Abstract
Autophagy is a highly regulated intracellular process for the degradation of cytoplasmic components, especially protein aggregates and damaged organelles. It is essential for maintaining healthy cells. Impaired or deficient autophagy is believed to cause or contribute to aging and age-related disease. In this study, we investigated the effects of age on autophagy in the kidneys of 3-, 12-, and 24-month-old Fischer 344 rats. The results revealed that autophagy-related gene (Atg)7 was significantly downregulated in kidneys of increasing age. The protein expression level of the autophagy marker light chain 3/Atg8 exhibited a marked decline in aged kidneys. The levels of p62/SQSTM1 and polyubiquitin aggregates, representing the function of autophagy and proteasomal degradation, increased in older kidneys. The level of 8-hydroxydeoxyguanosine, a marker of mitochondrial DNA oxidative damage, was also increased in older kidneys. Analysis by transmission electron microscope demonstrated swelling and disintegration of cristae in the mitochondria of aged kidneys. These results suggest that autophagic function decreases with age in the kidneys of Fischer 344 rats, and autophagy may mediate the process of kidney aging, leading to the accumulation of damaged mitochondria.
Keywords: Autophagy, Kidney, Aging, Mitochondria, Oxidative damage
Introduction
Autophagy is an evolutionarily conserved process present in a number of eukaryotic organisms, from yeast to nematodes to humans, by which cytoplasmic constituents are sequestered into double-membrane structures forming autophagosomes. These fuse with lysosomes, forming single-membrane structures called autolysosomes. Finally, the cytoplasmic components are degraded by acid hydrolases, and the degraded products are released into the cytosol and recycled into biological structures or to supply energy during periods of starvation (Levine and Klionsky 2004; Xie and Klionsky 2007; Mizushima et al. 2008). Autophagy is important in the normal turnover of cellular components as well as in response to such stresses as nutrient starvation or growth factor deprivation. A key function of autophagy is to remove damaged organelles, including mitochondria and aberrant macromolecules, to prevent further injury and cellular dysfunction. This may be of particular importance in the aging process.
Aging is characterized by the progressive accumulation of damaged and defective cellular components, progressive structural disorganization, diminishing functional capacity, decreasing adaptability, and increasing likelihood of disease and death (Rajawat et al. 2009; Terman and Brunk 2005). Many studies have demonstrated that age-related changes result from the accumulation of damaged macromolecules by physiologically produced reactive oxygen species (ROS; Harman 1956; Sohal and Weindruch 1996; Beckman and Ames 1998). The increase of ROS in the cytoplasm is a causative event of aging (Bergamini et al. 2004; Brunk and Terman 2002; Honda and Honda 2002; Sohal 2002; Blagosklonny 2008), and mitochondria are a key source of ROS. A popular hypothesis postulates that macromolecule oxidative damage caused by ROS progressively accumulates and is the possible cause of senescence-associated deleterious alterations (Rattan 2008). Therefore, cellular self-degradative systems, especially lysosome-mediated autophagy, may play a pivotal role in the control of aging (Ciechanover 2005; Ward 2002; Martinez-Vicente et al. 2005; Ravikumar and Rubinsztein 2006). Renal cells consume oxygen at a high rate and are highly dependent on aerobic metabolism for ATP production. Thus, they may be especially susceptible to aging because they generate more ROS and accumulate more damage during the oxidative phosphorylation process (Hengartner 2000). The mechanism of renal aging is not clear, and it is unknown whether autophagy changes with aging in the kidneys. In the present study, we investigated the effect of age on the expression of autophagy-related genes (Atg) in the kidneys of 3-, 12-, and 24-month-old male Fischer 344 rats.
Materials and methods
Animals
Male Fischer 344 rats (n = 30) were kept under a specific pathogen-free condition: 22 ± 1°C, 40% humidity, 12:12-h light/dark cycle, five males per cage, and free access to diet. All experiments involving animals were approved by the Institutional Animal Care and Use Committee at the Chinese PLA General Hospital and Military Medical Postgraduate College. Rats were anesthetized via intraperitoneal injection of sodium pentobarbital (40 mg/kg). There were three groups of rats: those that were 3 months (n = 10), 12 months (n = 10), and 24 months old (n = 10), corresponding to young, adult, and old, respectively. At 3, 12, and 24 months of age, the kidney tissues from each group of rats were removed and perfused with ice-cold, isotonic phosphate-buffered saline (PBS; pH 7.4) to remove any remaining blood. A portion of the tissue was placed into 10% neutral buffered formalin for immunohistochemistry staining. Another portion was immersed into OCT compound (Tissue-Tek; Sakura Finetek, Torrance, CA, USA) for immunofluorescence staining. The remaining tissue was immediately frozen in liquid nitrogen and stored at −80°C until further processing.
Senescence-associated β-galactosidase staining
Cryostat sections (4 μm) were mounted onto glass slides and fixed in 0.2% glutaraldehyde and 2% formaldehyde at room temperature for 15 min. Sections were washed in PBS and incubated in freshly prepared senescence-associated β-galactosidase (SA-β-gal) staining solution (1 mg/mL X-gal, 40 mM citric acid/sodium phosphate (pH 6.0), 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2) overnight at 37°C without CO2. Tissue sections were counterstained with eosin and examined under a microscope.
Western blot
Tissues were lysed in radioimmunoprecipitation assay buffer (50 mM Tris–Cl (pH 7.6), 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate (SDS), 0.5% deoxycholic acid, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 0.5 mM phenylmethylsulfonyl fluoride) for 30 min on ice prior to centrifugation at 12,000 rpm for 30 min at 4°C. Protein concentration was determined with the Pierce BCA assay kit (Lot #JK126465; Thermo Fisher Scientific, Rockford, IL, USA). A total of 60–100 μg protein was separated by 10–16% SDS–polyacrylamide gel electrophoresis, transferred to an nitrocellulose membrane, blocked with 5% skim milk for 1 h at room temperature, and probed with the following primary antibodies at 4°C overnight: rabbit polyclonal anti-light chain 3 (LC3) antibody (Sigma, St. Louis, MO, USA) at 1:2,000, mouse monoclonal anti-p16 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 1:200, goat polyclonal anti-beclin-1 (Santa Cruz Biotechnology) at 1:1,000, mouse monoclonal anti-ubiquitin (Mabtech, Nacka Strand, Sweden) at 1:1,000, and mouse monoclonal anti-p62 (Santa Cruz Biotechnology) at 1:200. Blots were subsequently probed with horseradish peroxidase-conjugated anti-mouse, anti-goat, or anti-rabbit IgG (Santa Cruz Biotechnology) at 1:1,000–5,000. Immunoreactive bands were visualized by enhanced chemiluminescence, and densitometry was performed using Quantity One software (Bio-Rad Laboratories, Hercules, CA, USA).
Immunofluorescence staining
Renal tissues were embedded in OCT compound. Cryostat sections (4 μm) were stained with rabbit polyclonal anti-LC3 antibody (Sigma) at 1:100 and mouse monoclonal anti-8-hydroxydeoxyguanosine (OHdG) antibody (Santa Cruz Biotechnology) at 1:50. Sections were incubated sequentially with rhodamine-conjugated AffiniPure goat anti-rabbit IgG followed by fluorescein-conjugated AffiniPure goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA, USA). The results were analyzed under an Olympus laser scanning confocal microscope.
RNA isolation and real-time quantitative PCR
Total RNA was isolated from renal tissues using TRIzol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Reverse transcription was performed using a TIANScript RT kit (Tiangen Biotech, Beijing, China). Amplification was performed in a Rotor-Gene 3000 (Corbett Research Pty Ltd, Sydney, Australia). Reaction contained 50 ng total DNA, 0.2 μM primers, and 12.5 μL 2× SYBR green buffer (TransGen, Beijing, China) in a final volume of 25 μL. Atg5 and Atg7 were used as markers for autophagy (McMullen et al. 2009). Primers were designed using the software package Primer Express 2.0 (Applied Biosystems, Carlsbad, CA, USA) based on GenBank nucleotide sequences as follows: Atg5 (accession no. AM087012): forward 5′-AGG CTC AGT GGA GGC AAC AG-3′, reverse 5′-CCC TAT CTC CCA TGG AAT CTT CT-3′; Atg7 (accession no. NM_001012097): forward 5′-GAC CTG GGC TCC TCA CTT TTT G-3′, reverse 5′-CCC TGG GCG GCT CAC TG-3′; and β-actin: forward 5′-GCG CTT TTG ACT CAA GGA TTT AA-3′, reverse 5′-GGG ATG TTT GCT CCA ACC AA-3′. Atg5, Atg7, and β-actin PCR was performed using the following cycling conditions: 95°C for 4 min, 40 cycles of denaturation at 95°C for 30 s, and extension at 72°C for 30 s. The annealing temperatures were 60°C for Atg7 and 55°C for Atg5 and β-actin. All samples were run in triplicate. The relative abundance of target mRNA was determined with the comparative cycle threshold method (Giulietti et al. 2001).
Immunohistochemistry staining
Kidneys were fixed in 10% formaldehyde at 4°C overnight and then processed for paraffin-embedding following standard procedures. Sections were prepared at 3 μm. For immunohistochemical analysis, tissue sections were subjected to antigen retrieval by microwaving or autoclaving for 10 or 15 min in 10 mM sodium citrate buffer (pH 6.0). Endogenous peroxidase was blocked by incubation with 3% hydrogen peroxide for 10 min. Sections were washed in PBS and incubated with 1.5% normal goat serum for 20 min, followed by incubation with a 1:50 dilution of mouse monoclonal anti-8-OHdG antibody (Santa Cruz Biotechnology) overnight at 4°C. Sections were washed three times with PBS and incubated with biotin-conjugated goat anti-mouse IgG (Invitrogen) for 30 min at room temperature. Sections were again washed with PBS and incubated with streptavidin-conjugated peroxidase (Invitrogen) for 30 min at room temperature. Sections were washed for a final time in PBS and incubated with DAB (Invitrogen) followed by examination under a microscope.
