Abstract
A defining feature of many cancers is deregulated translational control. Typically, this occurs at the level of recruitment of the 40S ribosomes to the 5′-cap of cellular messenger RNAs (mRNAs), the rate-limiting step of protein synthesis, which is controlled by the heterotrimeric eukaryotic initiation complex eIF4F. Thus, eIF4F in particular, and translation initiation in general, represent an exploitable vulnerability and unique opportunity for therapeutic intervention in many transformed cells. In this article, we discuss the development, mode of action and biological activity of a number of small-molecule inhibitors that interrupt PI3K/mTOR signaling control of eIF4F assembly, as well as compounds that more directly block eIF4F activity.
Pharmacological agents that block eukaryotic translation initiation factor eIF4F are potential cancer inhibitors. Some interrupt PI3K/mTOR signaling control of eIF4F assembly; others more directly block eIF4F activity.
It would seem fitting to finish this collection with an article on the topic of “emerging therapeutics in mRNA translation” because much of our current thinking into the molecular biology of this rich field owes a great debt to inhibitors of protein synthesis. The experimental use of antibiotics that block the ribosome at key enzymatic steps has been instrumental in defining general features of translation and identifying paradigms of translational control. From a therapeutic perspective, however, it has only been those antibiotics that inhibit prokaryotic protein synthesis that have had any medical success, contributing significantly to the treatment of bacterial infectious disease (Chambers 2001a,b). It thus had seemed that the use and potential of eukaryotic protein synthesis inhibitors were limited to tools in basic research. This view has changed during the last 20 years of research. We now have a more profound understanding of the complex regulatory apparatus that the eukaryotic cell uses to control messenger RNA (mRNA) translation and, with the emergence of several novel compounds that target steps other than elongation, a greater appreciation of the rewiring of translation factors that occurs in human disease, and most notably, cancer (Silvera et al. 2010). This latter and exciting development forms the basis of this article. For a more complete description of other known protein synthesis inhibitors we refer the reader to Pelletier and Peltz (2007).
TARGETING TRANSLATION INITIATION AS AN ANTINEOPLASTIC APPROACH
The earliest, most varied and most widely studied inhibitors of eukaryotic protein synthesis are those that target the elongation step of mRNA translation (Pestka 1977; Vazquez 1979). Most of them act either by impairing peptidyl transferase function, impeding translocation, or interfering with aminoacyl-tRNA (transfer RNA) binding or accommodation. Despite their structural diversity and nuanced specificities, most eukaryotic elongation inhibitors have shown limited therapeutic value especially when compared to their prokaryotic counterparts, most likely owing to nonspecific toxicity derived from blocking global protein synthesis in nontransformed cells at the doses tested. Even with these apparent limitations, there has been renewed interest in using translation elongation inhibitors as antineoplastics. For example, homoharringtonine (omacetaxine mepesuccinate or HHT), a tree-derived alkaloid from the conifer Cephalotaxus harringtonia, has shown promise in several clinical trials for myelodysplastic syndrome and in phase II clinical trials in patients with gleevec-resistant chronic myelogenous leukemia and in acute myeloid leukemia (AML) (Kantarjian et al. 2001; Kim et al. 2011). HHT is thought to inhibit peptide chain elongation by binding the A site of the ribosome, blocking aminoacyl-tRNA binding, and halting peptide chain elongation (Gurel et al. 2009). Recent experiments have provided some understanding of the possible mechanism of HHT’s mode of action. On treatment of leukemic cells with HHT, the short-lived antiapoptotic protein Mcl-1 was observed to be rapidly degraded, an effect that was solely attributed to the acute loss in overall protein synthesis and that was reversed upon proteasome inhibition (Tang et al. 2006; Robert et al. 2009; Chen et al. 2011a). This, and perhaps the degradation of other short-lived prosurvival factors, explains some of the synergistic effects observed with different inhibitors of translation elongation in the Burkitt’s-like Eμ-Myc lymphoma mouse model and would warrant greater study and further evaluation of these drugs in combination therapies (Robert et al. 2009).
Other elongation inhibitors have had less clinical success when applied toward the treatment of cancer owing to unacceptable toxicities or poor pharmacological properties limiting their therapeutic window (Dumez et al. 2009). Not surprisingly, the current trend in the development of therapeutic agents that disrupt translation has thus drifted away from inhibitors of elongation to those that target initiation, potentially shifting the pharmacological response from global protein synthesis to a more selective (and possibly more cancer cell-dependent) translationally regulated effect.
eIF4F and Tumorigenesis
The eIF4F translation complex is currently at the forefront of the development of pharmacological agents that block initiation. Briefly, eIF4F is a heterotrimeric complex composed of eIF4E, the m7GpppN cap-binding protein that anchors the complex to the 5′-end of the mRNA; eIF4A, an RNA DEAD box helicase that is thought to unwind RNA secondary structure surrounding the cap and increase the efficiency of ribosome binding; and eIF4G, a large scaffolding protein that bridges eIF4F to the 43S ribosomal complex (see Lorsch et al. 2012). Given eIF4E’s limited abundance, the recruitment of the 43S ribosomal complex to the mRNA by eIF4F is thought to be rate limiting for translation initiation (Duncan et al. 1987). The interaction of eIF4F with the cap structure is affected by mRNA proximal secondary structure with increased structure diminishing the efficiency of the eIF4E-cap interaction and/or imposing a structural barrier to the weak eIF4A helicase activity (Pelletier and Sonenberg 1985b; Lawson et al. 1986). Consequently, altering the levels of eIF4F can influence which mRNAs are more readily translated (Lawson et al. 1986, 1988), correlating with the degree of secondary structure in the 5′-untranslated regions (5′-UTRs) of the mRNA—the greater the thermal stability, the more poorly the transcript is translated, presumably by obstructing efficient ribosome loading and 48S complex formation (Pelletier and Sonenberg 1985a,b; Lawson et al. 1986; Babendure et al. 2006). Increasing the amounts of cellular eIF4E can thus disproportionately stimulate the expression of messages that were once outcompeted by unstructured mRNAs, encoding proteins that are often progrowth and prosurvival in nature. In fact, this is thought to be the underlying mechanism behind eIF4E’s tumorigenic properties, the selective increase in translation of a limited set of oncogenic and metastatic transcripts (Lazaris-Karatzas et al. 1990; Ruggero et al. 2004; Wendel et al. 2004). The role that eIF4E plays in cancer has received much broader scientific and clinical interest of late since the discovery that its function is regulated by mTOR (mammalian target of rapamycin), a master regulator of cellular homeostasis and key signaling node often up-regulated in cancers, and that the chemotherapeutic rapamycin, its highly specific inhibitor, can block its activity (Kunz et al. 1993; Sabatini et al. 1994; see Dobson et al. 2012 for details).
Therapeutic Strategies Targeting Regulation of eIF4F Assembly
Rapalogs (Rapamycin Analogs)
mTOR is a member of the phosphoinositide 3-kinase (PI3K) -related protein kinase (PIKK) family and is the catalytic subunit of two functionally distinct complexes: mTORC1 and mTORC2—defined by the nature of interacting accessory proteins. mTORC2 integrates cell survival, proliferation, lipogenesis (as well as other catabolic processes), and cytoskeletal organization to growth factor signaling, primarily through phosphorylation of members of the AGC class of kinases upstream of mTORC1. mTORC1 serves as a regulator of eIF4F assembly and translation initiation, sensing the energy and nutrient status of a cell and transmitting either a growth or starvation response to the translation apparatus (Sengupta et al. 2010). Briefly, mTOR stimulates eIF4F formation through two mechanisms: (1) It phosphorylates eIF4E-binding proteins (4E-BPs; of which there are three, the most prominent being 4E-BP1 and 4E-BP2), which prevents them from disrupting the eIF4E/eIF4G interaction, and (2) mTOR signals the degradation of PDCD4, which interferes with the eIF4A/eIF4G interaction (Gingras et al. 1999b; Dorrello et al. 2006). Thus, active mTOR promotes eIF4F formation and translation initiation.
The identification of germline mutations in genes that encode negative regulators of mTOR signaling (e.g., Pten and Tsc1/2), the fact that rapamycin possesses antiproliferative activity against a number of cancer cell lines (Hidalgo and Rowinsky 2000), and the observation that a majority of human cancers arise owing to activated mTORC1 signaling (Yuan and Cantley 2008), emphasized the need to uncover the role of mTOR in human cancer. Importantly, it is well established that mTOR control of eIF4F assembly acts as a critical node for cancer cell survival and proliferation (Wendel et al. 2004; Mills et al. 2008).
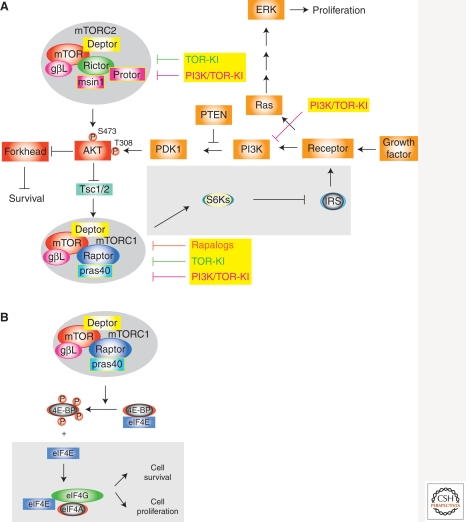
The rapalogs temsirolimus and everolimus have been clinically approved for the treatment of metastatic renal cell carcinoma, whereas temsirolimus has also been approved for the treatment of mantle cell lymphoma (Table 1). Currently, rapalogs are under investigation as cancer therapies in numerous clinical trials (www.clinicaltrials.gov). There are, however, limitations to rapamycin-based therapies. Among these is the presence of a p70S6K-IRS-1 (insulin receptor substrate-1) negative-feedback loop (Fig. 1A) (Harrington et al. 2005). The mTOR substrate p70S6K normally suppresses PI3K signaling by inactivating IRS-1 as well as platelet-derived growth factor receptor (PDGFR) and uncoupling PI3K from upstream growth factor signals (Harrington et al. 2005). Thus, exposure of tumor cells to rapamycin leads to inhibition of p70S6K activity and subsequent stimulation of upstream PI3K signaling—an unfavorable situation for cancer therapy because this activates several mTORC1-independent prosurvival and proliferative signals and is associated with treatment failure (O’Reilly et al. 2006). Additionally, the mitogen-activated protein kinase (MAPK) pathway can be activated upon mTORC1 inhibition via the p70S6K /PI3K/Ras pathway and indeed MAPK activation has been noted in human tumors following RAD001 treatment (Fig. 1A) (Carracedo et al. 2008).
Table 1.
Compounds that affect eIF4F assembly or activity
| Targets | Compound | Sponsor | Stage of development |
|---|---|---|---|
| mTORC1 | Rapamycin (Sirolimus) | Pfizer | FDA approved |
| RAD001 (Everolimus) | Novartis | FDA approved | |
| CCI-779 (Temsirolimus) | Pfizer | FDA approved | |
| AP23573 (Ridaforolimus) | Merck/ARIAD | Accepted for FDA approval | |
| TOR-KI | Torin1 | N/A | Ex vivo cell culture |
| INK128 | Intellikine | Phase I | |
| AZD8055 | AstraZeneca | Phase I/II | |
| AZD2014 | Phase I | ||
| OSI027 | OSI Pharmaceuticals | Phase I | |
| PI3K/TOR-KI | PI-103 | N/A | Ex vivo cell culture |
| NVPBEZ235 | Novartis | Phase I/II | |
| SF1126 | Semafore | Phase I | |
| GSK2126458 | Glaxo Smith Kline | Phase I | |
| XL765 | Exelixis | Phase I/II | |
| BGT226 | Novartis | Phase I/II | |
| GDC0980 | Genetech | Phase I | |
| PF04691502 | Pfizer | Phase I | |
| PKI587 | Pfizer | Phase I | |
| eIF4F | Cap analogs | N/A | In vitro studies |
| 4Ei-1 | N/A | In vitro studies | |
| 4EGI-1 | N/A | Ex vivo cell culture | |
| 4E1RCat | N/A | Active in preclinical models | |
| 4E2RCat | N/A | Active in preclinical models | |
| ISIS-EIF4ERx | