Abstract
Environmental epigenetics has an important role in regulating phenotype formation or disease etiology. The ability of environmental factors and exposures early in life to alter somatic cell epigenomes and subsequent development is a critical factor in how environment affects biology. Environmental epigenetics provides a molecular mechanism to explain long term effects of environment on the development of altered phenotypes and “emergent” properties, which the “genetic determinism” paradigm cannot. When environmental factors permanently alter the germ line epigenome, then epigenetic transgenerational inheritance of these environmentally altered phenotypes and diseases can occur. This environmental epigenetic transgenerational inheritance of phenotype and disease is reviewed with a systems biology perspective.
Keywords: Epigenetics, Transgenerational, Environmental exposures, Reproduction, Systems biology, Development
1. Introduction
Aristotle introduced the question if the embryo contain all its parts in little from the beginning, or is there a true formation of new structures as it develops (Peck, 1943). Since the emergence of neo-Darwinism the paradigm has been that genes are responsible for the formation of phenotypes and that they control development, with environmental or epigenetic actions being minimally important (Wake and Larson, 1987). According to this view, the environment is assumed to have only a permissive role in development (Gilbert, 2005). However, systemic or integrative biologists have recognized the importance of the role of environment in the developmental process and its importance in shaping phenotypes (Oster and Alberch, 1982; Nijhout et al., 1986). Although the role of genetics is crucial for the developmental process, an exclusively genetic approach ignores important environmentally dependent events that occur during development. In contrast, consideration of epigenetic processes provides the opportunity to understand how environmental signals regulate genome activity or interfere with developmental processes involved in the formation of the adult phenotype (Jaenisch and Bird, 2003; Jirtle and Skinner, 2007). The establishment of epigenetic patterns during development is a crucial process that links how environmental conditions influence development. The action of these environmental compounds induces the establishment of specific epigenetic patterns during key developmental periods that influence phenotypic variation, which in some cases lead to disease states (Jirtle and Skinner, 2007; Skinner et al., 2010).
The influence of environmental factors on biological processes has been appreciated and investigated for hundreds of years; however, the basic molecular mechanism of how the environment can influence long term gene regulation has only recently been addressed through epigenetic studies. Even in the early days of genetic knowledge, scientists such as Richard Goldschmidt, an integrative biologist, highlighted the fact that early developmental events could have as much importance as genetics in the origin of the adult phenotype (Goldschmidt, 1933). This message is still valid today when even after years of important discoveries in the field of genetics, the phenotypic states cannot be explained solely by observed changes in DNA sequence. Epidemiological studies have for decades suggested significant environmental impacts on biology that could not be explained solely by genetic features. Examples of such observations follow:
The association of disease states with single nucleotide polymorphisms (SNPs) in the “Genome-Wide Association Studies” (GWAS) has revealed that genetic components usually explain less than 20% of the phenotypic variance (Wallace, 2010). The importance of moving beyond studying SNPs to examine more complex chromosomal regulation and epigenetic changes is paramount (Yuan and Ferguson, 2011).
The premise that a detailed genetic knowledge would allow the causes of the majority of diseases to be determined, which was based on a reductionist view of genotype–phenotype correlations, is now proven to be false when the evidence suggests that the majority of diseases are complex traits (Dipple et al., 2001).
The importance of the developmental origins of disease is now well accepted, which has also been recently recognized by the World Health Organization (Godfrey et al., 2007).
It is known that the risk for common metabolic diseases varies by geography and ethnicity, therefore making regional environmental influences an important component in disease incidence (Wallace, 2010). Several disease mechanisms are now known to have an important epigenetic component, such as in allergy (Kuriakose and Miller, 2010), hepatic cancer (Pogribny et al., 2006), gastric cancer (Nan et al., 2005), asthma (Martino and Prescott, 2011), colorectal cancer (Choong and Tsafnat, 2011), prostate cancer (Perry et al., 2010), HIV latency (Hakre et al., 2011) or brain disorders (Kaminsky and Tsafnat, 2010).
Studies in monozygotic twins (same genetic composition) have revealed striking discordances in the prevalence of many diseases that so far have been thought to have a genetic association (Bell and Spector, 2011). Association of these diseases with epigenetic mechanisms is evident.
Several common diseases have had a dramatic increase in incidence in the past decade, with exposure to environmental factors accounting for 40% of deaths worldwide and with the majority of cancer being linked to environmental exposures (Pimentel et al., 2007).
Among all environmental compounds or toxicants that are associated with the onset of diseases very few have the ability to alter DNA sequence or promote mutations. For example, carcinogenic metals are weak mutagens (Martinez-Zamudio and Ha, 2011) and few endocrine disruptors have been found to have a mutagenic effect (Skinner et al., 2010; Guerrero-Bosagna and Valladares, 2007).
In regards to evolutionary biology, the random mutation hypothesis is not sufficient to explain the origin of phenotypes (Brisson, 2003; Lenski and Mittler, 1993). However, epigenetically induced genomic changes may play an important role in promoting specific types of mutation (Skinner et al., 2010; Guerrero-Bosagna et al., 2005), as for example CG to TG transitions (Sved and Bird, 1990).
These are just a few of the biological observations that suggest a significant impact of the environment on disease etiology, which cannot be easily explained through classic genetic mechanisms. The gaps in knowledge have been partially addressed due to the advent of epigenetics, a phenomenon described by Waddington in the 1940’s. His initial definition of epigenetics had a developmental basis, defining it as “the branch of biology which studies the causal interactions between genes and their products which bring phenotypes into being” (Jablonka et al., 2002). However, the specific molecular mechanisms involved in epigenetic modification of genome activity were not elucidated until more recently (1970’s through today). Currently, the study of epigenetics focuses on “molecular factors and processes around DNA that regulate genome activity independent of DNA sequence and that are mitotically stable” (Skinner et al., 2010). Epigenetic systems have been described in several organisms and include histone modifications, chromatin structure, non-coding RNA and DNA methylation and hydroxymethylation (Chen and Riggs, 2005; Craig, 2005; Iqbal et al., 2011; Kim, 2006; Margueron et al., 2005; Skinner and Guerrero-Bosagna, 2009; Wallace and Orr-Weaver, 2005). Epigenetics is a critical element in the regulation of genome activity, which cooperates with genetic factors in regulating physiological responses (Gilbert, 2005). Epigenetics also provides a mechanism that allows environmental factors to influence long-term regulation of gene expression later in life that underlies the process of phenotype formation. Environmentally induced epigenetic changes may have a wide range of phenotypic consequences leading to disease conditions such as cancer, reproductive defects and obesity (Anway et al., 2005, 2006; Cheng et al., 2004; Guerrero-Bosagna et al., 2008; Howdeshell et al., 1999; Newbold et al., 2006, 2008; Waterland et al., 2008; Yamasaki et al., 1992). Focus on these observations have lead to the defining of the area of environmental epigenetics, which has had a major role in the elucidating disease etiology.
2. Environmental epigenetic influences on meiosis and mitosis
Cell replication requires the conservation of epigenetic patterns to daughter cells. Environmental exposures can act on both somatic and germ cell replication, influencing the establishment or maintenance of specific epigenetic patterns. These epigenetic processes and programming are influenced by environmental factors. For example, dietary compounds have been implicated in the modulation of histone modifications (Delage and Dashwood, 2008) and even trace elements have been shown to alter DNA methylation during early development (Vahter, 2007; Waalkes et al., 2004). Since DNA and histone methylation are enzymatic reactions that requires methyl donors to proceed (Singal and Ginder, 1999), the environment can influence DNA methylation through modifying the activity of methyltransferases or through altering the availability of methyl donors, which are the substrate for the methylation reaction on histones or CpG dinucleotides. Two different classes of DNA methyltransferases (Dnmts) exist: maintenance and de novo methyltransferases. One important aspect of epigenetic mechanisms is their ability to be maintained even after mitotic cell divisions (Skinner, 2011a,b). The mechanism by which DNA methylation states are mitotically maintained is through the action of DNMT1, a methyltransferase that only acts upon hemi-methylated strands of DNA (Yoder et al., 1997). After each cell division, DNMT1 activity will generate newly synthesized DNA strands that are methylated according to the DNA methylation pattern of the paternal strand. In contrast, other methyltransferases such as DNMT3A or DNMT3B act upon unmethylated strands of DNA (Yokochi and Robertson, 2002), which is why they are called de novo methyltransferases. Each DNA methyltransferase acts at a different window of time during development (La Salle et al., 2004), with the action of de novo methyltransferases being crucial in critical windows during early development when DNA methylation patterns are established. However, once they are established, the action of DNMT1 is crucial to maintaining these methylation patterns through adulthood. Therefore, given the susceptibility of epigenetic mechanisms to alteration by environmental compounds, exposures can shift the normal differentiation process through a modification of the epigenome. These are exposures that occur at critical windows of development for a given cell type. The exact mechanism that explains how some regions are targeted by environmental compounds for DNA methylation changes and not others is still unknown. However, the mitotic stability of the epigenome explains how an early life exposure can impact adult onset disease and phenotypes (Skinner, 2011a,b).
