase expression pattern of human lung tissues
The polymerase chain reaction (PCR) condition and Taqman probes and primers for Nrf2, NAD(P)H quinone oxidoreductase 1 (NQO1), heme oxygenase-1 (HO-1) and reduced form of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were reported previously (31). Briefly, Taqman probes were from the Universal Probe Library (Roche, Indianapolis, IN): hNrf2 (#70), hNQO1 (#87), hHO-1 (#25) and hGAPDH (#25). Oligos used for primers were synthesized by Integrated DNA Technologies (Coralville, IA).
hNrf2: forward (acacggtccacagctcatc) and reverse (tgtcaatcaaatccatgtcctg)
hNQO1: forward (atgtatgacaaaggacccttcc) and reverse (tcccttgcagagagtacatgg)
hHO-1: forward (aactttcagaagggccaggt) and reverse (ctgggctctccttgttgc)
hGAPDH: forward (ctgacttcaacagcgacacc) and reverse (tgctgtagccaaattcgttgt).
The real-time PCR condition was as follows: 1 cycle of initial denaturation (95°C for 10 min), 40 cycles of amplification (95°C for 10 s and 60°C for 20 s) and a cooling program (50°C for 5 s). The GAPDH-normalized data are presented as the fold change of gene expression in the Keap1- or Nrf2-overexpressing cell line, compared with the control cell line. For measurement of NQO1 mRNA expression in normal and tumor tissues, we purchased a Real-time PCR array of first-strand complementary DNA (cDNA) from human lung tissues in 96-well plates from OriGene (Rockville, MD) (HLRT101). The cDNAs were prepared from normal lung tissues or lung cancer biopsy samples. Each sample represents a cDNA prepared from an individual person. Tumor tissues were graded into different stages according to the differentiation statues. The tissues included in this lung qPCR array include normal tissues, stage I, stage II and stage III cancer tissues. The cDNAs from one plate were used for measurement of NQO1 mRNA levels by real-time reverse transcription–PCR (RT-PCR) analysis. Same cDNAs in another plate were used for measurement of GAPDH. The data presented are relative NQO1 mRNA levels normalized to GAPDH. This experiment was conducted three times and the mean and standard deviation were calculated.
Chemicals, cell culture and establishment of stable cell lines
All chemicals including cisplatin, doxorubicin and etoposide were purchased from Sigma (Saint Louis, MO). A549, SH-SY5Y and MDA-MB-231 cells (American Type Culture Collection, Manassas, VA) were grown in the American Type Culture Collection-suggested medium with 10% fetal bovine serum at 37°C in a humidified incubator containing 5% CO2. The Keap1–chitin-binding domain (CBD) or hemagglutinin (HA)–Nrf2 cDNA was cloned into the cloning sites of the pBabe-puro vector or into a lentivector purchased from System Biosciences (Mountain View, CA) using the standard recombinant DNA technique. The retrovector containing 29mer short hairpin RNA (shRNA) for Nrf2 was reported previously (31). For establishment of stable cell lines harboring Keap1–CBD, HA–Nrf2 or Nrf2–shRNA, the retrovectors containing the cDNA of interest were first transfected into SD-3443, an amphotropic packaging cell line from American Type Culture Collection, whereas the lentivectors were transfected into 293TN cells alone with the packaging plasmids according to the manufacturer’s instruction. Viruses produced in the supernatant were collected and used to infect cancer cell lines in the presence of 2–8 μg/ml polybrene. At 48 h postinfection, cells were grown in the medium containing 1.5 μg/ml of puromycin for selection. Stable cell lines were established once all the cells in the negative control plate were killed. Stable cell lines were continuously grown in the medium containing 1.5 μg/ml of puromycin.
Transient transfection of small interfering RNA, antibodies and immunoblot analysis
The Nrf2–siRNA and the control siRNA were purchased from Qiagen (Valencia, CA), (HP-validated siRNA and all stars Neg. siRNA AF 488). Transient transfection of siRNA was performed using HiPerFect Transfection Reagent according to the manufacturer’s protocol (Qiagen). The antibodies for Keap1, Nrf2, α-tubulin, β-actin, NQO1, HO-1 (Santa Cruz Biotechnology, Santa Cruz, CA). CBD (New England Biolabs, Ipswich, MA) and HA (Covance, Berkeley, CA) were purchased from commercial sources. Cells were lysed in sample buffer [50 mM Tris–HCl (pH 6.8), 2% sodium dodecyl sulfate, 10% glycerol, 100 mM dithiothreitol and 0.1% bromophenol blue]. Following sonication, cell lysates were electrophoresed through sodium dodecyl sulfate–polyacrylamide gel and subjected to immunoblot analysis.
Cell death detection, the NQO1 enzymatic activity and the glutathione concentration
Cell viability was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay based on the functional change of mitochondria during cell death (31). For detection of apoptotic cell death, three different methods were used: (i) Cell Death Detection ELISAPLUS (Roche); (ii) In Situ Cell Death Detection (Roche) and (iii) Annexin V–Fluorescein Isothiocyanate Apoptosis Detection (Sigma). The NQO1 activity was measured as the dicoumarol-inhibitable fraction of Dichlorphenolindophenol (DCPIP) reduction as reported previously (31). The intracellular glutathione concentration was measured using QuantiChrom glutathione assay kit from BioAssay Systems (Hayward, CA). All the procedures were followed according to the manufacturer’s instructions. All the experiments described in this section were performed in triplicates to obtain means and standard deviations. The significant differences between groups were analyzed by two-sample two-way Student’s t-test, as indicated in figures and figure legends.
Results
The Nrf2-mediated defense response is upregulated in cancer tissues
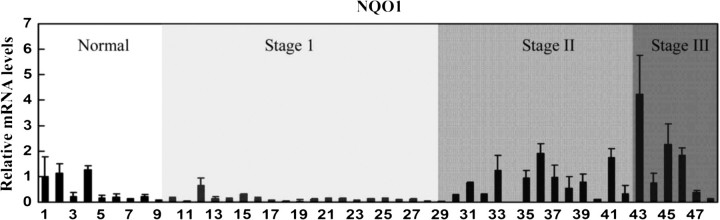
To investigate the possibility that certain cancer cells may acquire growth advantage by diminishing the negative regulation of Nrf2 by Keap1, we assessed activation of the Nrf2 signal pathway by measuring the mRNA expression profile of NQO1, an Nrf2-downstream target gene, in lung cancer tissues ranging from normal to more advanced cancer stages (stages I, II and III). The experiment was conducted three times with the same sets of cDNA samples from different plates. Results from the real-time RT-PCR analysis with the cDNAs showed a very striking pattern: there were elevated NQO1 expressions in tissues from stage II and stage III patients (Figure 1). Data shown in Figure 1 represent the relative mRNA of NQO1 normalized to GAPDH. Noticeably, there were three individuals in the normal group with elevated NQO1 levels. It is probably due to polymorphism of Nrf2, Keap1 or NQO1 genes. This result implicates that the Nrf2-dependent defensive pathway is probably activated in certain tumors to confer growth advantage.
Fig. 1.
The Nrf2-dependent defense response is upregulated in lung tumors and cancer cell lines. cDNAs from human lung tissues of normal and various stages of cancers were subjected to real-time reverse transcription–PCR for detection of NQO1 mRNA expression. The same set of samples in another plate was used for detection of GAPDH. The data represent relative mRNA levels of NQO1 normalized to GAPDH. The experiment was conducted three times and the standard deviation was shown.
Genetic knockdown of endogenous Nrf2 sensitizes lung carcinoma cells to chemotherapeutic drugs
It has been reported that Keap1 mRNA expression is downregulated in lung carcinoma A549 cells. In addition, there is a mutation in the Kelch domain of Keap1 (G333C) that destroys the function of Keap1, resulting in an elevated Nrf2 protein level in this cell line. We first confirmed the enhanced Nrf2 protein level and mRNA levels of NQO1 and HO-1 in A549 cells using immunoblot and real-time RT-PCR analysis. The Nrf2 protein level and mRNA levels for NQO1 and HO-1 in A549 cells were elevated more substantially than in other cell lines tested (data not shown). Based on these results, we chose the A549 cell line to test our hypothesis that knockdown of endogenous Nrf2 sensitizes A549 cells to chemotherapeutic drugs. Two approaches were used to knock down the endogenous Nrf2 level: transient transfection of Nrf2–siRNA and stable expression of Keap1. In one experimental setting, A549 cells were transiently transfected with a control siRNA and an siRNA specific for Nrf2. At 72 h posttransfection, the endogenous Nrf2 protein level and the mRNA levels of Nrf2, NQO1 and HO-1 were measured. Transfection of Nrf2–siRNA reduced the Nrf2 mRNA and protein levels >60% (Figure 2A left panel and Figure 2B). It also reduced NQO1 and HO-1 mRNA 60% and 40%, respectively (Figure 2A). To further confirm the repression of Nrf2 by Nrf2–siRNA, the intracellular NQO1 activities and the glutathione levels were measured. Transfection of Nrf2–siRNA significantly reduced the NQO1 activities and the glutathione levels (Figure 2C). Next, the functional significance of Nrf2 knockdown was tested by comparing the susceptibility of cells transfected with Nrf2–siRNA or control siRNA to chemotherapeutic drugs. Following 72 h siRNA transfection, cells were treated with cisplatin, doxorubicin or etoposide for 48 h, and cell viability was measured by the MTT assay. As shown in Figure 2D, reduced expression of endogenous Nrf2 by transfection of Nrf2–siRNA sensitized cells to all three chemotherapeutic agents tested.
Fig. 2.
