Abstract
The lens epithelium derived growth factor p75 (LEDGF/p75) is a transcription co-activator that promotes resistance to oxidative stress- and chemotherapy-induced cell death. LEDGF/p75 is also known as the dense fine speckles autoantigen of 70 kD (DFS70), and has been implicated in cancer, HIV-AIDS, autoimmunity, and inflammation. To gain insights into mechanisms by which LEDGF/p75 protects cancer cells against stress, we initiated an analysis of its interactions with other transcription factors and the influence of these interactions on stress gene activation. We report here that both LEDGF/p75 and its short splice variant LEDGF/p52 interact with MeCP2, a methylation-associated transcriptional modulator, in vitro and in various human cancer cells. These interactions were established by several complementary approaches: transcription factor protein arrays, pull down and AlphaScreen® assays, co-immunoprecipitation, and nuclear co-localization by confocal microscopy. MeCP2 was found to interact with the N-terminal region shared by LEDGF/p75 and p52, particularly with the PWWP-CR1 domain. Like LEDGF/p75, MeCP2 bound to and transactivated the Hsp27 promoter (Hsp27pr). LEDGF/p75 modestly enhanced MeCP2-induced Hsp27pr transactivation in U2OS cells, while this effect was more pronounced in PC3 cells. LEDGF/p52 repressed Hsp27pr activity in U2OS cells. Interestingly, siRNA-induced silencing of LEDGF/p75 in U2OS cells dramatically elevated MeCP2-mediated Hsp27pr transactivation, whereas this effect was less pronounced in PC3 cells depleted of LEDGF/p75. These results suggest that the LEDGF/p75-MeCP2 interaction differentially influences Hsp27pr activation depending on the cellular and molecular context. These findings are of significance in understanding the contribution of this interaction to the activation of stress survival genes.
Keywords: LEDGF/p75, MeCP2, protein-protein interactions, PWWP domain, transcription
Introduction
LEDGF/p75 is a stress response protein with increasing relevance to cancer, HIV-AIDS, autoimmunity, inflammation, and eye disease. LEDGF/p75 and its short splice variant p52 are derived from the same PSIP1 gene and were originally identified as transcription coactivators that interact with the RNA polymerase II transcription complex (1). LEDGF/p75 is also known as the dense fine speckled autoantigen of 70 kD (DFS70), which is targeted by autoantibodies in various human inflammatory conditions (2). Although initially proposed to be a growth factor for lens epithelial cells (3), subsequent studies revealed that LEDGF/p75 is a stress survival protein that protects against oxidative stress-induced cellular damage and death (4,5). As a key cellular co-factor for HIV-1 replication, LEDGF/p75 binds the HIV-1 integrase (IN) through its C-terminal integrase-binding domain (IBD) and tethers it to the chromatin, facilitating lentiviral integration to transcriptionally active regions of the host genome (6–8).
LEDGF/p75 is emerging as an oncoprotein in various human cancers. It is targeted by autoantibodies in patients with prostate cancer (PCa), and is overexpressed in prostate tumors and other human malignancies, including chemotherapy-resistant human acute myelogenic leukemia (9–12). In addition, it can be found as a fusion protein with NUP58 in leukemia patients (13). Its overexpression in tumor cells attenuates lysosomal cell death induced by stressors that trigger oxidative stress (e.g. certain chemotherapeutic drugs, tumor necrosis factor, and serum starvation) (5,10,12,14). LEDGF/p75 also enhanced the tumorigenic potential of HeLa cells in xenograft models (10).
LEDGF/p75 is thought to promote cell survival under stress by transcriptionally activating genes encoding protective proteins such as heat shock protein 27 (Hsp27), αB-crystallin, peroxiredoxin 6 (PRDX6), and vascular endothelial growth factor c (VEGF-c) (15–17). Presumably, LEDGF/p75 transactivates these stress genes by binding to heat shock elements (HSE) and stress elements (STRE) in their promoter regions (15–17). In leukemia cells, LEDGF/p75 interacts with the menin/MLL-HMT (mixed lineage leukemia histone methyltransferase) transcription complex to transactivate cancer-associated genes and facilitate leukemic transformation (18).
The transcriptional and pro-survival activities of LEDGF/p75 are attenuated by TGF-β1 (19), Bcl-2 (20), SUMOylation (21), and caspase-mediated cleavage (5). The pro-survival function of LEDGF/p75 is also negatively regulated by alternative splicing since its short splice variant p52 antagonizes its transcriptional activity and induces apoptosis in cancer cells (22). Although it is not clear how LEDGF/p75 is upregulated in the presence of stress, recent studies implicated transcription factors SP1 and STAT3 in this process (23,24).
LEDGF/p75 and p52 share amino (N)-terminal amino acids (aa 1–325); however, p52 has an intron-derived carboxyl (C)-terminal tail (CTT, aa 326–333) implicated in its pro-apoptotic activity (1,22). The N-terminal region shared by both proteins contains a PWWP domain (aa 1–93), a structural entity implicated in chromatin binding, HIV-integration, protein-protein interactions in the chromatin, transcriptional repression, and DNA methylation (25–30). The N-terminal region also has a positively charged domain (CR1) immediately after the PWWP domain that is followed by a nuclear localization signal (NLS) and two AT-hook (ATH) sequences that cooperate with the PWWP domain for chromatin binding (31,32). A second charged region (CR2), also designated the supercoiled-DNA recognition domain (SRD) (aa 200–336), facilitates LEDGF/p75 binding to active transcription sites (33). The C-terminus of LEDGF/p75 (aa 347–429) encompasses both the IBD and the autoepitope recognized by human anti-LEDGF/p75 autoantibodies (8,34). This region is involved in protein-protein interactions and binding to HSE in promoter regions (35–38). Both the N- and C-terminal regions of LEDGF/p75 contribute to its transcription and stress survival functions (5,38).
Understanding the mechanisms by which LEDGF/p75 promotes tumor cell resistance to cell death and chemotherapy requires detailed knowledge of its cellular functions, particularly its interactions with other cancer-associated proteins and its target genes. To date only a few cellular interacting partners of LEDGF/p75 have been identified. These are the PC4 transcription factor, menin/MLL, the Cdc7 activator of S-phase kinase (ASK), the pogZ transposase, and the myc-interacting protein JPO2 (18,35–37). Using transcription factor protein arrays we identified several candidate interacting partners of LEDGF/p75. Among these, methyl-CpG binding protein 2 (MeCP2) was of particular interest because, like LEDGF/p75, it has been linked to PCa (39). MeCP2 belongs to the family of methyl-CpG binding proteins, and is mutated in Rett syndrome, a childhood neurodevelopmental disease (40). MeCP2 is a transcription factor that is generally considered to bind methylated CpG islands in order to repress transcription. However, recent evidence indicates that MeCP2 binds DNA regardless of methylation status and acts as a transcriptional modulator that remodels chromatin to activate or repress genes depending on the cellular and molecular context (40–42). In this study we characterized the interaction between LEDGF/p75 and MeCP2 in vitro and in cancer cells. We provide evidence that the N-terminal PWWP-CR1 region of LEDGF/p75 binds to MeCP2 and influences its transcriptional function.
Materials and Methods
Cell culture, antibodies and plasmids
U2OS, PC3, 293T, HeLa cells were obtained from the American Type Culture Collection and cultured in McCoy’s 5A medium or RPMI 1640 (Gibco), supplemented with 2 mM L-glutamine and penicillin/streptomycin, and 10% fetal bovine serum. PC3 cells stably expressing LEDGF/p75 (14), were grown in RPMI 1640 with 10% (v/v) fetal bovine serum (FBS), 20 µg/µl of gentamicin, and 0.5 mg/ml of geneticin. Cells were grown with 5% CO2 at 37°C.
The following antibodies were used: mouse monoclonals anti LEDGF/p75-p52 (BD Biosciences), anti-β-actin (Sigma); rabbit polyclonals anti-LEDGF/p75 (Bethyl Laboratories), anti-MeCP2 (ProteinTech Group), anti-HA (Santa Cruz Biotechnology), anti-eGFP (produced in Z. Debyser’s laboratory); goat polyclonals anti-eGFP (produced in Z. Debyser’s laboratory), anti-GFP (Santa Cruz Biotechnology), anti-GST (Pharmacia Biotech), anti-Flag-HRP (Sigma) and rat monoclonal horseradish peroxidase (HRP)-conjugated anti-HA (Roche Diagnostics). Human antibodies to LEDGF/p75 were a gift from Dr. Eng M. Tan (Scripps Research Institute, La Jolla, CA).
Plasmid pET28a-dfs70 encoding His-LEDGF/p75 was a kind gift from Dr. Edward Chan (University of Florida, Gainesville). Plasmids pDEST-GST-MeCP2 and pcDNA-Flag-MeCP2 were a kind gift from Dr. Adrian Bird (University of Edinburgh, UK). Plasmids pKB6H-p52, pMal™-p2x-BRD4-Ct, and p-eGFP-BRD4-Ct were generated in Z. Debyser’s laboratory. Plasmids pCruzHA-LEDGF/p75, pCruzHA-p52, and pGL3-Hsp27pr-Luc were generated as described (22). Plasmid eGFP-p52 was cloned by replacing the LEDGF/p75 cDNA in p-eGFP-LEDGF/p75 vector with the LEDGF/p52 cDNA at XhoI and BamHI restriction sites.
Purification of recombinant LEDGF/p75, p52 and MeCP2
GST-tagged MeCP2 was produced from pDEST-MeCP2 in E. coli BL21 grown in the presence of sorbitol and betaine. Expression was induced in lysogeny broth (LB) medium at 37°C by addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells harvested 3 h after induction were lysed by sonication in core buffer (50 mM Tris HCl, 0.5 M NaCl, pH7.5, 10 mM EDTA, 10 mM EGTA, 1% Triton X-100, and 20 µg/ml lysozyme). The fusion protein, captured on glutathione agarose (Sigma) or glutathione sepharose beads (GE Healthcare Life Sciences), was eluted with 20 mM glutathione in core buffer. His-tagged LEDGF/p75 and p52 were expressed from pET28a-dfs70 and pKB6H52 in E. coli BL21, respectively. Expression was induced with 1 mM and 3 mM IPTG, respectively, at 37°C for 3 h. Bacteria were lysed by sonication in B-PER® bacterial protein extraction reagent (Thermo Scientific). The recombinant proteins were captured on nickel columns (Novagen) or TALON® His-Tag Purification Resins (Clontech), and washed with 20 mM HEPES, 0.5 M NaCl, 2 mM KCl, 1% NP-40, and 5 mM imidazole. Proteins were eluted with 20 mM HEPES, 137 mM NaCl, 2 mM KCl, 300 mM imidazole, and dialyzed using D-tube™ dialyzer (Novagen).
Transcription factor arrays
Two transcription factor protein arrays that were commercially available at the time we initiated these studies, Active Protein Array™ (Active Motif), and TranSignal Protein Arrays I–III (Panomics), were used for identifying interacting partners of LEDGF/p75 following the manufacturer’s instructions. These arrays contained 170 transcription factors and co-activators, as well as RNA polymerase II, spotted on membranes in duplicates or triplicates. Briefly, membranes were blocked with 5% milk in tris-buffered saline tween-20 (TBS-T) buffer for 1 h. Recombinant His-LEDGF/p75 was incubated overnight with the membranes and after washes with TBS-T, the membranes were probed with human anti-LEDGF/p75 autoantibody for 2 h. Following washes with TBS-T, the membranes were incubated with HRP-conjugated secondary antibodies, and protein interaction signals were detected by chemiluminescence (Amersham).
Pull down assays
GST or GST-MeCP2 proteins bound to glutathione beads were blocked in HEPES buffer (20 mM HEPES, pH 7.4, 2 mM DTT, 137 mM NaCl, 2 mM KCl, 5% glycerol) with 0.1% bovine serum albumin at 4°C for 1 h. His-LEDGF/p75 or His-p52 were then incubated with the beads at 4°C for 1 h in HEPES buffer. The beads were then collected by centrifugation at 5,000 rpm for 30 seconds at 4°C, the supernatant was discharged, and the beads were washed two times with 1 ml of HEPES buffer + 0.1% NP-40 followed by HEPES buffer + 0.5% NP-40 + 0.5 M NaCl. Bound proteins were eluted in SDS-PAGE sample buffer, separated by SDS-PAGE (10% Bis-Tris gel) and detected by immunoblotting.
Analysis of protein-protein interactions by AlphaScreen® assay
The AlphaScreen® assay was performed according to the manufacturer’s protocol (Perkin Elmer). Briefly, reactions were performed in 25 µl final volume in 384-well Optiwell™ microtiter plates. The reaction buffer contained 25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.1% Tween 20, 0.1% BSA. Varying concentrations of GST-MeCP2 in bacterial lysate and purified Flag-LEDGF/p75 were incubated in 15 µl reaction volume at 4°C for 1 h. The concentration of GST-MeCP2 in the lysates was estimated using BSA standards on SDS-PAGE gels stained with Coomassie Blue. Equal volume of bacterial lysate not expressing GST-MeCP2 was used as negative control. Subsequently, 5 µl of the diluted donor (glutathione) and acceptor (Flag) beads were added. After incubation for 1 h in the dark, light emission was measured in the EnVision® reader (Perkin Elmer) and analyzed using the EnVision® manager software.
Transient and Stable Transfection
293T cells were transfected in 10 cm3 plates by polyethyleneimine (PEI) transfection. Ten µg of plasmid DNA were used to transiently transfect cells at >50% confluency. U2OS and PC3 cells were transfected using TransIt® 2020 (Mirus) transfection reagent. Transfected cells were grown for 24–48 h before analysis. Stable PC3 clones overexpressing LEDGF/p75 were generated by transfecting cells with pcDNA-LEDGF/p75, or empty pcDNA vector for controls, and growing them in selection media containing geneticin (Calbiochem) as described (14).
Co-immunoprecipitation
Cells were collected 48 h post transfection and lysed in RIPA buffer (Santa Cruz Biotechnology). Antibodies were incubated with cell lysates for 1 h before protein A/G+ agarose beads (Santa Cruz Biotechnology) were added. The beads were collected by centrifugation and washed 3 times with core buffer (50 mM Tris pH 7.5, 300 mM NaCl, 1 mM MgCl2, 5% glycerol, complete protease inhibitor EDTA free (Roche) and 0.1% NP-40. 293T cells were transiently transfected with p-eGFP-LEDGF/p75 and pcDNA-Flag-MeCP2. Whole cell lysates were collected 24 h post transfection and lysed in core buffer with 1% Triton X-100. Antibody against GFP was incubated with cell lysate overnight before protein G sepharose beads (GE Healthcare) were added. Precipitation of Flag-MeCP2 was done using Flag-agarose beads. Immunoprecipitated proteins were detected by immunoblotting using appropriate antibodies.
Confocal microscopy
Cells were co-transfected with different combinations of plasmids encoding eGFP-tagged LEDGF/p75, HcRed-tagged p75 or p52, or Flag-tagged MeCP2. Cells were then fixed with 4% formaldehyde and permeabilized with 0.5% Triton X-100. Ectopically expressed Flag-MeCP2 was visualized by incubation with anti-Flag antibodies, followed by incubation with corresponding secondary antibodies labeled with either FITC or rhodamine. For visualization of endogenous LEDGF/p75 and MeCP2 proteins, fixed/permeabilized cells were co-incubated with human anti-LEDGF/p75 and rabbit anti-MeCP2 antibodies, both used at 1:200 dilution. After incubation with respective secondary antibodies labeled with FITC or rhodamine, cells were mounted with medium containing 4’,6-diamidino-2-phenylindole (DAPI) (Vectashield). Confocal microscopy was performed in a Zeiss LSM 710 NLO microscope with 63× oil immersion objective and appropriate filters. Images were analyzed using ImageJ software.
Luciferase-based Transcription Reporter Assays
Hsp27pr luciferase transcription reporter assays were performed as described previously (22). Briefly, cells were co-transfected with plasmids encoding the proteins of interest or empty vector, and pGL3-Hsp27pr. At 48 h post-transfection, cells were lysed and luciferase assays were performed using the Luciferase Assay System (Promega). Relative light units were obtained in a MicroLumatPlus Lb 96V luminometer (Berthold Tech) and luciferase values were normalized to protein concentration of lysates from cells co-transfected with the empty vectors and pGL3-Hsp27pr. Student’s’ t test analysis was performed using Microsoft Excel. Experiments were repeated at least three times.
LEDGF/p75 knockdown by RNA interference
Transient knockdown of LEDGF/p75 was carried out using synthetic siRNA oligos as described previously (43). The siLEDGF/p75 sequence corresponded to nucleotides 1340–1360 (5′- AGACAGCAUGAGGAAGCGAdTdT-3′) with respect to the first nucleotide of the start codon of the LEDGF/p75 open reading frame. Ambion Silencer® Negative Control siRNA #1 was used as scrambled control. siRNAs were transfected into U2OS cells using siQuest (Mirus). LEDGF/p75 knockdown was verified by immunoblotting.
Chromatin Immunoprecipitation Assays
U2OS cells were fixed in 1% formaldehyde for 10 minutes and subjected to chromatin immunoprecipitation (ChIP) assay using ChIP-IT Express Enzymatic kit (Active Motif, Carlsbad). Anti-LEDGF/p75 antibodies (A300-848A, Bethyl), anti-MeCP2 antibodies (07-013, Millipore) and rabbit IgG (Santa Cruz Biotechnology) were used to immunoprecipitate protein-chromatin complexes. PCR was performed using primers to amplify Hsp27 promoter: set A forward 5'- CGC TTA AGC ACC AGG GCC GG -3 and reverse 5'- CCG GCC CTG GTG CTT AAG CG -3’; set B forward 5'- CTGGGCTCAAGCACCAGACTC -3' and reverse 5'- CAAATGAATTCGAGAGCGCGACGC-3'; set C: forward 5’-CAGGGTTTTGCTCTGTAG CC-3’ and reverse 5’-CCACACGCGTGTGAGATAGAATGTG-3’; set D: forward 5’-CTCTGCCTTCTGGGGTTCAAG-3’ and reverse 5’-TTGAACCCCGGTGAGTAGAG-3’.
Results
Identification of MeCP2 as a candidate interacting partner of LEDGF/p75
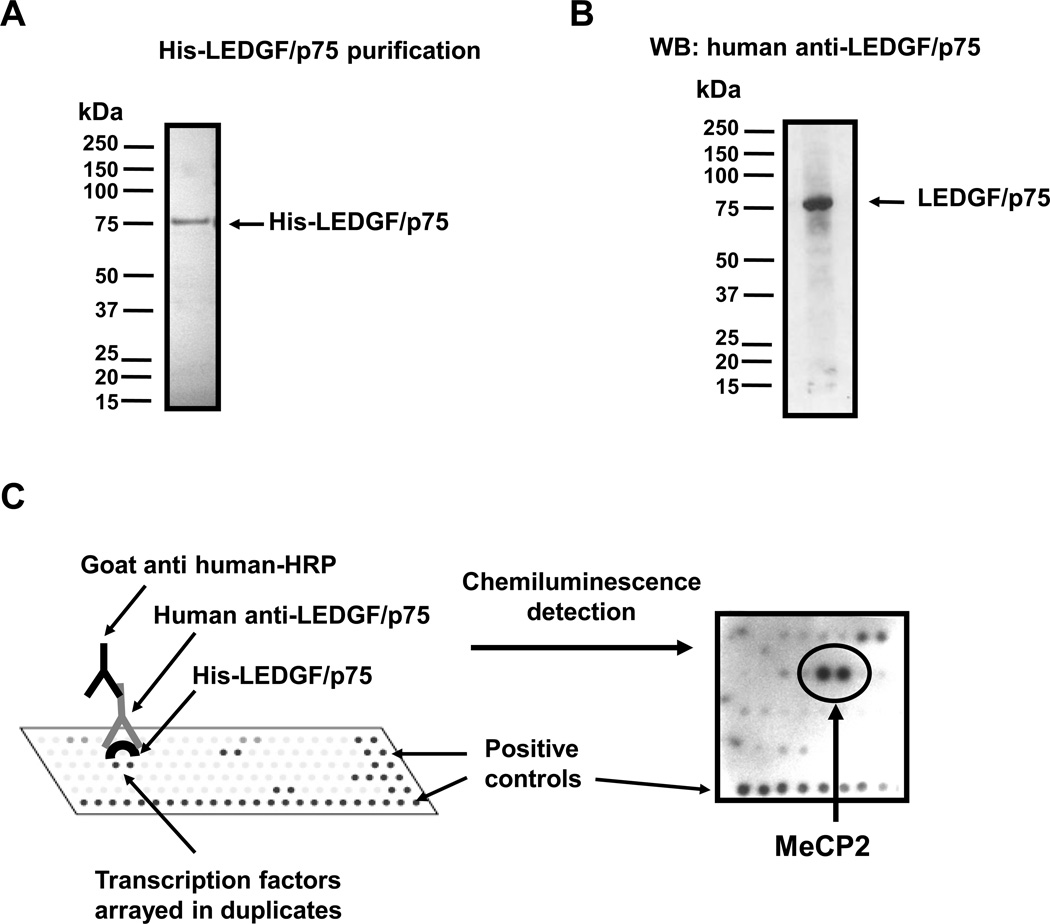
We hypothesized that LEDGF/p75 cooperates with transcription factors to facilitate the activation of stress genes. In order to identify novel cellular interacting partners of LEDGF/p75, we screened 170 different transcription factors for LEDGF/p75 binding using transcription factor protein arrays that were commercially available at the time we initiated these studies (Panomics and Active Motif). Purified recombinant His-tagged LEDGF/p75 (Fig. 1A) was incubated with recombinant transcription factors spotted on membranes, and protein-protein interactions were identified using a specific human autoimmune serum against LEDGF/p75 (Fig. 1B), followed by signal detection with chemiluminescence (Fig. 1C). We detected moderate to strong protein-protein interaction signals with 17 different transcription factors, with the strongest reactivity corresponding to the LEDGF/p75-MeCP2 interaction (Fig. 1C). One array (Active Motif) yielded strong signals with transcription factor PC4 and RNA polymerase II subunits (data not shown), consistent with the early report that LEDGF/p75 co-purifies with these proteins (1).
Figure 1. Identification of candidate interacting partners of LEDGF/p75 using transcription factor protein arrays.
(A) Coomassie blue stained SDS-PAGE gel showing purified His-LEDGF/p75. E. coli BL21 strain was transformed with pET28a-dfs70 encoding His-LEDGF/p75 and induced with IPTG. Lysate was passed through a nickel column to purify His-LEDGF/p75. (B) Immunoblot showing the specificity of the human autoantibody against LEDGF/p75 used as detection reagent in the transcription factor protein arrays. The autoantibody reacts specifically with LEDGF/p75 in a PC3 PCa cell lysate. (C) Transcription factor arrays were used to identify candidate interacting transcription factors of LEDGF/p75. Purified His-LEDGF/p75 was incubated with transcription factors spotted on membranes. Protein interactions were detected with human anti-LEDGF/p75 autoantibody and chemiluminescence. A section of the transcription factor array membrane containing MeCP2 is showed.
LEDGF/p75 interacts with MeCP2 in vitro
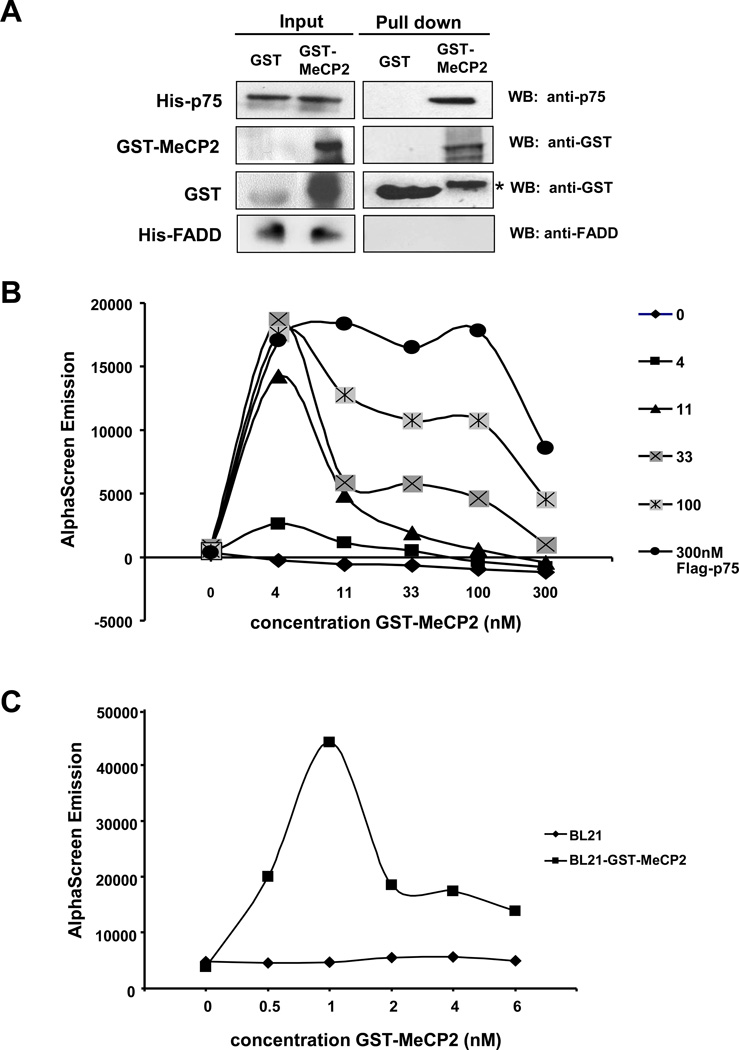
To confirm the interaction between LEDGF/p75 and MeCP2 in vitro, pull-down experiments were performed using recombinant proteins. Beads with bound GST-MeCP2 or GST were incubated with His-LEDGF/p75, and interactions were detected by immunoblotting using anti-GST antibody. His-LEDGF/p75 was pulled down with GST-MeCP2 but not with GST (Fig. 2A). A protein-protein interaction assay using the AlphaScreen® technology (Perkin-Elmer) was used for additional confirmation of the LEDGF/p75-MeCP2 interaction in vitro. To prevent false positive signals due to rapid degradation of purified recombinant GST-MeCP2, we used GST-MeCP2 induced in the E. coli BL21 bacterial lysate. Enhanced bacterial expression of GST-MeCP2 was attained in the presence of sorbitol and betaine. Cross titration of increasing concentrations of recombinant Flag-LEDGF/p75 and GST-MeCP2 demonstrated binding between both proteins over a wide range of concentrations (Fig 2B). Optimum binding was observed between 11 nM Flag-LEDGF/p75 and estimated 1 nM GST-MeCP2 (Fig 2C).
Figure 2. LEDGF/p75 interacts with MeCP2 in vitro.
(A) Pull down assays with His-LEDGF/p75 and GST-MeCP2. Recombinant His-LEDGF/p75 was incubated with GST or GST-MeCP2 bound to glutathione beads, and pulled down proteins were analyzed by immunoblotting using antibodies specific for GST or LEDGF/p75. His-FADD (Fas-associated protein with death domain), was used as negative control. Protein input was determined by immunoblotting of whole cell extracts. *Denotes degraded GST-MeCP2. (B) Cross titration for Flag-LEDGF/p75 and GST-MeCP2 interaction as measured by AlphaScreen® assay. Interaction was measured at different concentrations of Flag-LEDGF/p75 as indicated on the vertical legend, and GST-MeCP2 as indicated on the X-axis. (C) An estimated 1 nM MeCP2 was sufficient to interact with 11 nM LEDGF/p75. E.coli BL21 lysate not expressing GST-MeCP2 was used as a negative control. Results in (B) and (C) are representative of three independent measurements.
LEDGF/p75 interacts with MeCP2 in cellular models
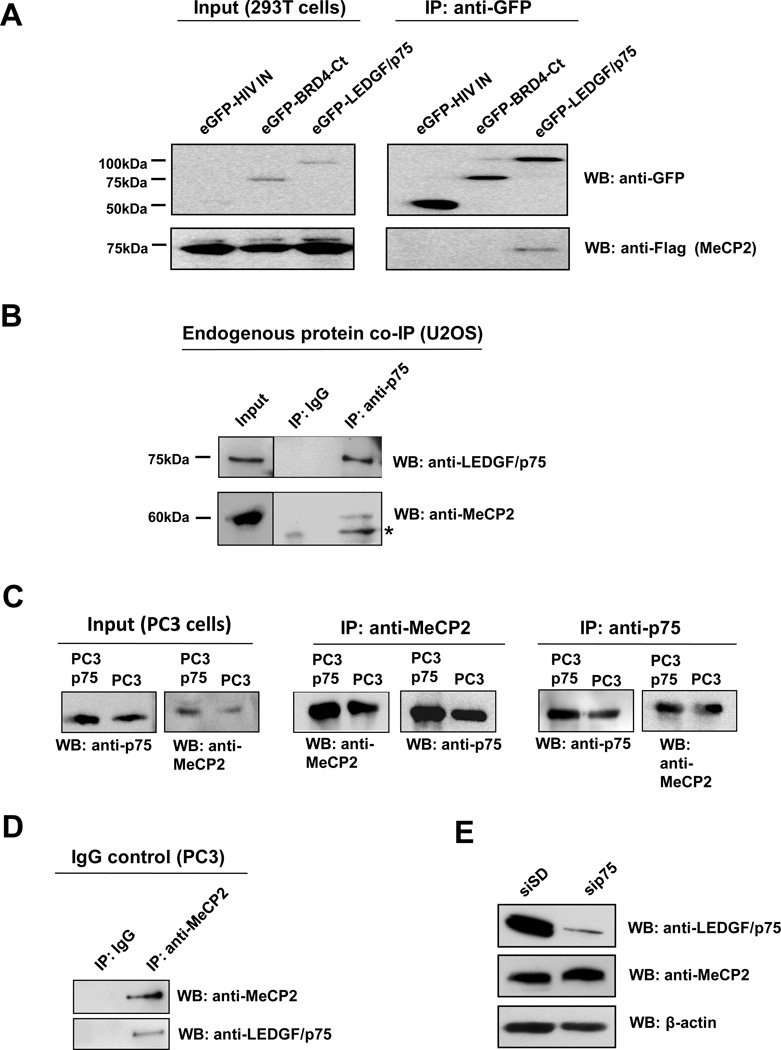
The LEDGF/p75-MeCP2 interaction was confirmed in a cellular system by co-immunoprecipitation (co-IP) assays. Plasmids encoding Flag-MeCP2 and different eGFP-tagged proteins were co-transfected in 293T cells (for high transfection efficiency). Expressed proteins were then immunoprecipitated with anti-GFP antibody and pulled down by protein G agarose beads. Immunoprecipitated proteins were analyzed by immunoblotting with antibodies against eGFP and Flag tags. Immunoblotting analysis showed MeCP2 interaction with eGFP-LEDGF/p75, but not with eGFP-HIV-IN or the irrelevant protein eGFP-BRD4-Ct (carboxyl terminal fragment of the bromodomain containing protein 4) (Fig 3A). To verify that LEDGF/p75 and MeCP2 interact endogenously in cancer cells, co-IP experiments were performed using U2OS osteosarcoma cells, which express high endogenous levels of LEDGF/p75 (22). U2OS cell lysates were incubated with antibody against LEDGF/p75, and the immunoprecipitated proteins were detected by immunoblotting. Endogenous LEDGF/p75 and MeCP2 were detected in immunoprecipitates of endogenous LEDGF/p75 but not in immunoprecipitates of an IgG control antibody (Fig 3B).
Figure 3. Co-immunoprecipitation of LEDGF/p75 and MeCP2.
(A) 293T cells co-transfected with Flag-MeCP2 and eGFP-LEDGF/p75, eGFP-HIV-IN, or eGFP-BRD4-Ct expression constructs were lysed 24 h post transfection. Proteins in cell lysates were immunoprecipitated (IP) with antibody against GFP, resolved by SDS-PAGE, and detected by immunoblotting using anti-GFP and anti-Flag antibodies. (B) Endogenous proteins in U2OS cell lysates were immunoprecipitated with mouse monoclonal anti-LEDGF/p75 antibody, as well as an irrelevant IgG as negative control. Immunoprecipitated proteins were detected by immunoblotting with rabbit anti-LEDGF/p75 and anti-MeCP2 antibodies. *Denotes degraded MeCP2. Protein input was determined by immunoblotting of whole cell extracts. (C) Proteins in lystates from PC3 cells or cells stably overexpressing LEDGF/p75 (PC3p75) were immunoprecipitated with rabbit anti-MeCP2 and mouse monoclonal anti-LEDGF/p75 antibodies. Immunoprecipitated proteins were detected by immunoblotting with rabbit anti-LEDGF/p75 and anti-MeCP2 antibodies. (D) Endogenous proteins in PC3 cell lysates were immunoprecipitated with rabbit anti-MeCP2 antibody, as well as an irrelevant IgG as negative control. Immunoprecipitated proteins were detected by immunoblotting with anti-LEDGF/p75 and anti-MeCP2 antibodies. (E) Immunoblot of total proteins from PC3 cells with (sip75) and without (siSD) siRNA-mediated knockdown, using rabbit antibodies to LEDGF/p75 and MeCP2.
The LEDGF/p75-MeCP2 interaction was also confirmed by reciprocal co-IP and immunoblotting in PC3 PCa cells (Fig. 3C). Whole cell lysates from PC3 cells stably overexpressing LEDGF/p75 (PC3p75) or from untransfected PC3 cells (PC3) were incubated with antibody against MeCP2, and the immunoprecipitated endogenous proteins were detected by immunoblotting with antibodies to MeCP2 or LEDGF/p75 (Fig. 3C, middle panel). In the reciprocal experiment, the PC3p75 and PC3 cells were incubated with antibody against LEDGF/p75, and the immunoprecipitated endogenous proteins were detected by immunoblotting with antibodies to MeCP2 or LEDGF/p75 (Fig. 3C, right panel). Endogenous LEDGF/p75 and MeCP2 were detected in immunoprecipitates of endogenous MeCP2 but not in immunoprecipitates of an IgG control antibody (Fig 3D). As an additional control, we sought to confirm that the anti-LEDGF/p75 and MeCP2 antibodies were recognizing different endogenous proteins in PC3 cells. For these experiments, LEDGF/p75 was knocked down in PC3 cells by transient transfection of siRNA oligos (sip75). Immunoblots revealed that endogenous LEDGF/p75 was dramatically reduced by sip75 but not by the control scrambled duplex oligos (siSD) (Fig. 3E). The endogenous levels of MeCP2 were not affected, indicating that the anti-LEDGF/p75 and anti-MeCP2 antibodies were recognizing different proteins.
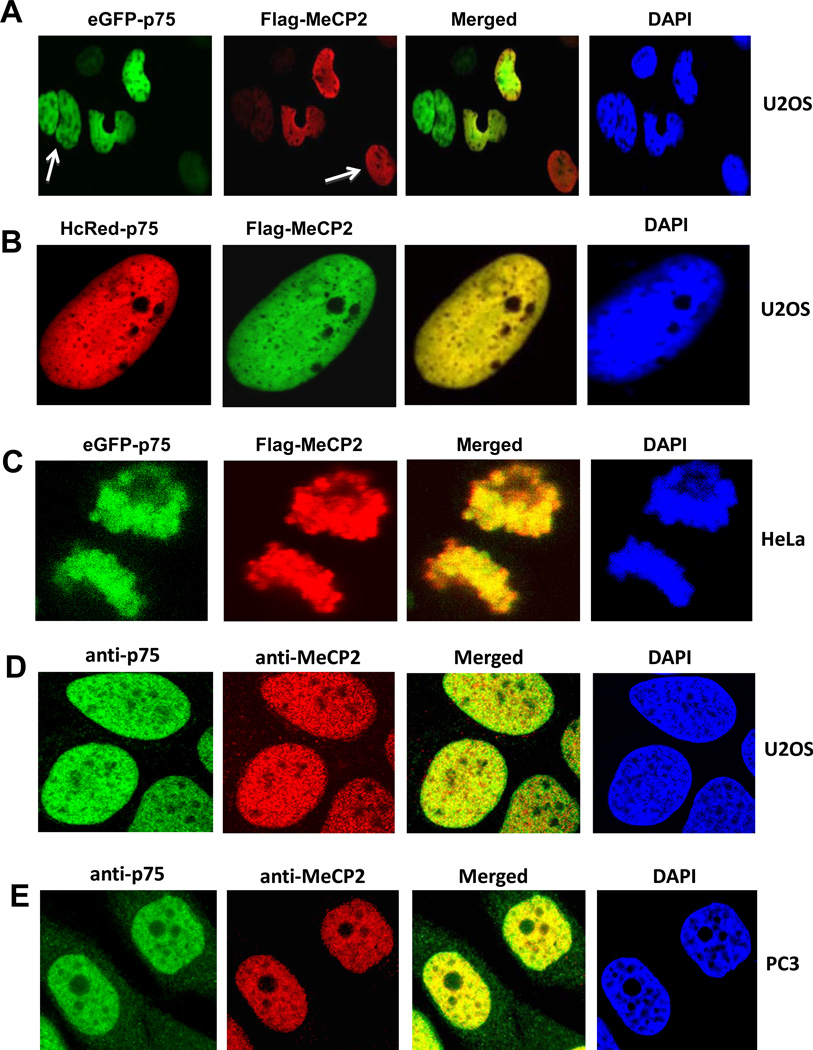
Confocal microscopy analysis was performed to examine the intracellular co-localization of LEDGF/p75 and MeCP2. U2OS and HeLa cells were transiently transfected with different combinations of tagged LEDGF/p75 and MeCP2 constructs. Fig. 4A shows that co-expression of eGFP-p75 and Flag-MeCP2 in U2OS cells gave an identical nuclear localization pattern. White arrows pointing to nuclei expressing only eGFP-p75 or only Flag-MeCP2 indicated that the observed co-localization was not due to bleeding of fluorescence from one channel into the other (Fig. 4A). The nuclear co-localization of LEDGF/p75 and MeCP2 could be better appreciated in nuclei of U2OS co-expressing HcRed-p75 and Flag-MeCP2, or in mitotic HeLa cells co-expressing eGFP-p75 and Flag-MeCP2 (Fig. 4B and 4C, respectively). The nuclear co-localization of endogenous LEDGF/p75 and MeCP2 was also established in U2OS and PC3 cells by co-incubating each cell line with both human antibody to LEDGF/p75 (green) and rabbit antibody to MeCP2 (red) (Fig. 4D,E). Both proteins displayed a distinctive nuclear speckled pattern that co-localized with DAPI-stained chromatin.
Figure 4. Nuclear co-localization of LEDGF/p75 and MeCP2 by confocal micrscopy.
U2OS cells were transiently co-transfected with plasmids encoding Flag-MeCP2 and eGFP-LEDGF/p75 (A), or the combination of HcRed-LEDGF/p75 and Flag-MeCP2 (B). HeLa cells were transiently co-transfected with plasmids encoding Flag-MeCP2 and eGFP-LEDGF/p75 (C). White arrows in panel A point to cells that were stained with eGFP-LEDGF/p75 but not with Flag-MeCP2, or viceversa, ruling out the possibility of bleeding from one channel into the other. Ectopically expressed MeCP2 was detected 48 h post-transfection using anti-Flag antibodies and visualized with FITC-labeled secondary antibody. Endogenous co-localization of LEDGF/p75 and MeCP2 was observed in U2OS and PC3 cells after co-incubation with human anti-LEDGF/p75 and rabbit anti-MeCP2 antibodies, followed by detection with corresponding secondary antibodies. Nuclei were stained with DAPI, and fluorescent signals were analyzed by confocal microscopy.
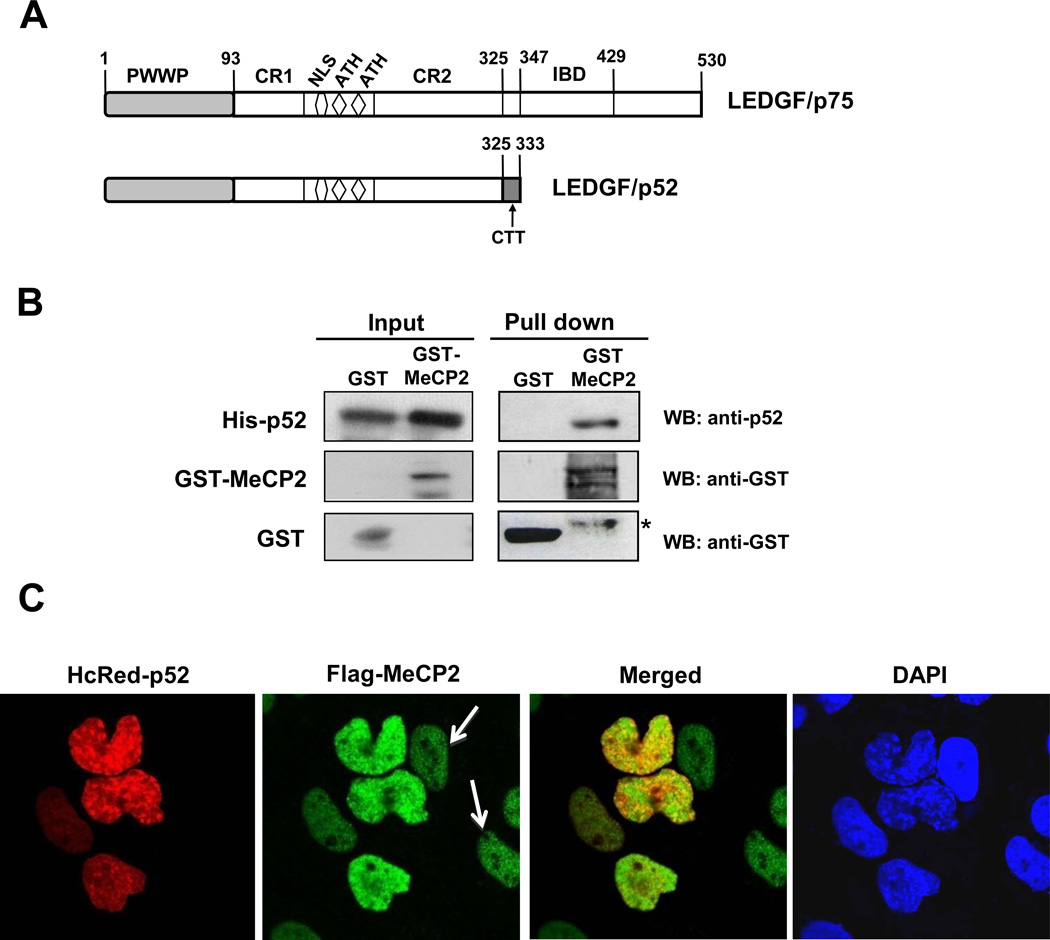
LEDGF/p52 also interacts with MeCP2
As mentioned above, LEDGF/p75 and p52 share their N-terminal region, which contains the PWWP domain, CR1 and CR2, NLS, and AT-hooks that collectively facilitate DNA binding (Fig. 5A). To determine if MeCP2 also binds to p52 we performed pull-down assays in which recombinant His-p52 was incubated with GST or GST-MeCP2 beads. Immunoblotting showed that His-p52 was pulled-down by GST-MeCP2 but not by GST (Fig 5B). Confocal microscopy showed partial co-localization of HcRed-p52 with Flag-MeCP2 in U2OS nuclei co-expressing these tagged proteins (Fig 5C). White arrows pointing to nuclei expressing only Flag-MeCP2 indicated that the observed co-localization was not due to the bleeding of fluorescence from one channel into the other (Fig. 5C). Taken together, these results indicated that p52 also interacts with MeCP2 and pointed to interaction of this protein with the N-terminal region of LEDGF/p75.
Figure 5. LEDGF/p52 interacts with MeCP2.
(A) Schematic domain structure of LEDGF/p75 and p52. (B) Pull down assay was performed as described in the legend of Fig. 2A using recombinant His-LEDGF/p52. *Denotes degraded MeCP2. Protein input was determined by immunoblotting of whole cell extracts. (C) LEDGF/p52 partially co-localizes with MeCP2 in the cell nucleus. White arrows point to cells that were stained with Flag-MeCP2 but not with HcRed-p52, ruling out the possibility of bleeding from one channel into the other. Co-localization assay was performed as described in the legend of Fig. 4.
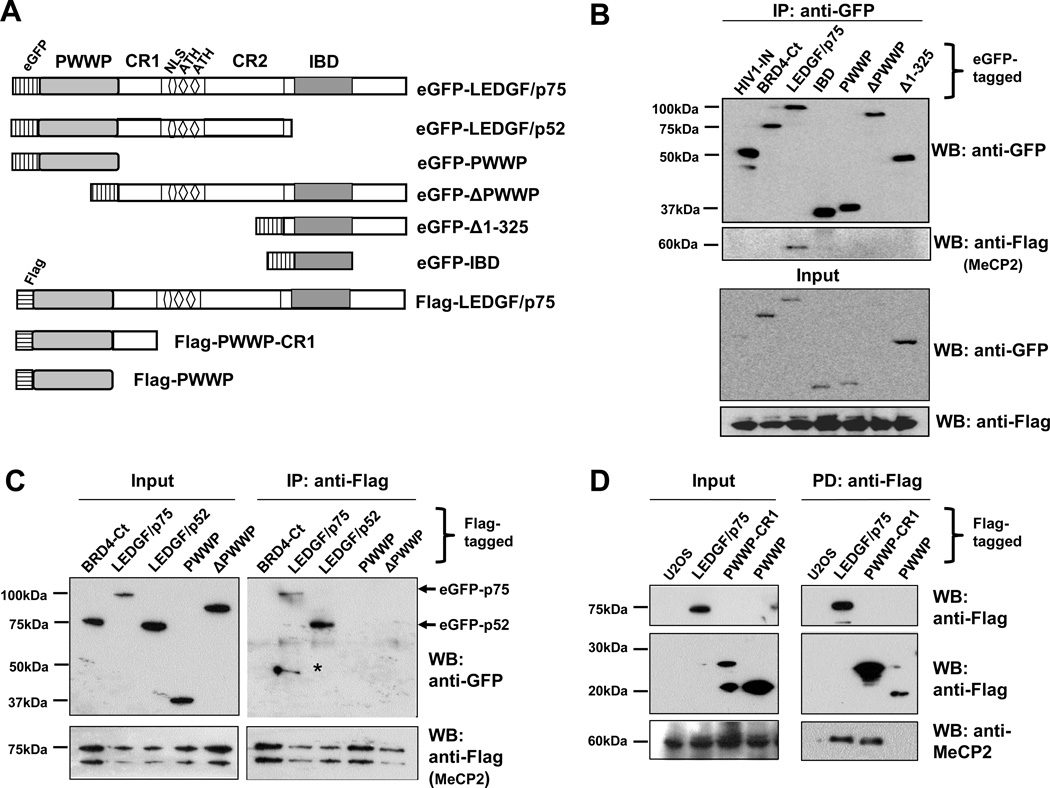
The N-terminal region of LEDGF/p75 mediates the interaction with MeCP2
In light of the fact that LEDGF/p75 and p52 share the same N-terminal region (aa 1–325), we sought to map the minimal interacting region of these proteins with MeCP2. To accomplish this, deletion constructs comprising different regions of LEDGF/p75 were used. Co-IP was performed in cells transiently co-transfected with plasmids encoding Flag-MeCP2 and one of the following eGFP-tagged constructs: LEDGF/p75 (aa 1–530), LEDGF/p52 (aa 1–333), PWWP domain (aa 1–93), ΔPWWP (aa 94–530), Δ1–325 (aa 325–530) or IBD (aa 347–429) (Fig 6A). Co-IP with anti-Flag antibody followed by detection with anti-GFP antibody showed that Flag-MeCP2 co-precipitated with both full length LEDGF/p75 and p52 but not with any of the truncated LEDGF/p75 constructs (Fig 6B,C).
Figure 6. The N-terminus of LEDGF/p75 interacts with MeCP2.
(A) Diagram of LEDGF/p75 deletion constructs used to map interaction regions. (B) Flag-MeCP2 binds to eGFP-LEDGF/p75 but not to eGFP-tagged truncated LEDGF/p75 constructs. 293T cells ectopically overexpressing the tagged proteins labeled in the blot were immunoprecipitated with GFP antibody and visualized by immunoblotting with antibodies to GFP and Flag. (C) Flag-MeCP2 binds to eGFP-tagged LEDGF/p75 and LEDGF/p52 but not to truncated constructs. Proteins ectopically overexpressed in U2OS cells were immunoprecipitated with Flag antibody and visualized with both anti-GFP and anti-Flag antibodies. *Denotes degradation product of LEDGF/p75. (D) Recombinant Flag-LEDGF/p75, Flag-PWWP-CR1 (aa 1–141) and Flag-PWWP (aa 1–101) were incubated with U2OS cell lysate. Proteins pulled down with anti-Flag affinity matrix were detected by immunoblotting. Endogenous MeCP2 in the cell lysate was pulled down by Flag-LEDGF/p75 and Flag-PWWP-CR1. Absence of recombinant proteins (U2OS only) served as negative control.
Since both LEDGF/p75 and p52 interacted with MeCP2, and the PWWP domain alone or ΔPWWP did not interact with MeCP2, we concluded that additional regions in the N-terminal portion of these proteins may be needed for MeCP2 binding. To identify these regions, deletion constructs consisting of the PWWP domain alone (aa 1–93), or in combination with its downstream CR1 region (aa 1–141), were used to examine MeCP2 binding. U2OS cell lysates containing endogenous MeCP2 were incubated with the following Flag-tagged recombinant proteins: LEDGF/p75, PWWP-CR1 (aa 1–141), or PWWP (aa 1–108) (Fig 6A). Pull-down was done using anti-Flag agarose beads. Immunoblotting analysis with anti-Flag antibody or anti-MeCP2 antibody showed that endogenous MeCP2 was pulled down with Flag-LEDGF/p75 and Flag-PWWP-CR1 but not with Flag-PWWP (Fig. 6D). This suggested that the extreme N-terminal region (aa 1–141) of LEDGF/p75 mediates its interaction with MeCP2.
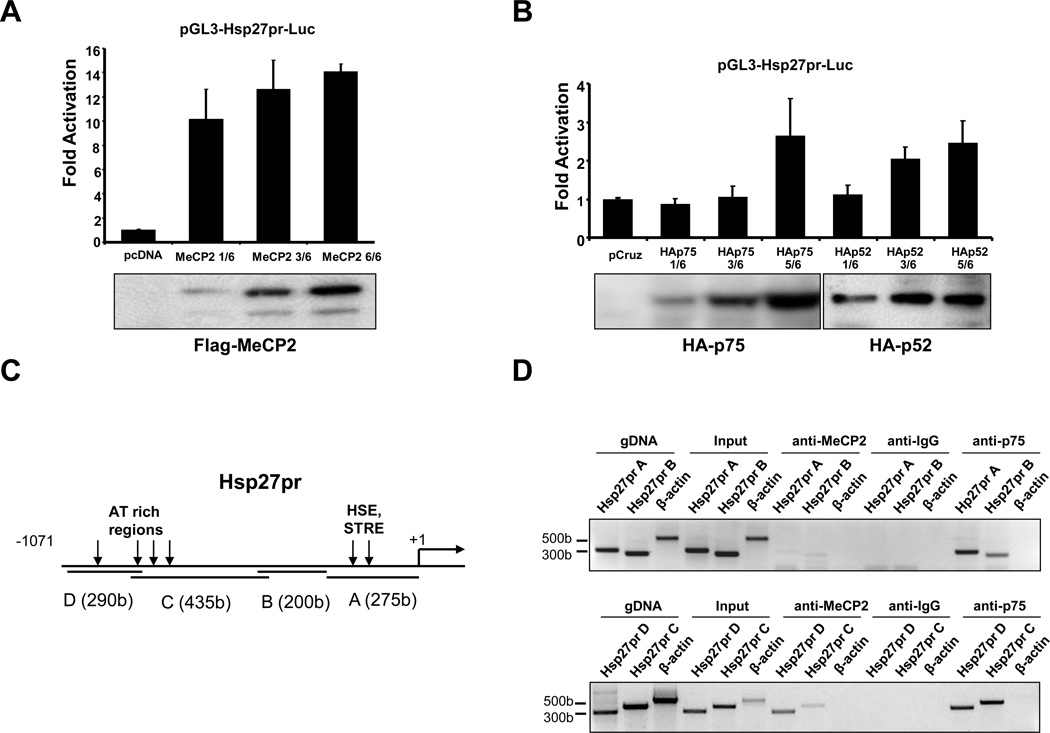
MeCP2 transactivates the Hsp27 promoter
The interaction of LEDGF/p75 and p52 with MeCP2 suggested that these proteins are part of a transcription complex that activates and regulates stress genes such as HSP27. To examine this, we first determined if MeCP2 transactivates the Hsp27 promoter (Hsp27pr) in luciferase reporter assays. U2OS cells were transiently co-transfected with pGL3-Hsp27pr-Luc and either pcDNA empty vector, pcDNA-Flag-MeCP2, pCruz empty vector, pCruz-HA-LEDGF/p75, or pCruz-HA-p52. Transient expression of Flag-MeCP2 transactivated Hsp27pr at higher levels (14 fold) (Fig 7A) than those induced by LEDGF/p75 and p52 (2–3 fold) (Fig 7B).
Figure 7. LEDGF/p75, LEDGF/p52, and MeCP2 transactivate Hsp27 promoter.
U2OS cells were co-transfected with pGL3-Hsp27pr-luc and the indicated amount of (A) empty pcDNA vector or pcDNA-Flag-MeCP2, or (B) pCruzHA, pCruzHA-LEDGF/p75, or pCruzHA-p52 (1=1.66 µg DNA). Promoter activity determined as luciferase light units/protein is expressed as fold activation compared to control activity (co-transfection of pGL3-Hsp27pr-luc with empty expression vectors). Data from each graph is representative of at least three independent experiments. Representative immunoblots corresponding to the reporter assays show protein expression. (C) Schematic diagram of Hsp27pr showing AT rich regions, and HSE and STRE sites. PCR primers targeted Hsp27pr regions A (bp −271 to +18), B (bp −480 to −220), C (bp −803 to −382), and D (bp −1071 to −781). (D) Chromatin immunoprecipitation analysis of MeCP2 and LEDGF/p75 binding to Hsp27pr. Formaldehyde fixed chromatin from U2OS cells was precipitated with non-specific IgG, or antibodies specific for MeCP2 or LEDGF/p75. PCR amplifications of immunoprecipitated DNA derived from U2OS cells were carried out with primer sets specific for Hsp27pr regions A to D. Hsp27pr primers amplified DNA fragments precipitated by LEDGF/p75 antibody or MeCP2 antibody but not by IgG. Primers that target human β-actin controlled for optimal enzymatic digestion of chromatin. Data represent the average of at least three independent experiments.
To establish if transactivation of Hsp27pr by LEDGF/p75 and MeCP2 was mediated by their binding to this promoter, chromatin immunoprecipitation (ChIP) assay was performed. Based on previous reports, LEDGF/p75 was predicted to bind HSE and STRE located in the proximal region (−185 to −111) of Hsp27pr (15), whereas MeCP2 was predicted to bind AT-rich repeats in the distal region (Fig. 6C) (44). ChIP assays using a specific MeCP2 antibody revealed binding of this protein to Hsp27pr regions C and D (bp −1071 to −382), located upstream of the HSE and STRE consensus sequences (region A, Fig. 7D). On the other hand, LEDGF/p75 bound to the entire Hsp27pr tested (bp −1071 to +18). ChIP with control rabbit anti-IgG did not produce any bands, while β-actin primers showed optimal enzymatic digestion of the chromatin. These results indicated that both LEDGF/p75 and MeCp2 bind to the Hsp27pr, with overlapping binding sites at the distal regions of the promoter.
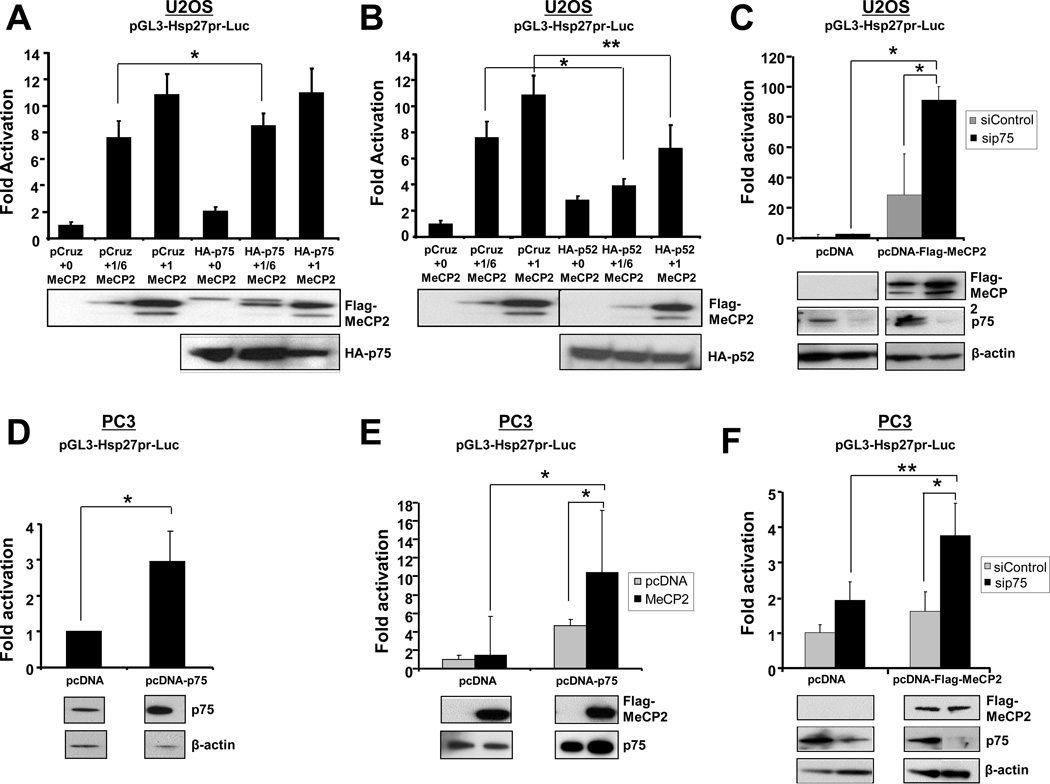
LEDGF/p75 and p52 modulate the transcriptional activity of MeCP2
We sought to determine if LEDGF/p75 and p52 modulate the ability of MeCP2 to transactivate Hsp27pr. Luciferase reporter assays were performed with co-expression of Flag-MeCP2 and HA-LEDGF/p75, or Flag-MeCP2 and HA-p52. Co-expression of HA-LEDGF/p75 and Flag-MeCP2 in U2OS cells enhanced Hsp27pr transactivation levels above those induced by Flag-MeCP2 alone, although this effect was marginal (8 fold vs 7 fold increase, respectively, P<0.05) and only at the low LEDGFp75/MeCP2 plasmid ratio (1/6) (Fig 7A). On the other hand, co-expression of Flag-HA-MeCP2 with HA-p52 significantly reduced Hsp27pr activity (~40%) (Fig. 8B), suggesting that p52 represses MeCP2 transcriptional activity.
Figure 8. LEDGF/p75 and p52 influence MeCP2-induced transactivation of Hsp27pr.
(A) U2OS cells were co-transfected with pGL3-Hsp27pr-luc, pCruzHA or pCruzHA-LEDGF/p75, and increasing amounts of pcDNA-Flag-MeCP2 (1=1.66 µg DNA). (B) U2OS cells were co-transfected with pGL3-Hsp27pr-luc, pCruzHA or pCruzHA-LEDGF/p52, and increasing amounts of pcDNA-Flag-MeCP2. (C) Transient knockdown of LEDGF/p75 in U2OS cells was achieved using specific siRNA oligos. Cells were then co-transfected with pcDNA-Flag-MeCP2 and pGL3-Hsp27pr-luc. Promoter activity determined as luciferase light units/protein is expressed as fold activation compared to control activity. (D) PC3 cells stably transfected with empty pcDNA vector or pcDNA-LEDGF/p75 were co-transfected with pGL3-Hsp27pr-luc and pMAX-GFP (transfection control). (E) PC3 cells stably transfected with empty pcDNA vector or pcDNA-LEDGF/p75 were co-transfected with pGL3-Hsp27pr-luc and pcDNA-Flag-MeCP2. (F) PC3 cells stably transfected with empty pcDNA vector or pcDNA-LEDGF/p75 were transfected with siRNA oligos to knockdown this protein. Cells were then co-transfected with pGL3-Hsp27pr-luc and pcDNA-Flag-MeCP2. Promoter activity determined as luciferase light units/GFP is expressed as fold activation compared to control activity. Data represent the average of at least three independent experiments. Representative immunoblots corresponding to the reporter assays show protein expression. All protein bands in panel F were from the same blot. *p<0.05; **, p<0.01.
LEDGF/p75 is a transcription co-activator of RNA polymerase II transcription (1), whereas MeCP2 participates in chromatin remodeling events leading to recruitment of RNA polymerase II to activate gene trancription (reviewed in 41,42). In light of this, we had expected that ectopic co-expression of HA-LEDGF/p75 and Flag-MeCP2 would result in a robust enhancement of Hsp27pr activity in U2OS cells. However, because neither synergistic nor additive effects were observed, we decided to transiently silence endogenous LEDGF/p75 using siRNA while overexpressing Flag-MeCP2. Surprisingly, U2OS cells overexpressing Flag-MeCP2 but depleted of endogenous LEDGF/p75 displayed robust and significant increase (3-fold) in Hsp27pr activation compared to cells transfected with control siRNAs, suggesting that LEDGF/p75 may repress MeCP2-driven Hsp27pr activity in these cells (Fig. 8C).
In view of the fact that both LEDGF/p75 and MeCP2 have been implicated in PCa cell growth and survival (9,11,14,39), we also examined the effects of their co-expression on Hsp27pr activation in PC3 cells. First, we evaluated Hsp27pr activation in PC3 cells stably overexpressing LEDGF/p75, and observed activation levels that were 3–5 fold above cells stably transfected with empty pcDNA vector (Fig. 8D,E). In contrast to what we observed in U2OS cells, transient Flag-MeCP2 overexpression in PC3 cells stably transfected with empty pcDNA vector did not significantly enhance Hsp27pr activation compared to cells without Flag-MeCP2 transfection (Fig. 8E, pcDNA). However, PC3 cells overexpressing both LEDGF/p75 (stably) and Flag-MeCP2 (transiently) significantly enhanced Hsp27pr activation compared to cells without Flag-MeCP2 transfection (Fig. 8E, pcDNA-p75). This enhancement was more robust (10-fold) than that observed in U2OS (see Fig. 7B). Interestingly, PC3 cells with transient Flag-MeCP2 overexpression but depleted of LEDGF/p75 also showed a significant increase in Hsp27pr activation, compared to cells transfected with control siRNAs (2-fold) or cells with without Flag-MeCP2 overexpression (1.75-fold) (Fig. 8F). However, this LEDGF/p75-mediated transcriptional repression effect was modest compared to that observed in U2OS cells.
Discussion
This study established MeCP2 as an interacting partner of LEDGF/p75 and p52 in vitro and in cellular systems using several complementary approaches. There was consistent agreement in the results obtained from all the different approaches, suggesting specific interaction between these proteins. The eleven to one nM ratio needed for Flag-LEDGF/p75 to bind GST-MeCP2 in the AlphaScreen assay suggested weak binding between the two proteins under the in vitro conditions of this assay. It should be noted that the concentration of GST-MeCP2 was approximate since this protein degraded rapidly when purified, which obliged us to use it in the bacterial lysates for the assays. Alternatively, LEDGF/p75 and MeCP2 may need other nuclear proteins or native chromatin for stronger interaction.
We examined whether the LEDGF-MeCP2 interaction influences Hsp27pr activation in luciferase reporter assays. Hsp27 is a target gene of LEDGF/p75 implicated in PCa resistance to cell death and chemotherapy (15,22,45,46). Overexpression of LEDGF/p75 upregulates Hsp27 transcript in cancer cells, correlating with its promoter transactivation (22). Our data indicated that binding of LEDGF/p75 to Hsp27pr was not limited to the region where HSE and STRE are located (bp −271 to +18), since binding was observed throughout the entire Hsp27pr region examined (bp −1071 to −220). This observation sheds some light into the question of whether LEDGF/p75 binding to promoter regions is mainly restricted to STRE and HSE, as observed by some investigators but not by others (15,32). Recently, studies using the DamID technology, focusing on the highly annotated ENCODE (encyclopedia of DNA Elements) region, revealed that LEDGF/p75 binding to DNA occurs primarily downstream of active transcription unit start sites, is not restricted to STREs, and correlates with active chromatin markers and RNA polymerase II binding (47). Nevertheless, STREs appear to be important for LEDGF/p75 mediated VEGF-c promoter transactivation (17). It is plausible that LEDGF/p75 binding to particular motifs in promoter regions could be influenced by interactions with specific transcription factors as well as the cellular type and microenvironment.
Our results revealed that ectopic MeCP2 overexpression robustly transactivated Hsp27pr in U2OS cells, surpassing the transactivation levels induced by LEDGF/p75 or p52. This suggested that Hsp27 is a transcriptional target gene of MeCP2, with LEDGF/p75 and p52 possibly playing a regulatory role. Alternatively, LEDGF/p75 and MeCP2 may play redundant roles in activating Hsp27pr, with the cellular and molecular context determining which of these proteins play a major role in the promoter activation. While our ChIP data suggested binding of MeCP2 to upstream regions of the Hsp27pr, further studies are needed to determine the exact binding sites, whether or not they require DNA methylation, and if MeCP2 could be activating or repressing different transcription factors in order to activate this promoter.
To our knowledge, this is the first report documenting MeCP2-induced activation of Hsp27, a key pro-survival gene in PCa (44,45). MeCP2 was shown to be critical for PCa cell growth and tumorigenicity, presumably by repressing tumor suppressing genes (39). However, it cannot be ruled out that MeCP2 also contributes to prostate tumorigenesis by transactivating stress survival genes such as Hsp27. In agreement with our results, MeCP2 has been shown to directly bind promoter sites of proteins and enhance their activation, as demonstrated by its association with transcription activator Creb1 on promoters of transcribed genes (42). It is possible that MeCP2 interacts with other transcription factors on Hsp27pr in a manner similar to its association with Creb1 at promoter sites, contributing to up-regulation of Hsp27pr activity in human cancer cells.
The co-expression of LEDGF/p52 with MeCP2 in U2OS cells resulted in markedly reduced transactivation of Hsp27pr, suggesting that p52 represses the transcriptional activity of MeCP2. Bueno et al. (21) recently reported that mutations impairing SUMOylation of LEDGF/p75 increase its transcriptional activity, but not that of p52, suggesting that these splice variants activate Hsp27pr via different molecular mechanisms. Considering the observed antagonistic functions of LEDGF/p75 (pro-survival) and p52 (pro-apoptotic) (22), it is possible that the latter may directly antagonize LEDGF/p75 and MeCP2 by disrupting their binding to Hsp27pr or their interaction with other transcription components. Binding of p52 to MeCP2 could also form a growth suppressor unit, down-regulating the activation of Hsp27pr, similar to that of the growth suppressor unit formed by interaction of JunD with menin (48). Alternatively, reduced Hsp27pr activity may be due to p52-induced apoptosis, which generates a dominant negative fragment p38 that interferes with the transcription of this gene (22).
Contrary to our expectations, we did not detect a robust cooperation between ectopically expressed MeCP2 and LEDGF/p75 on Hsp27pr activation in U2OS cells. Instead, we observed that transient silencing of LEDGF/p75 in this cell line induced a dramatic increase in MeCP2-driven Hsp27pr activity. However, in PC3 cells the cooperation between LEDGF/p75 and MeCP2 on Hsp27pr activation appeared to be stronger than in U2OS cells, whereas the enhancement of promoter activity by the LEDGF/p75 knockdown was weaker. These apparently contradictory results suggest the interesting possibility that LEDGF/p75 could be a modulator of MeCP2 transcriptional activity, enhancing or repressing this activity depending on the cellular and molecular context, or the microenvironment. LEDGF/p75 might be part of a feedback loop by competing with MeCP2 or blocking other transcription co-factors from promoter regulatory sites, as in the feedback loop observed between HSF1 and Hsp27 (49). This would be important to reduce spurious or excessive transcriptional activation of Hsp27 and other stress genes.
While the mechanism by which LEDGF/p75 modulates MeCP2 transcriptional activity is unknown, it should be emphasized that the PWWP-CR1 region of LEDGF/p75 appears to be responsible for binding to MeCP2. The PWWP domain of LEDGF/p75 has been shown to cooperate with the NLS and AT-hooks downstream the CR1 region to facilitate interaction with DNA/chromatin (31–33). Recently, this domain was demonstrated to lock into the chromatin cargo proteins such as HIV-IN that are bound to the IBD domain of LEDGF/p75 (30). PWWP domains belong to the Royal family of protein domains that regulate chromatin remodeling and function (29). In DNA methyltransferases, the PWWP domain is essential for heterochromatin targeting, possibly by facilitating interaction with methylated histones (25,26). The PWWP domain of hepatoma derived growth factor (HDGF), a cancer-related protein that shares sequence homology with LEDGF/p75, is required for binding to chromatin and the transcriptional co-repressor C-terminal binding protein (CtBP), which results in transcriptional repression of specific genes (27,28). Thus, PWWP domains are emerging as structural entities that bind to DNA, histones, and transcription factors to regulate gene expression.
It is possible that LEDGF/p75 could tether MeCP2 to transcriptionally active sites in the chromatin via the PWWP-CR1 region. However, given the documented role of PWWP domains in transcriptional repression, it would be tempting to speculate that this region may negatively regulate the activity of MeCP2 and other interacting transcription factors. Singh et al (38) provided evidence that the N-terminal region of LEDGF/p75 has negative transcriptional regulatory elements, and proposed that LEDGF/p75, like other transcription proteins, bear repressive as well as activation domains, with the repressive domain being the PWWP domain (38). Interestingly, chromosomal translocations in acute leukemia produce a NUP98-LEDGF/p75 fusion protein lacking the N-terminal region of LEDGF/p75 (13,18), suggesting the possibility that this PWWP-CR1 deficient protein induces deregulated oncogene transcription.
To our knowledge, this is the first report implicating the N-terminal region of LEDGF/p75 in protein-protein interactions. To date, all the interacting partners of LEDGF/p75, namely HIV-IN, JPO2, menin/MLL, ASK, and pogZ, have been shown to interact with the C-terminal region of the protein, particularly with the IBD domain. Through this interaction, some of these proteins are recruited to transcriptionally active sites in the chromatin (18,30). In the case of menin/MLL, the interaction with LEDGF/p75 actually leads to the activation of cancer-related genes involved in leukemogenesis (18).
In conclusion, we validated MeCP2 as a cellular interacting partner of both LEDGF/p75 and p52, and a transactivator of Hsp27pr. Follow-up studies should determine if rational mutation of specific sites in the PWWP-CR1 domain of LEDGF/p75 (e.g. Pro-Trp-Trp-Pro motif, sumoylation sites, etc.) results in loss of MeCP2 binding to LEDGF/p75 or Hsp27pr, or in deregulated stress promoter activity that is mediated by MeCP2 or other LEDGF/p75-interacting transcription factors. It should also be determined if the LEDGF/p75 PWWP domain is required for MeCP2 binding to chromatin, and if the complex between these proteins bind to methylated or demethylated promoter regions. Additional studies will be needed to define if LEDGF/p75, p52, and MeCP2 are part of a larger dynamic transcriptional complex that regulates the expression of stress survival genes depending on the cellular context and the microenvironment. Our results have implications for understanding the contribution of LEDGF/MeCP2 containing transcription complexes in the regulation of gene expression in cancer, HIV-AIDS, Rett syndrome, and other diseases in which these proteins have been implicated.
Acknowledgements
We thank Dr. Sean Wilson and Monica Rubalcava (Loma Linda University) for assistance with confocal microscopy, and Dr. Melissa McNeely, Dr. Antonio Gallo, Christine de Kogel, and Jonas Demeulemeester (Leuven) for the generation of plasmids and recombinant proteins, and technical assistance. We recognize the contribution of Melanie Mediavilla-Varela, Terry Brown-Bryan, Fabio Pacheco, Vidya Ganapathy, Papri Chatterjee, Evyn Adkins, Lauren Melendez, and Bobby Jimenez (LLU) for technical assistance in the initial stages of this work. Our thanks to Drs. Rik Gijsbers (Leuven), Penelope Duerksen-Hughes, Thomas Linkhart, and Nathan Wall (LLU) for excellent suggestions.
Grant support. This work was supported by grants NIH-NCMHD 5P20MD001632 (Casiano CA, De Leon M), NSF-DBI-0923559 (LLU School of Medicine-Advanced Imaging and Microscopy Facility), KU Leuven Research Council grant OT/09/047 (Debyser Z), and Flanders Research Foundation (FWO) grant G.0530.08 (Debyser Z).
References
- 1.Ge H, Si Y, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. Embo J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganapathy V, Casiano CA. Autoimmunity to the nuclear autoantigen DFS70 (LEDGF): what exactly are the autoantibodies trying to tell us? Arthritis Rheum. 2004;50:684–688. doi: 10.1002/art.20095. [DOI] [PubMed] [Google Scholar]
- 3.Singh DP, Ohguro N, Kikuchi T, Sueno T, Reddy VN, Yuge K, et al. Lens epithelium-derived growth factor: effects on growth and survival of lens epithelial cells, keratinocytes, and fibroblasts. Biochem Biophys Res Commun. 2000;267:373–381. doi: 10.1006/bbrc.1999.1979. [DOI] [PubMed] [Google Scholar]
- 4.Singh DP, Ohguro N, Chylack LT, Jr, Shinohara T. Lens epithelium-derived growth factor: increased resistance to thermal and oxidative stresses. Invest Ophthalmol Vis Sci. 1999;40:1444–1451. [PubMed] [Google Scholar]
- 5.Wu X, Daniels T, Molinaro C, Lilly MB, Casiano CA. Caspase cleavage of the nuclear autoantigen LEDGF/p75 abrogates its pro-survival function: implications for autoimmunity in atopic disorders. Cell Death Differ. 2002;9:915–925. doi: 10.1038/sj.cdd.4401063. [DOI] [PubMed] [Google Scholar]
- 6.De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J Virol. 2006;80:11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 8.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 9.Daniels T, Zhang J, Gutierrez I, Elliot ML, Yamada B, Heeb MJ, et al. Antinuclear autoantibodies in PCa: immunity to LEDGF/p75, a survival protein highly expressed in prostate tumors and cleaved during apoptosis. Prostate. 2005;62:14–26. doi: 10.1002/pros.20112. [DOI] [PubMed] [Google Scholar]
- 10.Daugaard M, Kirkegaard-Sorensen T, Ostenfeld MS, Aaboe M, Hoyer-Hansen M, Orntoft TF, et al. Lens epithelium-derived growth factor is an Hsp70-2 regulated guardian of lysosomal stability in human cancer. Cancer Res. 2007;67:2559–2567. doi: 10.1158/0008-5472.CAN-06-4121. [DOI] [PubMed] [Google Scholar]
- 11.Basu A, Rojas H, Banerjee H, Cabrera I, Perez K, Casiano CA. Expression of the stress oncoprotein LEDGF/p75 in major human cancers: a study of 21 tumor types. PloS ONE. 2012 doi: 10.1371/journal.pone.0030132. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang TS, Myklebust LM, Kjarland E, Gjertsen BT, Pendino F, Bruserud O, et al. LEDGF/p75 has increased expression in blasts from chemotherapy-resistant human acute myelogenic leukemia patients and protects leukemia cells from apoptosis in vitro. Mol Cancer. 2007;6:31. doi: 10.1186/1476-4598-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahuja HG, Hong J, Aplan PD, Tcheurekdjian L, Forman SJ, Slovak ML. t(9;11)(p22;p15) in acute myeloid leukemia results in a fusion between NUP98 and the gene encoding transcriptional coactivators p52 and p75-lens epithelium-derived growth factor (LEDGF) Cancer Res. 2000;60:6227–6229. [PubMed] [Google Scholar]
- 14.Mediavilla-Varela M, Pacheco FJ, Almaguel F, Perez J, Sahakian E, Daniels TR, et al. Docetaxel-induced PCa cell death involves concomitant activation of caspase and lysosomal pathways and is attenuated by LEDGF/p75. Mol Cancer. 2009;8:68. doi: 10.1186/1476-4598-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh DP, Fatma N, Kimura A, Chylack LT, Jr, Shinohara T. LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem Biophys Res Commun. 2001;283:943–955. doi: 10.1006/bbrc.2001.4887. [DOI] [PubMed] [Google Scholar]
- 16.Fatma N, Singh DP, Shinohara T, Chylack LT., Jr Transcriptional regulation of the antioxidant protein 2 gene, a thiol-specific antioxidant, by lens epithelium-derived growth factor to protect cells from oxidative stress. J Biol Chem. 2001;276:48899–48907. doi: 10.1074/jbc.M100733200. [DOI] [PubMed] [Google Scholar]
- 17.Cohen B, Addadi Y, Sapoznik S, Meir G, Kalchenko V, Harmelin A, et al. Transcriptional regulation of vascular endothelial growth factor C by oxidative and thermal stress is mediated by lens epithelium-derived growth factor/p75. Neoplasia. 2009;11:921–933. doi: 10.1593/neo.09636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma P, Fatma N, Kubo E, Shinohara T, Chylack LT, Jr, Singh DP. Lens epithelium-derived growth factor relieves transforming growth factor-beta1-induced transcription repression of heat shock proteins in human lens epithelial cells. J Biol Chem. 2003;278:20037–20046. doi: 10.1074/jbc.M212016200. [DOI] [PubMed] [Google Scholar]
- 20.Mao YW, Xiang H, Wang J, Korsmeyer S, Reddan J, Li DW. Human bcl-2 gene attenuates the ability of rabbit lens epithelial cells against H2O2-induced apoptosis through down-regulation of the alpha B-crystallin gene. J Biol Chem. 2001;276:43435–43445. doi: 10.1074/jbc.M102195200. [DOI] [PubMed] [Google Scholar]
- 21.Bueno MT, Garcia-Rivera JA, Kugelman JR, Morales E, Rosas-Acosta G, Llano M. SUMOylation of the lens epithelium-derived growth factor/p75 attenuates its transcriptional activity on the heat shock protein 27 promoter. J Mol Biol. 2010;399:221–239. doi: 10.1016/j.jmb.2010.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown-Bryan TA, Leoh LS, Ganapathy V, Pacheco FJ, Mediavilla-Varela M, Filippova M, et al. Alternative splicing and caspase-mediated cleavage generate antagonistic variants of the stress oncoprotein LEDGF/p75. Mol Cancer Res. 2008;6:1293–1307. doi: 10.1158/1541-7786.MCR-08-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desfarges S, Abderrahmani A, Hern‡ndez-Novoa B, Munoz M, Ciuffi A. LEDGF/p75 TATA-Less Promoter Is Driven by Transcription Factor Sp1. J Mol Biol. 2011;414:177–193. doi: 10.1016/j.jmb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Zammarchi F, de Stanchina E, Bournazou E, Supakorndej T, Martires K, Riedel E, Corben AD, Bromberg JF, Cartegni L. Antitumorigenic potential of STAT3 alternative splicing modulation. Proc Natl Acad Sci. 2011;108:17779–17784. doi: 10.1073/pnas.1108482108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Everett AD. Hepatoma-derived growth factor binds DNA through the N-terminal PWWP domain. BMC Mol Biol. 2007;8:101. doi: 10.1186/1471-2199-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Everett AD. Hepatoma-derived growth factor represses SET and MYND domain containing 1 gene expression through interaction with C-terminal binding protein. J Mol Biol. 2009;386:938–950. doi: 10.1016/j.jmb.2008.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation. FEBS Lett. 2000;473:1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 30.Gijsbers R, Vets S, De Rijck J, Ocwieja KE, Ronen K, Malani N, Bushman FD, Debyser Z. Role of the PWWP Domain of Lens Epithelium-derived Growth Factor (LEDGF)/p75 Cofactor in Lentiviral Integration Targeting. J Biol Chem. 2011;286:41812–41825. doi: 10.1074/jbc.M111.255711. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 32.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1653–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsutsui KM, Sano K, Hosoya O, Miyamoto T, Tsutsui K. Nuclear protein LEDGF/p75 recognizes supercoiled DNA by a novel DNA-binding domain. Nucleic Acids Res. 2011;39:5067–5081. doi: 10.1093/nar/gkr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa Y, Sugiura K, Watanabe A, Kunimatsu M, Mishima M, Tomita Y, Muro Y. Autoantigenicity of DFS70 is restricted to the conformational epitope of C-terminal alpha-helical domain. J Autoimmun. 2004;23:221–231. doi: 10.1016/j.jaut.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Bartholomeeusen K, Christ F, Hendrix J, Rain JC, Emiliani S, Benarous R, et al. Lens epithelium-derived growth factor/p75 interacts with the transposase-derived DDE domain of PogZ. J Biol Chem. 2009;284:11467–11477. doi: 10.1074/jbc.M807781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maertens GN, Cherepanov P, Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J Cell Sci. 2006;119:2563–2571. doi: 10.1242/jcs.02995. [DOI] [PubMed] [Google Scholar]
- 37.Hughes S, Jenkins V, Dar MJ, Engelman A, Cherepanov P. Transcription co-activator LEDGF interacts with the S-phase kinase Cdc7:ASK and stimulates its enzymatic activity. J Biol Chem. 2010;285:541–554. doi: 10.1074/jbc.M109.036491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh DP, Kubo E, Takamura Y, Shinohara T, Kumar A, Chylack LT, Jr, et al. DNA binding domains and nuclear localization signal of LEDGF: contribution of two helix-turn-helix (HTH)-like domains and a stretch of 58 amino acids of the N-terminal to the trans-activation potential of LEDGF. J Mol Biol. 2006;355:379–394. doi: 10.1016/j.jmb.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 39.Bernard D, Gil J, Dumont P, Rizzo S, Monte D, Quatannens B, et al. The methyl-CpG-binding protein MECP2 is required for PCa cell growth. Oncogene. 2006;25:1358–1366. doi: 10.1038/sj.onc.1209179. [DOI] [PubMed] [Google Scholar]
- 40.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 41.Adkins NL, Georgel PT. MeCP2: structure and function. Biochem Cell Biol. 2011;89:1–11. doi: 10.1139/O10-112. [DOI] [PubMed] [Google Scholar]
- 42.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandekerckhove L, Christ F, Van Maele B, De Rijck J, Gijsbers R, Van den Haute C, Witvrouw M, Debyser Z. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J Virol. 2006;80:1886–1896. doi: 10.1128/JVI.80.4.1886-1896.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 45.Rocchi P, Jugpal P, So A, Sinneman S, Ettinger S, Fazli L, et al. Small interference RNA targeting heat-shock protein 27 inhibits the growth of prostatic cell lines and induces apoptosis via caspase-3 activation in vitro. BJU Int. 2006;98:1082–1089. doi: 10.1111/j.1464-410X.2006.06425.x. [DOI] [PubMed] [Google Scholar]
- 46.Andrieu C, Taieb D, Baylot V, Ettinger S, Soubeyran P, De-Thonel A, et al. Heat shock protein 27 confers resistance to androgen ablation and chemotherapy in PCa cells through eIF4E. Oncogene. 2010;29:1883–1896. doi: 10.1038/onc.2009.479. [DOI] [PubMed] [Google Scholar]
- 47.De Rijck J, Bartholomeeusen K, Ceulemans H, Debyser Z, Gijsbers R. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res. 2010;38:6135–6147. doi: 10.1093/nar/gkq410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agarwal SK, Guru SC, Heppner C, Erdos MR, Collins RM, Park SY, et al. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell. 1999;96:143–152. doi: 10.1016/s0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- 49.Brunet Simioni M, De Thonel A, Hammann A, Joly AL, Bossis G, Fourmaux E, et al. Heat shock protein 27 is involved in SUMO-2/3 modification of heat shock factor 1 and thereby modulates the transcription factor activity. Oncogene. 2009;28:3332–3344. doi: 10.1038/onc.2009.188. [DOI] [PubMed] [Google Scholar]