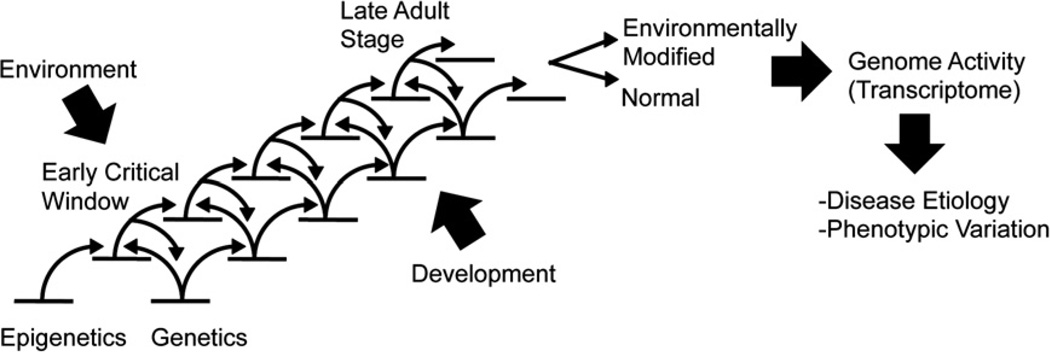
An altered epigenome induced by an environmental factor impacts the resulting phenotype, which creates an abnormal state of cellular or tissue differentiation. These phenotypic alterations may appear long after the exposure to environmental compounds, Fig. 1. When a group of cells is influenced early during development by an environmental exposure, they develop an altered epigenome that persists throughout the life of the individual, since the epigenome is maintained after mitosis. This would explain how early life events have consequences on the adult phenotype formation and disease onset, which is called the developmental origin of diseases (Godfrey et al., 2007; Hanson and Gluckman, 2008). After the individual has become an adult, the possible changes in the epigenome are limited to the process of maintenance of epigenetic marks rather than to the establishment of them. Therefore, as suggested by Waddington, as individuals age they become ‘canalized’ towards more limited possibilities of phenotypes (Waddington, 1959). Depending on when environmental exposures occur during development Fig. 1, different cell types and numbers can be affected in terms of their epigenomes and adult phenotypes. This occurs because the level of linkage between cell types, called ‘connectance’, varies during development, having consequences on phenotypic expression and plasticity (Amzallag, 2000). For instance, different windows of sensitivity exist for the morphogenesis of diverse abnormalities, where, for example, upper limbs abnormalities arise after exposures early in the prenatal development, while external genitalia defects arise after exposures during late prenatal and early postnatal development (Moore and Persaud, 1993). Environmental exposures on somatic cells and associated epigenetic alterations could be the basis for the majority of altered phenotypes and disease states that are observed. However, since these somatic cell effects are confined to one ontogeny, the changes produced will not be able to be transmitted to the next generation, which only occurs through alteration of the germ line. The critical role of the germ line in transmitting components of heredity has previously been described by the geneticist August Weissman in the late 19th century (Weissman, 1892). Therefore, environmentally induced epigenetic changes in somatic cells cannot be considered as an intrinsic (Guerrero-Bosagna and Skinner, 2009) transgenerational phenomena, unless modifications in the germ line epigenome are produced and perpetuated. Germ line epigenomic changes, in turn, can be induced when the methylation resetting window is targeted during early development. The germ line development has a relatively narrow critical window when the epigenome can be altered by an environmental exposure (Jirtle and Skinner, 2007; Skinner et al., 2010). In the event the germ line epigenome is modified, impacts on the generation derived from that germ line can occur and produce a transgenerational effect (Jirtle and Skinner, 2007; Skinner et al., 2010; Anway et al., 2005).
Fig. 1.
Epigenic and genetic cascade of events involved in development and environmental impacts.
3. Epigenetic transgenerational inheritance
One of the periods most sensitive to environmental exposures is fetal gonadal sex determination, when the germ line is undergoing epigenetic programming and DNA re-methylation (Skinner et al., 2010; Anway et al., 2005; Guerrero-Bosagna and Skinner, 2009). When an environmental factor promotes a permanent change of the gamete epigenome, this altered genome and associated phenotypes can be transgenerationally transmitted to subsequent generations and progeny (Skinner et al., 2010). The endocrine disruptor vinclozolin is a fungicide commonly used in agriculture that has anti-androgenic activity (Wong et al., 1995) and has been experimentally used to induce epigenetic transgenerational phenotypes. It was initially shown that a developmental exposure to vinclozolin can affect embryonic testis development and later cause an increase in spermatogenic cell apoptosis in the adult (Uzumcu et al., 2004). Interestingly, this spermatogenic defect was transgenerationally transmitted from the affected F1–F4 generations (Anway et al., 2005, 2006a,b) and hypothesized to be due to permanently altered DNA methylation of the male germ-line (Anway et al., 2005). One of the reasons for this hypothesis is that the pattern of gene expression was found to be altered in the fetal testis (Clement et al., 2007; Clement et al., 2010) and similar for all generations studied (F1–F3) Anway et al., 2008. Transgenerational transcriptome pattern changes were also identified in prostate (Anway and Skinner, 2008) and brain (Skinner et al., 2008), with each tissue having a unique set of differentially expressed genes. Recently, vinclozolin-induced epigenetic transgenerational alterations have been detected in sperm DNA using a genome-wide promoter approach (Guerrero-Bosagna et al., 2010). Therefore, the transgenerational alteration of the sperm epigenome induced by vinclozolin appears to promote transgenerational alterations in transcriptomes of adult somatic cells in different tissues. It remains to be established how this altered sperm epigenome relate to the somatic effects in the transcriptome and if epigenetic somatic changes are also produced.
In addition to the spermatogenic cell defects detected (Anway et al., 2005), transgenerational effects on the development of other adult onset disease states are also observed as the animals age, which includes the development of tumors, prostate and kidney diseases, and immune abnormalities (Anway et al., 2006a,b). Recent observations also suggest transgenerational effects on behaviors such as sexual selection and anxiety (Skinner et al., 2008; Crews et al., 2007). Therefore, a number of different altered adult phenotypes that are transgenerationally transmitted have been identified after a developmental exposure to vinclozolin, together with the identification of transcriptome somatic effects and germ line epigenetic effects. However, the experimental evidence of a transgenerational effect comes only with the generation that has had no germ line or somatic cell direct environmental exposure. For example, if a F0 gestating female is affected by an environmental factor, then the F1 fetus will have somatic and germ line effects and the F2 generation will have germ line effects, since the developing F2 generation germ cells in the F1 fetus are directly exposed when the F0 mother was exposed. Therefore, only the F3 generation will have truly transgenerational effects, since it was not directly exposed (Skinner, 2011a,b, 2008). Although these germ line epigenetic transgenerational effects might be limited by the time of the developmental exposure or the action of specific compounds, they can still have profound effects on biology, from disease etiology to evolution.
A number of studies from different laboratories have identified environmentally induced epigenetic transgenerational inheritance of adult onset disease phenotypes, Table 1. These include the actions of environmental endocrine disruptors and toxicants such as bis-phenol A (BPA) that promoted testis abnormalities observed in the F3 generation (Salian et al., 2009), dioxin that promoted uterus abnormalities observed in the F3 generation (Bruner-Tran and Osteen, 2010), and vinclozolin that promoted imprinted gene DNA methylation changes observed in the F3 generation (Stouder and Paoloni-Giacobino, 2010). Pharmaceutical agents such as thyroxine and morphine have been shown to promote behavioral abnormalities that were observed from the F1–F3 generations (Vyssotski, 2011). Recently, chemotherapy was shown to induce transgenerational effects such as despair-like behaviors, delivery complications, reduced primordial follicle pool and early lost of reproductive capacity (Kujjo et al., 2011). In addition to environmental toxicants, nutritional alterations have also been shown to induce epigenetic transgenerational inheritance of disease states (Waterland et al., 2008; Kaati et al., 2007). This includes caloric restriction promoting metabolic disease phenotypes observed in the F3 generation (Waterland et al., 2008), as well as high fat diets promoting transgenerational adult onset metabolic disease and obesity (Dunn and Bale, 2011; Pentinat et al., 2010). It is becoming increasingly evident in several organism models that a variety of environmental factors such as stress (Matthews and Phillips, 2010), endocrine disruptors (Walker and Gore, 2011) or cell culture conditions (Lee et al., 2009) are able to influence phenotype formation in a transgenerational manner, Table 1. Genetic mutations have been shown to produce epigenetic changes that are transgenerationally transmitted (Xing et al., 2007), which compliments the sequence of events leading to disease and phenotype formation (Nadeau, 2009). In addition to effects in laboratory animals, studies in human populations suggest that past exposures could have an important role in current disease states, which would have occurred through epigenetic inheritance (Pembrey et al., 2006; Heijmans et al., 2008). Given this recent evidence, it is anticipated that the majority of environmental exposure that can affect fetal gonadal development may promote epigenetic transgenerational inheritance and be a crucial component of adult phenotype formation. The impacts of environmental epigenetics are significant to biological sciences and a previously underappreciated factor in understanding phenotype formation and disease etiology. The future challenges will be to identify the biological mechanisms involved in this process. Future questions to be experimentally addressed will be:
Do environmental stimuli lead to patterned changes in DNA methylation or to stochastic changes?
If certain sites are differentially and persistently targeted for a DNA methylation change, is there any common genomic feature that would explain this susceptibility?
How do germ line epigenetic changes correlate with somatic tissue epigenetic changes latter on during development?
Table 1.
Examples Epigenetic Transgenerational Inheritance.
| Environmentally induced transgenerational inheritance | Reference |
|---|---|
| Vinclozolin induced epigenetic transgenerational adult onset disease in rats (F1–F4) | Anway et al. (2005) |
| Transgenerational response in longevity to nutrition (F0–F2) | Kaati et al. (2007) |
| Tumor susceptibility in Drosophila (F1–F3) | Xing et al. (2007 |
| Nutrition induced transgenerational obesity in mice (F1–F3) | Waterland et al. (2008) |
| BPA-induced transgenerational testicular abnormality (F1–F3) | Salian et al. (2009) |
| Stem cell culture induced adult onset disease (F0–F4) | Lee et al. (2009) |
| Dioxin induced transgenerational uterine abnormality (F1–F4) | Bruner-Tran and Osteen (2010) |
| Stress induced behavior alterations (F0–F2) | Matthews and Phillips (2010) |
| Transgenerational glucose intolerance (F0–F2) | Pentinat et al. (2010) |
| Transgenerational effects of morphine or thyroxine on hypocampus, birth weight and behavior (F0–F3) | Vyssotski (2011) |
| Transgenerational effects of chemotherapy in mice (F0–F6) | Kujjo et al. (2011) |
| Transgenerational effects of obesity on female body size (F0–F3) | Dunn and Bale (2011) |
4. Epigenetics, reductionism and systems biology
Systems biology focuses on the integration of molecular to physiological level understanding of the emergence of biological processes. Emergence is a concept that was initially proposed by Paul Weiss (Weiss, 1939), who stated that “phenotypes, and mechanisms that underlies them are dependent upon, and subordinate to, the law which rules the complex as a unit”. In spite of the concept that a true understanding of how organisms operate arises only when taking this integrative approach, the recent history of biology has been mainly written from a reductionist perspective. The genetic reductionism of the past years has left many gaps of knowledge, which are mainly due to the absence of a consideration of emergent properties (Soto et al., 2009). Emergent properties will only be adequately interrogated when organism systems as a whole and not only as a collection of components are included as the focus of research (Cornish-Bowden et al., 2004). The recent technological development of biological tools and the advent of the ‘omics’ era has allowed scientists the possibility of taking this systems biology approach, however, at the expense of the difficulties in integrating and managing enormous datasets (Joyce and Palsson, 2006). Methodologically, systems biology integrates knowledge from different biological components and sets of data into understanding the system as a whole (Ideker, 2004). This integration of data into a systemic approach has been the focus of developmental biology since its origins, by focusing on coordinated morphological changes (e.g. morphogenesis, connectance) that consider the emergent properties of the developmental process (Theissen, 2006). Since epigenetics has a developmental origin, the study of epigenetic mechanisms is inherently a systems biology approach because it includes analysis of top-down and bottom-up relationships between the whole and the parts, as well as integration of sets of data from different levels of analysis. The study of systems biology brings back the old dichotomy of parts versus the whole that dates from the Aristotelian saying “the whole is more than the sum of its parts” (Bostock, 2003). Systems biology re-emphasizes that emergent properties are only visible when considering the whole as a unit and lost when the focus is exclusive on the parts. Epigenetics is an essential molecular component to consider for taking this integrative approach. Future experiments should be designed to look into integrating genome wide data sets of several molecular components that influence the formation of phenotypes, rather than a reductionist approach considering selected specific genes. Research must not omit the integration of groups of genes and environmental parameters in order to study emergent phenotypes.
5. Conclusions
It is becoming increasingly accepted that environmental compounds can produce changes in the genome that in spite of not altering DNA sequence can produce important and permanent alterations in the phenotype. In addition, the ability of environmental exposures to influence generational effects has significant impacts in our understanding of the basic regulation of biology. A number of different experimental approaches need to be considered in order to understand how the environment can produce long lasting effects not only on individuals exposed, but on their subsequent progeny for generations to come. Environmental epigenetics provides the molecular understanding of how environmental factors promote immediate and long term effects on the individual exposed (Skinner et al., 2010). However, the transgenerational epigenetic mechanism requires the actions of environmental compounds during germ line differentiation to influence epigenetic programming. Although the actions of most environmental factors can easily produce alterations in the somatic cell epigenome, only in the event the germ line is epigenetically modified will transgenerational inheritance of disease or phenotypes be intrinsically promoted.
The suggestion that environmental factors can interfere with early development to induce epigenetic transgenerational disease is a new paradigm in disease etiology. The current paradigm for the causal factors in disease etiology involves genetic mutations or chromosomal abnormalities as the initial triggering mechanism. However, only a small percentage of nearly all disease states are associated with known genetic mutations. Epigenetics provides a molecular mechanism that is anticipated to have a significant role in disease etiology because epigenetic systems respond to environmental factors and directly influence phenotype formations and the onset of diseases, Fig. 1. Prenatal and early postnatal exposures are likely more critical in disease etiology than the adult exposures, which are more resistant to epigenetic change due to increased levels of cellular and tissue differentiation versus the sensitivity of epigenetic systems during early stages of development.
Reductionist approaches taken so far to explain the emergence of phenotypic features have fallen short in explaining the emergent properties that arise from the intricate cellular connections of a developmental system. The study of developmental epigenetics as being the link between environmental and genetic features will provide a new framework to understand the factors influencing phenotype formation. Such a focus is in the realm of systems biology. Considering the system requires an understanding of environmental actions on the biology. The amount of toxicants in our environment and the increasing evidence for the correlation of exposures with human diseases requires the inclusion of environmental epigenetics in future systems biology investigations. In addition to elucidate disease etiology, environmental epigenetics will also impact other areas of biology such as evolutionary studies, in which the environment will play a fundamental role in explaining the induction of new phenotypic variation, which before where primarily explained to be originated from random genetic modifications.
References
- Amzallag GN. Connectance in sorghum development: beyond the genotype–phenotype duality. Biosystems. 2000;56:1–11. doi: 10.1016/s0303-2647(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Anway MD, Skinner MK. Transgenerational effects of the endocrine disruptor vinclozolin on the prostate transcriptome and adult onset disease. Prostate. 2008;68:517–529. doi: 10.1002/pros.20724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006a;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Memon MA, Uzumcu M, Skinner MK. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J. Androl. 2006b;27:868–879. doi: 10.2164/jandrol.106.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic programming of the embryonic testis transcriptome. Genomics. 2008;91:30–40. doi: 10.1016/j.ygeno.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet. 2011;27:116–125. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock D. Aristotle Metaphysics: Books Z and H. New York, NY: Oxford University Press; 2003. [Google Scholar]
- Brisson D. The directed mutation controversy in an evolutionary context. Crit. Rev. Microbiol. 2003;29:25–35. doi: 10.1080/713610403. [DOI] [PubMed] [Google Scholar]
- Bruner-Tran KL, Osteen KG. Developmental exposure to TCDD reduces fertility and negatively affects pregnancy outcomes across multiple generations. Reprod. Toxicol. 2010;31:344–350. doi: 10.1016/j.reprotox.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, Riggs AD. Maintenance and regulation of DNA methylation patterns in mammals. Biochem. Cell Biol. 2005;83:438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- Cheng RY, Hockman T, Crawford E, Anderson LM, Shiao YH. Epigenetic and gene expression changes related to transgenerational carcinogenesis. Mol. Carcinog. 2004;40:1–11. doi: 10.1002/mc.20022. [DOI] [PubMed] [Google Scholar]
- Choong MK, Tsafnat G. Genetic and epigenetic biomarkers of colorectal cancer. Clin. Gastroenterol. Hepatol. 2011 doi: 10.1016/j.cgh.2011.04.020. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Clement TM, Anway MD, Uzumcu M, Skinner MK. Regulation of the gonadal transcriptome during sex determination and testis morphogenesis: comparative candidate genes. Reproduction. 2007;134:455–472. doi: 10.1530/REP-06-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement TM, Savenkova MI, Settles M, Anway MD, Skinner MK. Alterations in the developing testis transcriptome following embryonic vinclozolin exposure. Reprod. Toxicol. 2010;30:353–364. doi: 10.1016/j.reprotox.2010.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A, Cardenas ML, Letelier JC, Soto-Andrade J, Abarzua FG. Understanding the parts in terms of the whole. Biol. Cell. 2004;96:713–717. doi: 10.1016/j.biolcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Craig JM. Heterochromatin – many flavours, common themes. Bioessays. 2005;27:17–28. doi: 10.1002/bies.20145. [DOI] [PubMed] [Google Scholar]
- Crews D, Gore AC, Hsu TS, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. Transgenerational epigenetic imprints on mate preference. Proc. Natl. Acad. Sci. USA. 2007;104:5942–5946. doi: 10.1073/pnas.0610410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage B, Dashwood RH. Dietary manipulation of histone structure and function. Annu. Rev. Nutr. 2008;28:347–366. doi: 10.1146/annurev.nutr.28.061807.155354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipple KM, Phelan JK, McCabe ER. Consequences of complexity within biological networks: robustness and health, or vulnerability and disease. Mol. Genet. Metab. 2001;74:45–50. doi: 10.1006/mgme.2001.3227. [DOI] [PubMed] [Google Scholar]
- Dunn GA, Bale TL. Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology. 2011;152:2228–2236. doi: 10.1210/en.2010-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF. Mechanisms for the environmental regulation of gene expression: ecological aspects of animal development. J. Biosci. 2005;30:65–74. doi: 10.1007/BF02705151. [DOI] [PubMed] [Google Scholar]
- Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD, Hanson MA. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr. Res. 2007;61:5R–10R. doi: 10.1203/pdr.0b013e318045bedb. [DOI] [PubMed] [Google Scholar]
- Goldschmidt R. Some aspects of evolution. Science. 1933;78:539–547. doi: 10.1126/science.78.2033.539. [DOI] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Valladares L. Endocrine disruptors, epigenetically induced changes, and transgenerational transmission of characters and epigenetic states. In: Gore EAC, editor. Endocrine-Disrupting Chemicals: From Basic Research to Clinical Practice. Totowa, NJ: Humana Press, Inc.; 2007. pp. 175–189. 2007. [Google Scholar]
- Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One. 2010;5:e13100. doi: 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna CM, Skinner MK. Epigenetic transgenerational effects of endocrine disruptors on male reproduction. Semin. Reprod. Med. 2009;27:403–408. doi: 10.1055/s-0029-1237428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Sabat P, Valladares L. Environmental signaling and evolutionary change: can exposure of pregnant mammals to environmental estrogens lead to epigenetically induced evolutionary changes in embryos? Evol. Dev. 2005;7:341–350. doi: 10.1111/j.1525-142X.2005.05033.x. [DOI] [PubMed] [Google Scholar]
- Guerrero-Bosagna CM, Sabat P, Valdovinos FS, Valladares LE, Clark SJ. Epigenetic and phenotypic changes result from a continuous pre and post natal dietary exposure to phytoestrogens in an experimental population of mice. BMC Physiol. 2008;8:17. doi: 10.1186/1472-6793-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakre S, Chavez L, Shirakawa K, Verdin E. Epigenetic regulation of HIV latency. Curr. Opin. HIV AIDS. 2011;6:19–24. doi: 10.1097/COH.0b013e3283412384. [DOI] [PubMed] [Google Scholar]
- Hanson MA, Gluckman PD. Developmental origins of health and disease: new insights. Basic Clin. Pharmacol. Toxicol. 2008;102:90–93. doi: 10.1111/j.1742-7843.2007.00186.x. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howdeshell KL, Hotchkiss AK, Thayer KA, Vandenbergh JG, vom Saal FS. Exposure to bisphenol A advances puberty. Nature. 1999;401:763–764. doi: 10.1038/44517. [DOI] [PubMed] [Google Scholar]
- Ideker T. Systems biology 101 – what you need to know. Nat. Biotechnol. 2004;22:473–475. doi: 10.1038/nbt0404-473. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Matzke M, Thieffry D, Van Speybroeck L. The genome in context: biologists and philosophers on epigenetics. Bioessays. 2002;24:392–394. doi: 10.1002/bies.10071. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce AR, Palsson BO. The model organism as a system: integrating ‘omics’ data sets. Nat. Rev. Mol. Cell Biol. 2006;7:198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- Kaati G, Bygren LO, Pembrey M, Sjostrom M. Transgenerational response to nutrition, early life circumstances and longevity. Eur. J. Hum. Genet. 2007;15:784–790. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]
- Kaminsky Z, Tochigi M, Jia P, Pal M, Mill J, Kwan A, Ioshikhes I, Vincent JB, Kennedy JL, Strauss J, Pai S, Wang SC, Petronis A. A multi-tissue analysis identifies HLA complex group 9 gene methylation differences in bipolar disorder. Mol. Psychiatry. 2010 doi: 10.1038/mp.2011.64. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- Kujjo LL, Chang EA, Pereira RJ, Dhar S, Marrero-Rosado B, Sengupta S, Wang H, Cibelli JB, Perez GI. Chemotherapy-induced late transgenerational effects in mice. PLoS One. 2011;6:e17877. doi: 10.1371/journal.pone.0017877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose JS, Miller RL. Environmental epigenetics and allergic diseases: recent advances. Clin. Exp. Allergy. 2010;40:1602–1610. doi: 10.1111/j.1365-2222.2010.03599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Mertineit C, Taketo T, Moens PB, Bestor TH, Trasler JM. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev. Biol. 2004;268:403–415. doi: 10.1016/j.ydbio.2003.12.031. [DOI] [PubMed] [Google Scholar]
- Lee J, Kanatsu-Shinohara M, Ogonuki N, Miki H, Inoue K, Morimoto T, Morimoto H, Ogura A, Shinohara T. Heritable imprinting defect caused by epigenetic abnormalities in mouse spermatogonial stem cells. Biol. Reprod. 2009;80:518–527. doi: 10.1095/biolreprod.108.072330. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Mittler JE. The directed mutation controversy and neo-Darwinism. Science. 1993;259:188–194. doi: 10.1126/science.7678468. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Martinez-Zamudio R, Ha HC. Environmental epigenetics in metal exposure. Epigenetics. 2011;6:820–827. doi: 10.4161/epi.6.7.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino D, Prescott S. Epigenetics and prenatal influences on asthma and allergic airways disease. Chest. 2011;139:640–647. doi: 10.1378/chest.10-1800. [DOI] [PubMed] [Google Scholar]
- Matthews SG, Phillips DI. Minireview: transgenerational inheritance of the stress response: a new frontier in stress research. Endocrinology. 2010;151:7–13. doi: 10.1210/en.2009-0916. [DOI] [PubMed] [Google Scholar]
- Moore KL, Persaud TNN. Before we are born: essentials of embryology and birth defects. Philadelphia, PA: W.B. Saunders; 1993. [Google Scholar]
- Nadeau JH. Transgenerational genetic effects on phenotypic variation and disease risk. Hum. Mol. Genet. 2009;18:R202–R210. doi: 10.1093/hmg/ddp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan HM, Song YJ, Yun HY, Park JS, Kim H. Effects of dietary intake and genetic factors on hypermethylation of the hMLH1 gene promoter in gastric cancer. World J. Gastroenterol. 2005;11:3834–3841. doi: 10.3748/wjg.v11.i25.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold RR, Padilla-Banks E, Jefferson WN. Adverse effects of the model environmental estrogen diethylstilbestrol are transmitted to subsequent generations. Endocrinology. 2006;147:S11–S17. doi: 10.1210/en.2005-1164. [DOI] [PubMed] [Google Scholar]
- Newbold RR, Padilla-Banks E, Jefferson WN, Heindel JJ. Effects of endocrine disruptors on obesity. Int. J. Androl. 2008;31:201–208. doi: 10.1111/j.1365-2605.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- Nijhout FH, Wray GA, Kremen C, Teragawa K. Ontogeny, phylogeny and evolution of form: an algorithmic approach. Syst. Zool. 1986;35:445–457. [Google Scholar]
- Oster G, Alberch P. Evolution and bifurcation of developmental programs. Evolution. 1982;36:444–459. doi: 10.1111/j.1558-5646.1982.tb05066.x. [DOI] [PubMed] [Google Scholar]
- Peck AI. Aristotle: The Generation of Animals. Cambridge, MA: Cambridge Harvard University Press; 1943. [Google Scholar]
- Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J. Sex-specific, male-line transgenerational responses in humans. Eur. J. Hum. Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- Pentinat T, Ramon-Krauel M, Cebria J, Diaz R, Jimenez-Chillaron JC. Transgenerational inheritance of glucose intolerance in a mouse model of neonatal overnutrition. Endocrinology. 2010;151:5617–5623. doi: 10.1210/en.2010-0684. [DOI] [PubMed] [Google Scholar]
- Perry AS, Watson RW, Lawler M, Hollywood D. The epigenome as a therapeutic target in prostate cancer. Nat. Rev. Urol. 2010;7:668–680. doi: 10.1038/nrurol.2010.185. [DOI] [PubMed] [Google Scholar]
- Pimentel D, Cooperstein S, Randell H, Filiberto D, Sorrentino S, Kayes B, Nicklin C, Yagi J, Brian J, O’hern J, Habas A, Weinstein C. Ecology of increasing diseases: population growth and environmental degradation. Hum. Ecol. 2007;35:653–668. doi: 10.1007/s10745-007-9128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, Ross SA, Wise C, Pogribna M, Jones EA, Tryndyak VP, James SJ, Dragan YP, Poirier LA. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat. Res. 2006;593:80–87. doi: 10.1016/j.mrfmmm.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Salian S, Doshi T, Vanage G. Impairment in protein expression profile of testicular steroid receptor coregulators in male rat offspring perinatally exposed to Bisphenol A. Life Sci. 2009;85:11–18. doi: 10.1016/j.lfs.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Singal R, Ginder GD. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- Skinner MK. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod. Toxicol. 2008;25:2–6. doi: 10.1016/j.reprotox.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res. C Embryo Today. 2011a;93:51–55. doi: 10.1002/bdrc.20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Guerrero-Bosagna C. Environmental signals and transgenerational epigenetics. Epigenomics. 2009;1:111–117. doi: 10.2217/epi.09.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics. 2011;6:838–842. doi: 10.4161/epi.6.7.16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One. 2008;3:e3745. doi: 10.1371/journal.pone.0003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol. Metab. 2010;21:214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto AM, Rubin BS, Sonnenschein C. Interpreting endocrine disruption from an integrative biology perspective. Mol. Cell Endocrinol. 2009;304:3–7. doi: 10.1016/j.mce.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2010;139:373–379. doi: 10.1530/REP-09-0340. [DOI] [PubMed] [Google Scholar]
- Sved J, Bird A. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. USA. 1990;87:4692–4696. doi: 10.1073/pnas.87.12.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G. The proper place of hopeful monsters in evolutionary biology. Theory Biosci. 2006;124:349–369. doi: 10.1016/j.thbio.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Uzumcu M, Suzuki H, Skinner MK. Effect of the anti-androgenic endocrine disruptor vinclozolin on embryonic testis cord formation and postnatal testis development and function. Reprod. Toxicol. 2004;18:765–774. doi: 10.1016/j.reprotox.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Vahter ME. Interactions between arsenic-induced toxicity and nutrition in early life. J. Nutr. 2007;137:2798–2804. doi: 10.1093/jn/137.12.2798. [DOI] [PubMed] [Google Scholar]
- Vyssotski D. Transgenerational epigenetic compensation. Evolocus. 2011;1:1–6. [Google Scholar]
- Waalkes MP, Liu J, Chen H, Xie Y, Achanzar WE, Zhou YS, Cheng ML, Diwan BA. Estrogen signaling in livers of male mice with hepatocellular carcinoma induced by exposure to arsenic in utero. J. Natl. Cancer Inst. 2004;96:466–474. doi: 10.1093/jnci/djh070. [DOI] [PubMed] [Google Scholar]
- Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183:1654–1655. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Wake DB, Larson A. Multidimensional analysis of an evolving lineage. Science. 1987;238:42–48. doi: 10.1126/science.238.4823.42. [DOI] [PubMed] [Google Scholar]
- Walker DM, Gore AC. Transgenerational neuroendocrine disruption of reproduction. Nat. Rev. Endocrinol. 2011;7:197–207. doi: 10.1038/nrendo.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Bioenergetics and the epigenome: interface between the environment and genes in common diseases. Dev. Disabil. Res. Rev. 2010;16:114–119. doi: 10.1002/ddrr.113. [DOI] [PubMed] [Google Scholar]
- Wallace JA, Orr-Weaver TL. Replication of heterochromatin: insights into mechanisms of epigenetic inheritance. Chromosoma. 2005;114:389–402. doi: 10.1007/s00412-005-0024-6. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Travisano M, Tahiliani KG, Rached MT, Mirza S. Methyl donor supplementation prevents transgenerational amplification of obesity. Int. J. Obes. (Lond) 2008;32:1373–1379. doi: 10.1038/ijo.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P. Principles of Development. New York, NY: Holt; 1939. [Google Scholar]
- Weissman A. The Germ Plasm: A Theory of Heredity. New York, NY: Scribners; 1892. [Google Scholar]
- Wong C, Kelce WR, Sar M, Wilson EM. Androgen receptor antagonist versus agonist activities of the fungicide vinclozolin relative to hydroxyflutamide. J. Biol. Chem. 1995;270:19998–20003. doi: 10.1074/jbc.270.34.19998. [DOI] [PubMed] [Google Scholar]
- Xing Y, Shi S, Le L, Lee CA, Silver-Morse L, Li WX. Evidence for transgenerational transmission of epigenetic tumor susceptibility in Drosophila. PLoS Genet. 2007;3:1598–1606. doi: 10.1371/journal.pgen.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Loktionov A, Tomatis L. Perinatal and multigenerational effect of carcinogens: possible contribution to determination of cancer susceptibility. Environ. Health Perspect. 1992;98:39–43. doi: 10.1289/ehp.929839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- Yokochi T, Robertson KD. Preferential methylation of unmethylated DNA by mammalian de novo DNA methyltransferase Dnmt3a. J. Biol. Chem. 2002;277:11735–11745. doi: 10.1074/jbc.M106590200. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Ferguson LR. Nutrigenetics and prostate cancer-2011 and beyond. J. Nutrigenet. Nutrigenomics. 2011;4:121–136. doi: 10.1159/000327902. [DOI] [PubMed] [Google Scholar]