Transient knockdown of Nrf2 by siRNA sensitizes A549 lung carcinoma cells to chemotherapeutic agents. (A) mRNAs were extracted from A549 cells transiently transfected with Nrf2–siRNA or control siRNA for 72 h. After conversion of mRNAs into cDNAs, same amounts of cDNAs were subjected to real-time reverse transcription–PCR analysis for detection of relative mRNA levels of Nrf2, NQO1, HO-1 and GAPDH. The data presented were normalized to GAPDH. (B) The Nrf2 protein levels were compared in cells transfected with Nrf2–siRNA or control siRNA. (C) The NQO1 enzymatic activity and the intracellular glutathione level of the transfected cells were measured by reduction of DCPIP and QuantiChrom glutathione assay kit, respectively. (D) A549 cells, transfected with Nrf2–siRNA or control siRNA for 72 h, were treated with the indicated doses of cisplatin, doxorubicin and etoposide for 48 h. Cell viability was determined by the MTT assay. The data are presented as means ± SDs. *P < 0.05.
To further confirm the result obtained using transient siRNA knockdown, we utilized the Nrf2-negative regulator Keap1 to reduce the endogenous Nrf2 protein level. In this experimental setting, a stable expression approach was chosen to bypass the possible toxicity introduced by transfection reagents. A stable cell line expressing Keap1 was established using a lentivirus system. Expression of Keap1–CBD was detected by an anti-CBD antibody (Figure 3A, left panel). Ectopic expression of Keap1–CBD significantly decreased the endogenous Nrf2, NQO1 and HO-1 protein levels (Figure 3A, left panel), whereas expression of α-tubulin was unchanged (Figure 3A, left panel). Next, the enzymatic activity of NQO1 was measured by the DCPIP reduction assay. The NQO1 activity was reduced to 50% in the Keap1-overexpressing cell line, compared with that in the vector-expressing cell line (Figure 3A, middle panel). Since Nrf2 plays a critical role in maintaining cellular redox homeostasis, we measured the intracellular glutathione levels in these two cell lines. Stable overexpression of Keap1 significantly decreased the intracellular levels of glutathione to 30% (Figure 3A, right panel). Taken together, these results demonstrate that stable overexpression of Keap1 inhibits the Nrf2-mediated signal pathway.
Fig. 3.
Stable knockdown of Nrf2 by overexpression of Keap1 increases the susceptibility of A549 lung carcinoma cells to chemotherapeutic agents. (A) Two A549-derived cell lines stably expressing the control vector or Keap1–CBD were established using a lentivirus-based approach. Cell lysates from these two cell lines were subjected to immunoblot analysis with antibodies against CBD, Nrf2, NQO1, HO-1 and tubulin (left panel). The NQO1 enzymatic activity was measured by reduction of DCPIP (middle panel). The intracellular glutathione level of the two cell lines was determined with QuantiChrom glutathione assay kit. (B) Cell viability was assessed by the MTT assay following 48 h treatment with the indicated doses of cisplatin, doxorubicin and etoposide. (C) Cells growing on glass slide were treated with different doses of cisplatin for 48 h and labeled with fluorescein-deoxyuridine triphosphate using the In Situ Cell Death Detection kit. Pictures were taken under fluorescent microscope (left panel). Thirteen fields of each slide were counted to get the total number of apoptotic cells (table). (D) The two cell lines were treated with doxorubicin for 48 h; apoptotic cell death was measured using Cell Death Detection ELISAPLUS kit. (E) Cells were treated with cisplatin, doxorubicin and etoposide for 48 h and stained with Annexin V–fluorescein isothiocyanate and propidium iodide using the Annexin V–Fluorescein Isothiocyanate Apoptosis Detection Kit, followed by flow cytometric analysis. The data are presented as means ± SDs. *P < 0.05.
When these two stable cell lines were compared for their survival after 48 h treatment with cisplatin, doxorubicin or etoposide, the Keap1-overexpressing cell line displayed greater sensitivity to these drugs (Figure 3B). Since the MTT cell toxicity assay measures both necrotic and apoptotic cell death, next we measured only apoptotic cell death using three different methods. (i) In Situ Cell Death Detection kit was used for detection of apoptotic cell death on a single-cell level. As shown in Figure 3C, the Keap1-overexpressing cell line had more apoptotic cells following 48 h treatment with cisplatin. The table lists the total number of apoptotic cells counted. The number of apoptotic cells detected by this method is probably underestimated since many apoptotic cells were detached and washed away during the assay. Nevertheless, more apoptotic cells were observed in the Keap1-overexpressing cell line after drug treatment. (ii) Cell Death Detection ELISAPLUS kit quantitatively detects mono- and oligonucleosomes in the cytoplasmic fraction of cell lysates. This kit was used to assess apoptotic cell death following doxorubicin treatment. The Keap1-overexpressing cells showed more apoptotic cell death (Figure 3D). (iii) Annexin V–fluorescein isothiocyanate/propidium iodide dual staining combined with flow cytometric analysis was used to assess apoptotic cell death after 48 h treatment with all three drugs. The Keap1-overexpressing cell line had more apoptotic cells compared with the vector-expressing cell line (Figure 3E). Taken together, we have used two genetic approaches to reduce the endogenous Nrf2 protein levels: by transient transfection of Nrf2–siRNA and by stable overexpression of Keap1. As a consequence of the compromised Nrf2-dependent protective response, these cancer cells are more susceptible to all three chemotherapeutic drugs tested, strongly supporting our hypothesis that inhibition of the Nrf2-mediated protective response increases the efficacy of chemotherapeutic agents.
Stable overexpression of Nrf2 enhances the resistance of human breast adenocarcinoma and neuroblastoma cells to chemotherapeutic drugs
To generalize the notion that repression of the Nrf2-mediated response sensitizes cancer cells to chemotherapeutic drugs, we further investigated the effects of Nrf2 on cell survival in response to cancer drugs in other cancer cell types. The results shown in Figure 4 were obtained using a breast carcinoma cell line MDA-MB-231. This cell line was first utilized to establish different stable cell lines expressing (i) vector; (ii) HA–Nrf2; (iii) Keap1–CBD; (iv) shRNA control vector and (v) Nrf2–shRNA vector. Once the stable cell lines were established, all cell lines were tested for the Nrf2 protein level and mRNA levels of NQO1 and HO-1 to assess activation or repression of the Nrf2 signal pathway. Surprisingly, stable overexpression of Keap1 or Nrf2–shRNA did not inhibit the Nrf2 protein level and its downstream gene expression significantly (data not shown), which probably reflects the low endogenous Nrf2 level in the MDA-MB-231 cell line. Fortunately, stable overexpression of Nrf2 increased the mRNA expression of Nrf2, NQO1 and HO-1 (Figure 4A). The ectopically expressed HA–Nrf2 was also detected (Figure 4B). This Nrf2-overexpressing cell line allowed us to test our hypothesis from the opposite perspective, that is upregulation of the Nrf2 signal pathway increases cellular resistance to cancer drugs. The MTT toxicity assay clearly showed increased resistance of this cell line to all three drugs tested (Figure 4C).
Fig. 4.
Stable overexpression of Nrf2 enhances resistance of MDA-MB-231 breast adenocarcinoma cells to chemotherapeutic agents. (A) Two MDA-MB-231-derived cell lines stably expressing the control vector or HA–Nrf2 were established using a retrovirus-based approach. Relative amounts of mRNAs for Nrf2, NQO1 and HO-1 were compared in these two cell lines. (B) Nrf2 overexpression was confirmed by immunoblot analysis with anti-HA and anti-tubulin antibodies. (C) The susceptibility of these two cell lines to cisplatin, doxorubicin and etoposide was assessed using the MTT assay after 48 h treatment (55). The data are presented as means ± SDs. *P < 0.05.
A neuroblastoma cell line SH-SY5Y was also used for making stable cell lines expressing Nrf2, Keap1 and Nrf2–shRNA. Similar to what was observed with stable cell lines derived from MDA-MB-231, the cell line stably transfected with Nrf2–shRNA or Keap1 did not display any changes in the Nrf2-mediated response as judged by the endogenous Nrf2 protein level and the mRNA level of NQO1 or HO-1 (data not shown). However, the Nrf2-overexpressing cell line showed activation of the Nrf2-mediated response as indicated by overexpression of Nrf2 mRNA and protein, and transcriptional upregulation of NQO1 and HO-1 (Figure 5A). Furthermore, both the NQO1 enzymatic activity and the glutathione level were higher in the Nrf2-overexpressing cell line compared with those in the control cell line (Figure 5A). These data indicate the establishment of a cell line with the Nrf2 pathway upregulated. Next, this cell line was used to test the consequence of Nrf2 upregulation in protecting cancer cells from drug-induced toxicity. The Nrf2-overexpressing cell line displayed increased resistance to cisplatin-induced toxicity as measured by the MTT assay (Figure 5B). Also, apoptotic cell death was significantly reduced in the Nrf2-overexpressing cell line, as measured by both quantitative Cell Death Detection ELISAPLUS (Figure 5C) and by the In Situ Cell Death Detection method (Figure 5D).
Fig. 5.
Stable overexpression of Nrf2 enhances resistance of SH-SY5Y neuroblastoma cells to chemotherapeutic agents. (A) Two SH-SY5Y-derived cell lines stably expressing the control vector or HA–Nrf2 were established using a lentivirus-based approach. Relative amounts of mRNAs for Nrf2, NQO1 and HO-1 were measured (top three panels of first row). The elevated protein levels of Nrf2 and NQO1 were confirmed by immunoblot analysis (left panels of second row). The NQO1 enzymatic activity and the intracellular glutathione level were measured (middle and right panels of second row). (B) Cell viability of these two cell lines in response to 48 h cisplatin treatment was measured by the MTT assay. (C) Apoptotic cell death in response to cisplatin treatment was also measured using Cell Death Detection ELISAPLUS. (D) Following cisplatin treatment, cells were labeled with fluorescein-deoxyuridine triphosphate using the In Situ Cell Death Detection kit (left panel). The number of apoptotic cells was counted under fluorescent microscope and the average number of apoptotic cells per field is listed in the table. The data are presented as means ± SDs. *P < 0.05.
Nrf2 upregulation by tert-butylhydroquinone enhances the resistance of neuroblastoma cells to chemotherapeutic drugs
The results shown so far were obtained through genetic modulation of the Nrf2-mediated response; it would be more desirable to use small chemicals to manipulate the Nrf2 signal pathway. Ideally, small chemicals that specifically inhibit the Nrf2-dependent defense response can be used as adjuvants to chemotherapy to increase the efficacy of chemotherapeutic agents. However, chemicals that specifically inhibit the Nrf2 signal pathway have not been identified yet. Therefore, we used an inverse approach to prove that activation of Nrf2 by tBHQ, a well-studied Nrf2 activator, increases resistance of SH-SY5Y cells to chemotherapeutic agents. As shown in Figure 6, pretreatment with 40 μM tBHQ for 24 h significantly increased the Nrf2 protein level (Figure 6A) and the cell viability in response to treatment with cisplatin (Figure 6B), doxorubicin (Figure 6C) or etoposide (Figure 6D). Put together, our data demonstrate that modulation of the Nrf2-mediated cellular defense response is an effective means to manipulate the sensitivity of cancer cells to chemotherapeutic drugs.
Fig. 6.
SH-SY5Y cells were pretreated with 40 μM of tBHQ for 24 h. The induction of Nrf2 by tBHQ was detected by immunoblot analysis (A). The pretreated cells were then treated with the indicted doses of (B) cisplatin, (C) doxorubicin or (D) etoposide in the presence of 5 μM tBHQ for 48 h, followed by the MTT assay. The data are presented as means ± SDs. *P < 0.05.
Discussion
In this study, we have demonstrated the dark side of Nrf2. Since the discovery of Nrf2 and its important role in antioxidant response, Nrf2 has been reviewed as a ‘good’ transcription factor that is essential in protecting us from oxidative stress-related diseases. Many studies in the field have been focused on identification of Nrf2 activators to boost the Nrf2-dependent defense response for disease prevention. Here, we provide another scenario where activation of Nrf2 is ‘bad’ for cancer patients during the course of chemotherapy, as evidenced by our finding that modulation of the Nrf2-mediated response affects survival of cancer cells in response to chemotherapeutic agents. Stable overexpression of Nrf2, and therefore upregulation of the Nrf2-downstream effects, resulted in enhanced resistance of cancer cells to chemotherapeutic agents including cisplatin, doxorubicin and etoposide. Inversely, downregulation of the Nrf2 protein and its downstream response by overexpression of Keap1 or transient transfection of Nrf2–siRNA rendered cancer cells more susceptible to these drugs. In addition to genetic means of Nrf2 modulation, upregulation of Nrf2 by the small chemical tBHQ also enhanced the resistance of cancer cells, indicating the feasibility of using small chemical inhibitors of Nrf2 as adjuvants to chemotherapy to increase the efficacy of chemotherapeutic agents. Furthermore, this notion was tested in three different cancer cell lines such as lung carcinoma, breast adenocarcinoma and neuroblastoma with three different anticancer drugs, demonstrating that the strategy of using Nrf2 inhibitors to increase efficacy of chemotherapeutic agents is not specific to certain cancer types or anticancer drugs and thus can be applied during the course of chemotherapy to treat many cancer types. Based on these results, identification of small chemicals that potently and specifically inhibit the Nrf2-dependent response is extremely important and such chemicals can be used to increase the efficacy of anticancer drugs.
The data presented here were obtained using three cancer cell lines, neuroblastoma SH-SY5Y, breast adenocarcinoma MDA-MB-231 and non-small cell lung carcinoma A549. Except A549, in which Keap1 is mutated and underexpressed (28), the status of Keap1 in MDA-MB-231 and SH-SY5Y has not been reported. We observed that the basal expression level of Nrf2 was high in A549, but low in MDA-MB-231 and SH-SY5Y (data not shown). Interestingly, stable overexpression of Keap1 or Nrf2–shRNA failed to knock down Nrf2 further in MDA-MB-231- and SH-SY5Y-derived cell lines, while stable overexpression of Nrf2 gave phenotypes as shown in Figures 4 and 5. Based on the report that Keap1 is mutated in some non-small cell lung cancers, along with our data showing the high Nrf2-downstream gene expression (NQO1 mRNA) in most stage II and stage III cancer tissues, it is possible that inhibition of Nrf2 expression during chemotherapy may be more effective in these tumors in which Keap1 is mutated/underexpressed and Nrf2 is upregulated. Further studies investigating the correlation between the status of Keap1 and the efficacy of Nrf2 inhibition on cancer cell sensitivity to chemotherapeutic agents are important. Herein, identification of potent Nrf2 inhibitors and investigation of Keap1 status in different tumors are two research directions to be focused on.
Upregulation of the Nrf2-mediated survival pathway indeed protects cancer cells from all three different chemotherapeutic agents tested. Etoposide kills cells by increasing the topoisomerase II-induced DNA fragmentation, which triggers programed cell death (32). Similar to etoposide, doxorubicin is also classified as a topoisomerase II poison. However, doxorubicin exhibits a wide spectrum of cytotoxicity, possibly due to other mechanisms of action including generation of free radicals (33). Cisplatin attacks DNA by forming cisplatin–DNA adducts that inhibit DNA replication and transcription (34). Since Nrf2 regulates not only antioxidants but also a global cellular-defensive response, we reasoned that the Nrf2-dependent protection against chemotherapeutic agents should not be specific to ones that kill cancer cells by generation of reactive oxygen species. Although the number of anticancer drugs tested in this study is only limited to three, the sensitivity changes due to manipulation of the Nrf2 pathway were observed in response to all three drugs. These results indicate that the strategy of inhibiting Nrf2 during chemo treatment could be applied to broad range of anticancer drugs.
Our results clearly indicate that the Nrf2-dependent defense response helps survival of cancer cells during treatment with chemotherapeutic agents. It is unclear to what extent the Nrf2-dependent protection accounts for the drug resistance phenomena observed previously in many cancer types. Drug resistance during chemotherapy is the major obstacle to the successful treatment of many cancers including neuroblastoma, breast cancers and lung cancers. During chemotherapy, a strong initial response is frequently followed by the appearance of multidrug-resistant variants.
Several mechanisms are thought to account for the drug resistance phenotype. The multiple drug resistance-associated proteins have been reported to be responsible for decreased intracellular drug accumulation (35). Increased levels of cellular thiols, facilitated detoxification of drugs and rapid DNA repair were found to be associated to drug resistance (34,36,37). A recent report identified a function of p53 in determining multidrug sensitivity on neuroblastoma (38). In addition, upregulation of activating transcription factor 4 (ATF4) was reported to increase cisplatin resistance in human cancer cell lines (39). Interestingly, many genes reported to play roles in drug resistance seemingly have a functional link with Nrf2. For instance, Nrf2 was initially identified as a key regulator of phase II-detoxifying enzymes through antioxidant response elements in the promoter region of these genes. In addition, Nrf2 regulates many of the key enzymes that are important in maintaining cellular redox homeostasis, such as gamma-glutamylcysteine synthetase (γ-GCS), xCT cysteine/glutamate exchange transport and thioredoxin (40–44). γ-GCS is a rate-limiting enzyme controlling glutathione biosynthesis, whereas xCT cysteine/glutamate exchange transport facilitates the uptake of cysteine that is needed for glutathione synthesis. Thioredoxin is another antioxidant protein that works in concert with glutathione to maintain intercellular redox homeostasis. In addition, many members of transporters including the multidrug resistance-associated protein 1-6 (MRP 1-6), have been reported as Nrf2 target genes (45–47). ATF4 was reported to upregulate HO-1 transcription by forming heterodimer with Nrf2 (48). p53 and Nrf2 were found to work cooperatively in protecting against N-nitrosobutyl (4-hydroxybutyl) amine-induced bladder cancer (49). Collectively, these results implicate that upregulation of Nrf2 is responsible, at least in part, for drug resistance observed during the course of chemotherapy.
Consistent with the notion that Nrf2 protects cancer cells from killing by anticancer drugs, a recent paper reported that Nrf2-knockout murine embryonic fibroblasts were more sensitive to cisplatin treatment (50). Transient knockdown of Nrf2 with Nrf2–siRNA decreased survival of human ovarian cancer cells under cisplatin treatment (50). In analog to Nrf2, expression of HO-1 in many tumor tissues was also upregulated (51,52). Furthermore, it has been shown that decreased expression of HO-1 by either chemical inhibitor or HO-1 siRNA sensitized cells to cytotoxicity of cisplatin, whereas upregulation of HO-1 attenuates the toxicity induced by cisplatin or by photodynamic therapy (53–56). Since Nrf2 regulates many genes including HO-1, targeting Nrf2 may be more effective than targeting HO-1. In conclusion, our data demonstrate that inhibition of Nrf2 sensitizes cells to chemotherapeutic drugs, suggesting that Nrf2 inhibitors may be used concomitantly to increase the efficacy of anticancer therapy.
Identification of the dark side of Nrf2, that is upregulation of Nrf2 in cancer cells provides them with a growth advantage under detrimental environments, may generate a concern in using Nrf2 activators for the purpose of chemoprevention. However, it seems worth mention that induction of the Nrf2-dependent defensive response by Nrf2 activators in normal tissue is transient because the negative regulator Keap1 is only inhibited temporarily and partially. However, in cancer tissues, dysregulation of Nrf2 by Keap1 due to mutation of Keap1 results in strong and persistent induction of Nrf2. Previously, it was shown that induction of Nrf2 by tBHQ reached a peak following 18–24 h treatment and was gradually diminished after 24 h (57). Thus, it would probably be safe to use Nrf2-activating chemopreventive drugs every other day. In the case of cancer patients undergoing treatment with chemotherapeutic drugs, it will be wise to temporally inhibit the Nrf2-dependent cytoprotection using Nrf2 inhibitors to enhance patients’ response to anticancer drugs. In the present work, we provide the strong evidence that response of cancer cells to chemotherapeutic drugs can be elevated by inhibiting the Nrf2-dependent pathway. Identification of small chemicals that specifically inhibit Nrf2, along with animal studies and human clinical trials, is needed to test the efficacy of coadministration of Nrf2 inhibitors during chemotherapy.
Funding
American Cancer Society (RSG-07-154-01-CNE); National Institutes of Health (ES015010-01).
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ARE
antioxidant response element
- CBD
chitin-binding domain
- cDNA
complementary DNA
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HA
hemagglutinin
- HO-1
heme oxygenase-1
- Keap1
Kelch-like ECH-associated protein1
- mRNA
messenger RNA
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- Nrf2
NF-E2-related factor 2
- NQO1
NAD(P)H quinone oxidoreductase 1
- PCR
polymerase chain reaction
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- tBHQ
tert-butylhydroquinone
References
- 1.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 2.Venugopal R, et al. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl Acad. Sci. USA. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itoh K, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang DD, et al. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi M, et al. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Sun Z, et al. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell. Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JM, et al. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J. Biol. Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 9.Thimmulappa RK, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 10.Rangasamy T, et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J. Clin. Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho HY, et al. Nrf2 defends the lung from oxidative stress. Antioxid. Redox Signal. 2006;8:76–87. doi: 10.1089/ars.2006.8.76. [DOI] [PubMed] [Google Scholar]
- 12.Kwak MK, et al. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 13.Enomoto A, et al. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 14.Chan K, et al. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl Acad. Sci. USA. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rangasamy T, et al. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J. Exp. Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iizuka T, et al. Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes Cells. 2005;10:1113–1125. doi: 10.1111/j.1365-2443.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 17.Hu X, et al. Accelerated ovarian failure induced by 4-vinyl cyclohexene diepoxide in Nrf2 null mice. Mol. Cell. Biol. 2006;26:940–954. doi: 10.1128/MCB.26.3.940-954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki Y, et al. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol. Appl. Pharmacol. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 19.Ramos-Gomez M, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl Acad. Sci. USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iida K, et al. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004;64:6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 21.Braun S, et al. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol. Cell. Biol. 2002;22:5492–5505. doi: 10.1128/MCB.22.15.5492-5505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho HY, et al. The transcription factor NRF2 protects against pulmonary fibrosis. FASEB J. 2004;18:1258–1260. doi: 10.1096/fj.03-1127fje. [DOI] [PubMed] [Google Scholar]
- 23.Suh JH, et al. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc. Natl Acad. Sci. USA. 2004;101:3381–3386. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii Y, et al. Transcription factor Nrf2 plays a pivotal role in protection against elastase-induced pulmonary inflammation and emphysema. J. Immunol. 2005;175:6968–6975. doi: 10.4049/jimmunol.175.10.6968. [DOI] [PubMed] [Google Scholar]
- 25.Shih AY, et al. Induction of the Nrf2-driven antioxidant response confers neuroprotection during mitochondrial stress in vivo. J. Biol. Chem. 2005;280:22925–22936. doi: 10.1074/jbc.M414635200. [DOI] [PubMed] [Google Scholar]
- 26.Yates MS, et al. Chemopreventive promise of targeting the Nrf2 pathway. Drug News Perspect. 2007;20:109–117. doi: 10.1358/dnp.2007.20.2.108343. [DOI] [PubMed] [Google Scholar]
- 27.Kensler TW, et al. Effects of glucosinolate-rich broccoli sprouts on urinary levels of aflatoxin-DNA adducts and phenanthrene tetraols in a randomized clinical trial in He Zuo township, Qidong, People's Republic of China. Cancer Epidemiol. Biomarkers Prev. 2005;14:2605–2613. doi: 10.1158/1055-9965.EPI-05-0368. [DOI] [PubMed] [Google Scholar]
- 28.Singh A, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padmanabhan B, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Stacy DR, et al. Increased expression of nuclear factor E2 p45-related factor 2 (NRF2) in head and neck squamous cell carcinomas. Head Neck. 2006;28:813–818. doi: 10.1002/hed.20430. [DOI] [PubMed] [Google Scholar]
- 31.Wang XJ, et al. Nrf2 protects human bladder urothelial cells from arsenite and monomethylarsonous acid toxicity. Toxicol. Appl. Pharmacol. 2007;225:206–213. doi: 10.1016/j.taap.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bromberg KD, et al. A two-drug model for etoposide action against human topoisomerase IIalpha. J. Biol. Chem. 2003;278:7406–7412. doi: 10.1074/jbc.M212056200. [DOI] [PubMed] [Google Scholar]
- 33.Swift LP, et al. Doxorubicin-DNA adducts induce a non-topoisomerase II-mediated form of cell death. Cancer Res. 2006;66:4863–4871. doi: 10.1158/0008-5472.CAN-05-3410. [DOI] [PubMed] [Google Scholar]
- 34.Selvakumaran M, et al. Enhanced cisplatin cytotoxicity by disturbing the nucleotide excision repair pathway in ovarian cancer cell lines. Cancer Res. 2003;63:1311–1316. [PubMed] [Google Scholar]
- 35.Fujii R, et al. Active efflux system for cisplatin in cisplatin-resistant human KB cells. Jpn. J. Cancer Res. 1994;85:426–433. doi: 10.1111/j.1349-7006.1994.tb02376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 1994;54:4313–4320. [PubMed] [Google Scholar]
- 37.Chaney SG, et al. DNA repair: enzymatic mechanisms and relevance to drug response. J. Natl Cancer Inst. 1996;88:1346–1360. doi: 10.1093/jnci/88.19.1346. [DOI] [PubMed] [Google Scholar]
- 38.Xue C, et al. p53 determines multidrug sensitivity of childhood neuroblastoma. Cancer Res. 2007;67:10351–10360. doi: 10.1158/0008-5472.CAN-06-4345. [DOI] [PubMed] [Google Scholar]
- 39.Tanabe M, et al. Activating transcription factor 4 increases the cisplatin resistance of human cancer cell lines. Cancer Res. 2003;63:8592–8595. [PubMed] [Google Scholar]
- 40.Wild AC, et al. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 41.Chan JY, et al. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim. Biophys. Acta. 2000;1517:19–26. doi: 10.1016/s0167-4781(00)00238-4. [DOI] [PubMed] [Google Scholar]
- 42.Shih AY, et al. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J. Neurosci. 2003;23:3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YC, et al. Thioredoxin-dependent redox regulation of the antioxidant responsive element (ARE) in electrophile response. Oncogene. 2003;22:1860–1865. doi: 10.1038/sj.onc.1206369. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki H, et al. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 2002;277:44765–44771. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]
- 45.Aleksunes LM, et al. Induction of Mrp3 and Mrp4 transporters during acetaminophen hepatotoxicity is dependent on Nrf2. Toxicol. Appl. Pharmacol. 2007;226:74–83. doi: 10.1016/j.taap.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maher JM, et al. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab. Dispos. 2005;33:956–962. doi: 10.1124/dmd.105.003798. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi A, et al. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem. Biophys. Res. Commun. 2003;310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 48.He CH, et al. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 49.Iida K, et al. Nrf2 and p53 cooperatively protect against BBN-induced urinary bladder carcinogenesis. Carcinogenesis. 2007;28:2398–2403. doi: 10.1093/carcin/bgm146. [DOI] [PubMed] [Google Scholar]
- 50.Cho JM, et al. Role of the Nrf2-antioxidant system in cytotoxicity mediated by anticancer cisplatin: implication to cancer cell resistance. Cancer Lett. 2008;260:96–108. doi: 10.1016/j.canlet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 51.Was H, et al. Overexpression of heme oxygenase-1 in murine melanoma: increased proliferation and viability of tumor cells, decreased survival of mice. Am. J. Pathol. 2006;169:2181–2198. doi: 10.2353/ajpath.2006.051365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jozkowicz A, et al. Heme oxygenase-1 in tumors: is it a false friend? Antioxid. Redox Signal. 2007;9:2099–2117. doi: 10.1089/ars.2007.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim HR, et al. Suppression of Nrf2-driven heme oxygenase-1 enhances the chemosensitivity of lung cancer A549 cells toward cisplatin. Lung Cancer. 2007;60:47–56. doi: 10.1016/j.lungcan.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 54.Loboda A, et al. Janus face of Nrf2-HO-1 axis in cancer-friend in chemoprevention, foe in anticancer therapy. Lung Cancer. 2007;60:1–3. doi: 10.1016/j.lungcan.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 55.Nowis D, et al. Heme oxygenase-1 protects tumor cells against photodynamic therapy-mediated cytotoxicity. Oncogene. 2006;25:3365–3374. doi: 10.1038/sj.onc.1209378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim HJ, et al. Heme oxygenase-1 attenuates the cisplatin-induced apoptosis of auditory cells via down-regulation of reactive oxygen species generation. Free Radic. Biol. Med. 2006;40:1810–1819. doi: 10.1016/j.freeradbiomed.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 57.Wang XJ, et al. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents. Cancer Res. 2006;66:10983–10994. doi: 10.1158/0008-5472.CAN-06-2298. [DOI] [PubMed] [Google Scholar]