Transmission electron microscopy
Kidneys were cut into small tissue blocks (1 mm3) and fixed in 2.5% glutaraldehyde with 0.01 mol/L phosphate buffer at 4°C, followed by 2% osmium tetroxide. They were then dehydrated in a series of graded ethanol solutions. Ethanol was substituted with propylene oxide and then embedded in epoxy resin. Ultrathin sections were double-stained with uranyl acetate and lead and examined under a JEM1200EX transmission electron microscope (JEOL) at 80 kV.
Statistical analysis
All data analyses were performed using SPSS 13.0 (SPSS, Chicago, IL, USA) software; data are expressed as mean ± SD. Comparisons among groups were made using analysis of variance. p < 0.05 was considered statistically significant.
Results
Analysis of kidney functions in aged rats
We observed changes in metabolic parameters and renal functions in the three groups of rats. Body weight, body length, abdominal perimeter, and kidney weight in the 12- and 24-month-old rats increased significantly compared with those in the 3-month-old rats. The level of serum urea nitrogen and the urine protein/creatinine ratio increased markedly in the older rats. The level of serum glucose was unchanged among the three groups. The levels of triglycerides increased significantly in the aged rats (Table 1).
Table 1.
Effects of age on metabolic parameters and renal functions in three groups of rats
| Parameter | 3-month-old rats | 12-month-old rats | 24-month-old rats |
|---|---|---|---|
| Body weight (g) | 236.3 ± 12.28 | 466.86 ± 32.62* | 617.8 ± 57.34* |
| Body length (cm) | 20.67 ± 0.58 | 24.5 ± 0.58* | 28 ± 0.71* |
| Abdominal perimeter (cm) | 14.67 ± 0.58 | 22.25 ± 0.96* | 25.2 ± 1.92* |
| Kidney weight (g) | 1.67 ± 0.58 | 3.250 ± 0.5* | 5.01 ± 1.01* |
| Serum urea nitrogen (mmol/L) | 5.73 ± 0.47 | 6.51 ± 0.40 | 6.86 ± 1.10* |
| Serum creatinine (mol/L) | 27.15 ± 2.60 | 25.62 ± 1.94 | 25.8 ± 2.04 |
| Serum glucose (mmol/L) | 7.09 ± 1.02 | 5.745 ± 0.29 | 6.09 ± 1.31 |
| Triglycerides (mmol/L) | 1.20 ± 0.32 | 2.32 ± 0.46* | 3.22 ± 0.82* |
| Cholesterol (mmol/L) | 1.55 ± 0.16 | 2.73 ± 0.28 | 3.11 ± 0.36 |
| Urine protein/urine creatinine ratio (mg/mmol) | 146.01 ± 22.72 | 164.81 ± 38.31* | 256 ± 49.39* |
Data are presented as means ± SD (n = 10)
*p < 0.05 vs 3-month-old group
Changes in senescence markers in aged rat renal tissues
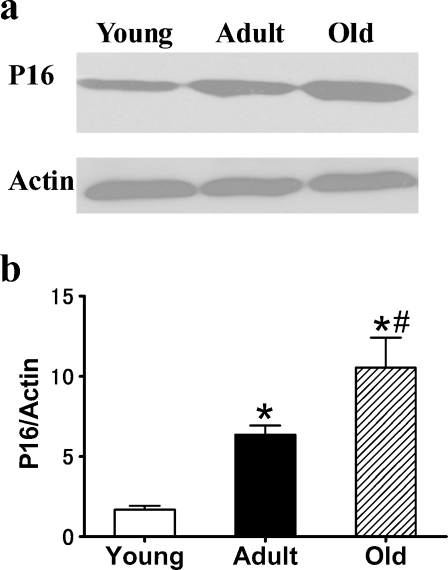
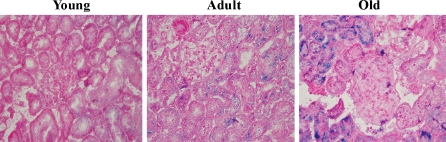
p16 is a robust biomarker and a possible effector of mammalian aging (Krishnamurthy et al. 2004). The expression of p16 was measured in 3-, 12-, and 24-month-old rat kidneys. The results showed that the expression of p16 increased significantly in the 12- and 24-month-old tissues (Fig. 1) compared with the 3-month-old tissue. We also determined the expression of another well-defined in vivo marker of senescence, SA-β-gal, which has a high pH galactosidase activity detectable in senescent cells and tissues (Dimri et al. 1995; Flores and Blasco 2009). As shown in Fig. 2, the positive rate of SA-β-gal staining was markedly elevated in the kidneys of 24-month-old rats compared with 3-month-old rats.
Fig. 1.
Expression of senescent biomarker p16 in the kidneys of 3-, 12-, and 24-month-old Fischer 344 rats. a Western blot results for p16 protein. b Quantitative analysis of band density for p16. Protein expression data are presented as mean ± SD (n = 10). *p < 0.05 vs young; #p < 0.05 vs adult
Fig. 2.
Senescence-associated β-galactosidase staining results for renal tissues from 3-, 12-, and 24-month-old Fischer 344 rats. Magnification, ×400. Blue precipitation in the cytoplasm was observed in the senescent cells
Changes in autophagy-related proteins/genes during the renal aging process
Autophagic function is usually measured by quantifying autophagy-related genes/proteins. Among the 31 Atg proteins identified, Atg1–10 are involved in autophagosome formation and are markers for the formation of the isolation membrane. In this study, we evaluated the changes in the expression of autophagy proteins/genes (beclin-1/Atg6, LC3/Atg8, Atg5, and Atg7) during the renal aging process.
LC3/Atg8
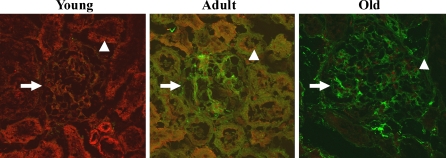
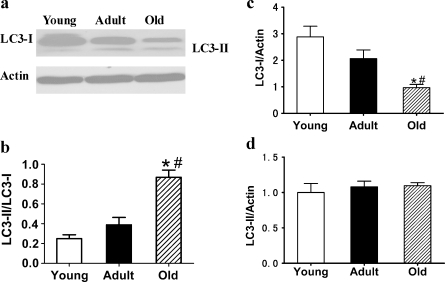
The microtubule-associated protein 1 LC3, a mammalian homolog of yeast Atg8, is required for the formation of autophagosomes, especially the expansion of early autophagosomes (Eskelinen 2005). Pre-LC3 is cleaved into its cytosolic form LC3-I by Atg4. LC3-I is then activated by Atg7 and converted into its membrane-bound form, LC3-II, localized in pre-autophagosomes and autophagosomes (Maiuri et al. 2007). LC3-I and LC3-II are general markers of autophagic membranes. The LC3-II/LC3-I ratio is correlated with autophagic flux (Kadowaki and Karim 2009). We first observed the expression of LC3 by immunofluorescence staining. The results showed that the expression of LC3 (including LC3 dots) was decreased in 24-month-old kidneys compared with 3-month-old kidneys (Fig. 3). In the photos of kidneys from young rats, the expression of LC3 (red) is the most prominent, and the expression of 8-OHdG (green) is the least among the three groups. In the adult ones, the expression of LC3 (red) is decreased and 8-OHdG (green) is increased significantly compared with the young ones, and their colocalization is shown as yellow. In the photos of kidneys from old rats, the expression of LC3 (red) is the least and the expression of 8-OHdG (green) is the most prominent among the three groups. Western blot analysis revealed that the expression of LC3-I was significantly decreased in 24-month-old kidneys and that there was no marked change in LC3-II expression among the three groups (Fig. 4), leading to a significantly increased LC3-II/LC3-I ratio in the 24-month-old kidneys. These results indicate that autophagy declines in the kidneys of aged rats.
Fig. 3.
Dual immunofluorescence staining results for LC3 proteins (red), 8-OHdG (green), and the colocalization (yellow) of LC3 and 8-OHdG in the kidneys of 3-, 12-, and 24-month-old Fischer 344 rats were scanned by a laser confocal microscope. Magnification, ×600. Central glomeruli are indicated by an arrow. Peripheral tubules are indicated by an arrowhead
Fig. 4.
a Expression of LC3-I and LC3-II proteins in the renal tissues of 3-, 12-, and 24-month-old Fischer 344 rats was quantified by Western blot analysis. b Ratio of LC3-II band and LC3-I band was analyzed. c, d Quantitative analysis of band density for LC3-I and LC3-II. Protein expression data are presented as mean ± SD (n = 10). *p < 0.05 vs young; #p < 0.05 vs adult
Beclin-1/Atg6
Beclin-1, the mammalian homolog of yeast Atg6, plays an important role in the induction and formation of the pre-autophagosome structure by associating with a multimeric complex of autophagy regulatory proteins (Atg14, Vps34/class 3 PI3 kinase, and Vps15; He and Klionsky 2009). We found that beclin-1 expression increased slightly but not significantly in 24-month-old kidneys (data not shown).
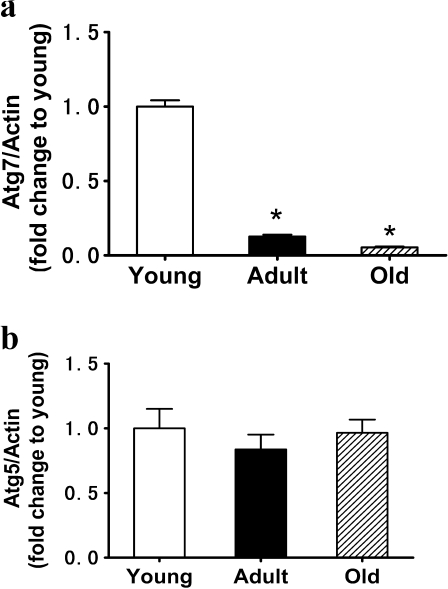
Atg7 and Atg5
Atg7 is required for the formation and expansion of autophagosomes by initiating the conjugation of Atg12 to Atg5 and LC3 to phosphatidylethanolamine. We examined the mRNA level of Atg7 at 3, 12, and 24 months in rat kidneys. The results demonstrated that the mRNA level of Atg7 was significantly decreased in the kidneys of 24-month-old rats (Fig. 5a). Atg5 is necessary for the formation of the autophagosome membrane through interaction with Atg12. We found that the mRNA level of Atg5 was not affected by age (Fig. 5b).
Fig. 5.
Expression of Atg7 (a) and Atg5 (b) mRNA in tissue extracts of kidneys of 3-, 12-, and 24-month-old Fischer 344 rats was measured by quantitative PCR. Data are presented as mean ± SD (n = 10). *p < 0.05 vs young
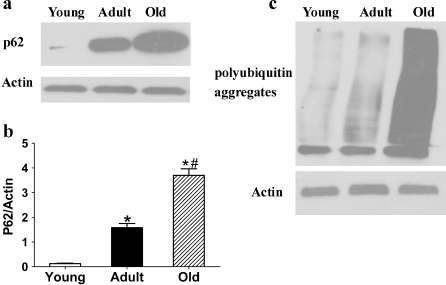
p62/SQSTM1 and polyubiquitin aggregates
p62/SQSTM1, a ubiquitin-binding protein, interacts with LC3 (Pankiv et al. 2007) in the autophagic degradation of ubiquitinated protein aggregates in mammalian cells. Polyubiquitin, an intracellular protein aggregate, is a signal for degradation by the ubiquitin–proteasome system (Bjørkøy et al. 2006). Experiments have demonstrated that p62/SQSTM1 plays a major role in the degradation of ubiquitin substrates. It may act as a critical ubiquitin chain-targeting factor that shuttles substrates for proteasomal degradation (Seibenhener et al. 2004). p62/SQSTM1 may be involved via LC3 in linking polyubiquitinated protein aggregates to the autophagy machinery, which would have a protective effect on cell survival (Bjørkøy et al. 2005). Therefore, p62 is considered another marker in monitoring autophagy flux in certain settings (Mizushima and Yoshimori 2007). Our results showed that the expression of p62/SQSTM1 and polyubiquitin aggregates was significantly increased in 24-month-old rat kidneys (Fig. 6).
Fig. 6.
a, c Expression of p62/SQSTM1 and polyubiquitin aggregates in tissue extracts of kidneys of 3-, 12-, and 24-month-old Fischer 344 rats was quantified by Western blot. b Quantitative analysis of band density for p62/SQSTM1. Protein expression data are presented as mean ± SD (n = 10). *p < 0.05 vs young; #p < 0.05 vs adult
Analysis of mitochondrial oxidative damage in older rat kidneys
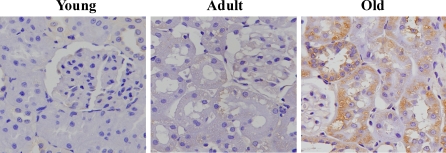
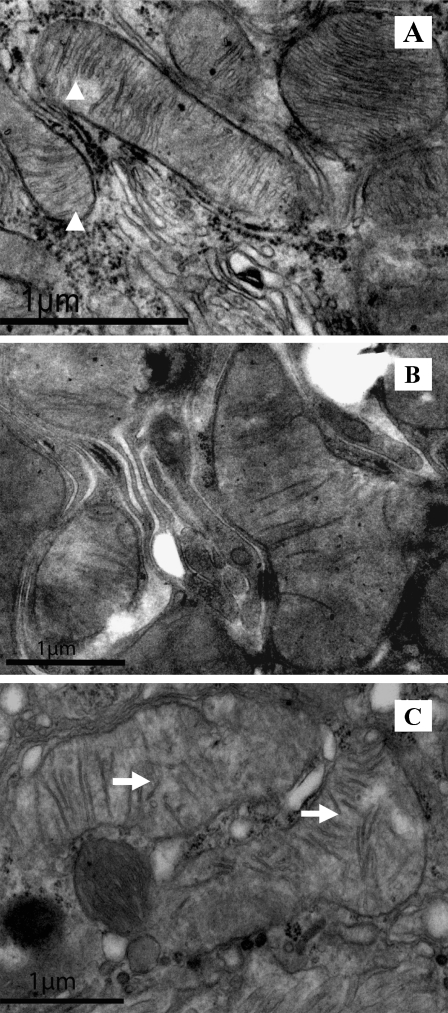
Impaired autophagic homeostasis may result in the accumulation of oxidative damage with age, ultimately leading to aging. Therefore, in this study, oxidative damage for mitochondria in renal tissues was evaluated by analyzing the expression of 8-OHdG, a sensitive biomarker for mitochondrial DNA (mtDNA) oxidative damage (Shigenaga et al. 1989; Richter et al. 1988; Fraga et al. 1990). We found that the level of 8-OHdG was elevated in the kidneys of 24-month-old rats compared with the 3-month-old rats, indicating that oxidative damage in mtDNA increased significantly during the renal aging process (Figs. 3 and 7). A significant accumulation of 8-OHdG was observed in the cytoplasm of the tubular cells of the aged kidneys. We further observed the changes in the mitochondrial structures using transmission electron microscopy. The results showed that the renal cells of 12-month-old rats showed little mitochondrial oxidative damage, whereas the mitochondria in the renal cells of the 24-month-old rats exhibited swelling and disintegration or disruption of mitochondrial cristae (Fig. 8).
Fig. 7.
Detection of 8-OHdG level by immunohistochemistry staining
Fig. 8.
Analysis of mitochondrial structures by transmission electron microscopy in the renal tissues of 3-, 12-, and 24-month-old Fischer 344 rats. Arrowheads indicate regular mitochondria; arrows indicate damaged mitochondria. a Three-, b 12-, and c 24-month-old Fischer 344 rats
Discussion
Autophagy plays an important role in degrading excess long-lived proteins and damaged organelles, especially mitochondria, in the cytoplasm of eukaryotic cells (Levine and Klionsky 2004). Growing evidence indicates that autophagy is essential for maintaining cellular homeostasis and physiological function in tissues and organs (Rajawat et al. 2009). For example, Atg5 and Atg7 knockout mice are unable to form autophagosomes, have abnormal heart function, and die shortly after birth (Komatsu et al. 2007a; Kuma et al. 2004). Recent studies have demonstrated an intimate relationship between autophagy and the aging process (Hara et al. 2006; Komatsu et al. 2005; Nakai et al. 2007). Although it is known that autophagy decreases with age in the liver (Wohlgemuth et al. 2007), the effects of aging on autophagy in the mammalian kidneys have not yet been elucidated.
In the present study, we examined the changes in expression of autophagy-related proteins/genes Atg5, beclin-1/Atg6, Atg7, LC3/Atg8, and p62/SQSTM1 and polyubiquitin aggregates in aging kidneys. The expansion of the membrane to form early autophagosomes is modulated by Atg7, LC3/Atg8, and Atg9, which control the size of the autophagosomes and the amplitude of autophagy. LC3/Atg8 is crucial in the sequestration of large mitochondria and protein aggregates. In this study, we found that expression of Atg7 and LC3/Atg8 was significantly decreased in aged rat kidneys, whereas there was no marked change in the expression of Atg5. These results are consistent with a previous study that reported a decrease in Atg7 expression in older rat hearts (Wohlgemuth et al. 2007). Isolated mitochondria obtained from Atg7−/− skeletal muscle cells exhibit a significant defect in mitochondrial respiration, indicating that there is a close relationship between autophagy, oxidative stress, and aging (Wu et al. 2009a). Similarly, gene expression of Atg5 and Atg7 and protein abundance of LC3 decrease in the extraocular muscle of aged rats (McMullen et al. 2009). In addition, we also indicated that protein expression of beclin-1/Atg6 was slightly but not significantly increased in the aged rat kidneys, which is in line with the earlier report (Kume et al. 2010). Previous studies have found that beclin-1 has a significant increase in the aged rat heart (Wohlgemuth et al. 2007) and skeletal muscle (Wohlgemuth et al. 2010). Changes in beclin-1 are tissue and cell context dependent. The roles of beclin-1/Atg6 are many; for example, it suppresses tumors and is involved in apoptosis. Its specific role in autophagy is defined by direct interactions with proteins that can either promote or inhibit autophagy (Cao and Klionsky 2007; Pattingre et al. 2008). The LC3-II/LC3-I ratio has been used to evaluate the extent of autophagy. We have shown that this ratio is significantly increased in older rat kidneys. Although this increased ratio could be interpreted as indicative of elevated autophagy, it may also be caused by a decreased LC3-I level due to a lack of conversion from proLC3 to LC3-I, or by decreased LC3-II degradation by Atg4 after fusion of autophagosomes and lysosomes.
Accumulating data show that in eukaryotic cells, short-lived proteins are degraded by the ubiquitin–proteasome system, whereas long-lived proteins and damaged organelles are degraded by lysosome-mediated autophagy. The latter plays a pivotal role in controlling aging (Ciechanover 2005; Ward 2002; Martinez-Vicente et al. 2005; Ravikumar and Rubinsztein 2006). Oxidative stress and other conditions lead to misfolding and aggregation of proteins such as polyglutamine and polyalanine expansions, which are degraded by autophagy (Ravikumar et al. 2002, 2004). Autophagy is generally thought of as a nonspecific bulk degradation mechanism. However, a recent study suggested that p62/SQSTM1 may link the recognition of polyubiquitinated protein aggregates to the autophagy machinery (Bjørkøy et al. 2006). A previous study found that when autophagy function is impaired, the expression of p62 is increased (Komatsu et al. 2007b). Therefore, p62/SQSTM1 may be seen as a further marker of the function of autophagy. In this study, we observed an increased level of p62/SQSTM1 and polyubiquitin aggregates in the older rat kidneys (Fig. 6), which is consistent with a previous report (Komatsu et al. 2007b). It has been demonstrated that chaperone-mediated autophagy was decreased in acute diabetes mellitus (Sooparb et al. 2004). Therefore, it is possible that factors other than aging (e.g., diabetes) could lead to the development of renal pathology, and autophagy plays a role in diabetic nephropathy independent of age. However, in our study, the level of serum glucose was unchanged among the three groups, which rules out diabetes as a factor (Table 1).
Many studies suggest that age-related damage is associated with the formation of mitochondrial ROS. Mitochondria are the primary sites of ROS generation, which may be one reason why they are more affected by age than other organelles. mtDNA is more vulnerable than nuclear DNA because it is not protected by histones, it contains a much larger proportion of expressed genes, and it is marked by less efficient repair, at least for some types of lesions (Brunk and Terman 2002). Under normal circumstances, the damaged mitochondria are degraded by autophagy. If the impaired mitochondria are not removed, they can generate more ROS, which further aggravates the oxidative damage, forming a vicious cycle (Cadenas and Davies 2000; Brunk and Terman 2002). Therefore, mitochondria play a central role in tissue damage and the regulation of aging.
Autophagy is the only intracellular degradative mechanism for the removal of damaged mitochondria (referred to as mitophagy). Recent studies have found that when autophagy is suppressed, abnormally large mitochondria accumulate in cultured cardiomyocytes, indicating that dysfunctional, senescent mitochondria accumulate in aging postmitotic cells because they are insufficiently removed for degradation. It has been postulated that those dysfunctional mitochondria may undergo further oxidative damage, resulting in increased ROS production and decreased energy production (Terman et al. 2003). These results suggest that deficiency in autophagy leads to an accumulation of damaged mitochondria (Wohlgemuth et al. 2007; Kume et al. 2010; Tong et al. 2010). There is growing evidence that the efficiency of the degradative system involved in mitochondrial turnover declines with age. The decreased effectiveness of autophagy with increasing age might lead to further mitochondrial damage by ROS and a decrease in ATP production, accelerating the aging process. In this study, we found that the expression of 8-OHdG, a sensitive biomarker for mtDNA oxidative damage, is significantly increased, a finding that is in accordance with a previous report (Miyazawa et al. 2009). Our results also reveal swelling mitochondria in aging rat renal tissues, consistent with these data. Deficiency and dysfunction in autophagy are implicated in the pathogenesis and progression of some renal diseases, for example, glomerulosclerosis, renal ischemia/reperfusion injury, cyclosporine nephrotoxicity, and cisplatin nephrotoxicity (Suzuki et al. 2008; Cheng et al. 2008; Wu et al. 2009b; Yang et al. 2008; Hartleben et al. 2010). Our results are helpful in understanding the role of autophagy in the onset and development of kidney aging and kidney disease and in exploring new therapies that could delay age-related renal disease.
Acknowledgments
This work was supported by a grant (no. 2007CB507403) from the National Basic Research Program of China to X.C., grants (no. 30870920, 30270505, and 30070288) from the National Natural Science Foundation of China to X.Y.B., and a grant (no. 2011CB964904) from the National Key Scientific Program of China to X.Y.B.
Contributor Information
Xue-Yuan Bai, Phone: +86-10-66935462, FAX: +86-10-68130297, Email: xueyuan_bai@yahoo.com.cn.
Xiangmei Chen, Phone: +86-10-66935462, FAX: +86-10-68130297, Email: xmchen301@126.com.
References
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bergamini E, Cavallini G, Donati A, Gori Z. The role of macroautophagy in the ageing process, anti-ageing intervention and age-associated diseases. Int J Biochem Cell Biol. 2004;36:2392–2404. doi: 10.1016/j.biocel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2:138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Aging: ROS or TOR. Cell Cycle. 2008;7:3344–3354. doi: 10.4161/cc.7.21.6965. [DOI] [PubMed] [Google Scholar]
- Brunk UT, Terman A. The mitochondrial–lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur J Biochem. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/S0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- Cheng CH, Chang HR, Chiang CW, Shu KH, Chou MC. Possible mechanism by which rapamycin increases cyclosporine nephrotoxicity. Transplant Proc. 2008;40:2373–2375. doi: 10.1016/j.transproceed.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Peacocke M, Campisi J, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, Peacocke M, Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- Flores I, Blasco MA. A p53-dependent response limits epidermal stem cell functionality and organismal size in mice with short telomeres. PLoS ONE. 2009;4:4934–4943. doi: 10.1371/journal.pone.0004934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci USA. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hartleben B, Gödel M, Meyer-Schwesinger C, Liu S, Ulrich T, Köbler S, Wiech T, Grahammer F, Arnold SJ, Lindenmeyer MT, Cohen CD, Pavenstädt H, Kerjaschki D, Mizushima N, Shaw AS, Walz G, Huber TB. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest. 2010;120:1084–1096. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. Oxidative stress and life span determination in the nematode Caenorhabditis elegans. Ann NY Acad Sci. 2002;959:466–474. doi: 10.1111/j.1749-6632.2002.tb02117.x. [DOI] [PubMed] [Google Scholar]
- Kadowaki M, Karim MR. Cytosolic LC3 ratio as a quantitative index of macroautophagy. Methods Enzymol. 2009;452:199–213. doi: 10.1016/S0076-6879(08)03613-6. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Sovak G, Cuervo AM. Protein degradation and aging. Exp Gerontol. 2005;40:622–633. doi: 10.1016/j.exger.2005.07.005. [DOI] [PubMed] [Google Scholar]
- McMullen CA, Ferry AL, Gamboa JL, Andrade FH, Dupont-Versteegden EE. Age-related changes of cell death pathways in rat extraocular muscle. Exp Gerontol. 2009;44:420–425. doi: 10.1016/j.exger.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa M, Ishii T, Yasuda K, Noda S, Onouchi H, Hartman PS, Ishii N. The role of mitochondrial superoxide anion (O2−) on physiological aging in C57BL/6J mice. J Radiat Res. 2009;50:73–83. doi: 10.1269/jrr.08097. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Rajawat YS, Hilioti Z, Bossis I. Aging: central role for autophagy and the lysosomal degradative system. Ageing Res Rev. 2009;8:199–213. doi: 10.1016/j.arr.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Rattan SI. Increased molecular damage and heterogeneity as the basis of aging. Biol Chem. 2008;389:267–272. doi: 10.1515/BC.2008.030. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Rubinsztein DC. Role of autophagy in the clearance of mutant huntingtin: a step towards therapy? Mol Aspects Med. 2006;27:520–527. doi: 10.1016/j.mam.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenaga MK, Gimeno CJ, Ames BN. Urinary 8-hydroxy-2-deoxyguanosine as a biologic marker of in vivo oxidative DNA damage. Proc Natl Acad Sci USA. 1989;86:9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS. Role of oxidative stress and protein oxidation in the aging process. Free Radic Biol Med. 2002;33:37–44. doi: 10.1016/S0891-5849(02)00856-0. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- Suzuki C, Isaka Y, Takabatake Y, Tanaka H, Koike M, Shibata M, Uchiyama Y, Takahara S, Imai E. Participation of autophagy in renal ischemia/reperfusion injury. Biochem Biophys Res Commun. 2008;368:100–106. doi: 10.1016/j.bbrc.2008.01.059. [DOI] [PubMed] [Google Scholar]
- Terman A, Brunk UT. The aging myocardium: roles of mitochondrial damage and lysosomal degradation. Heart Lung Circ. 2005;14:107–114. doi: 10.1016/j.hlc.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Terman A, Dalen H, Eaton JW, Neuzil J, Brunk UT. Mitochondrial recycling and aging of cardiac myocytes: the role of autophagocytosis. Exp Gerontol. 2003;38:863–876. doi: 10.1016/S0531-5565(03)00114-1. [DOI] [PubMed] [Google Scholar]
- Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, Kelleher RJ, 3rd, Shen J. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proc Natl Acad Sci USA. 2010;107:9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WF. Protein degradation in the aging organism. Prog Mol Subcell Biol. 2002;29:35–42. doi: 10.1007/978-3-642-56373-7_3. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, Dunn WA., Jr Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res. 2007;10:281–292. doi: 10.1089/rej.2006.0535. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Seo AY, Marzetti E, Lees HA, Leeuwenburgh C. Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise. Exp Gerontol. 2010;45:138–148. doi: 10.1016/j.exger.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Quijiao C, Chen E, Liu H, Cao L, Fergusson MM, Rovira II, Gutkind S, Daniels MP, Komatsu M, Finkel T. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging. 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Hsiao TY, Chien CT, Lai MK. Ischemic conditioning by short periods of reperfusion attenuates renal ischemia/reperfusion induced apoptosis and autophagy in the rat. J Biomed Sci. 2009;16:19. doi: 10.1186/1423-0127-16-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yang C, Kaushal V, Shah SV, Kaushal GP. Autophagy is associated with apoptosis in cisplatin injury to renal tubular epithelial cells. Am J Physiol Renal Physiol. 2008;294:F777–F787. doi: 10.1152/ajprenal.00590.2007. [DOI] [PubMed] [Google Scholar]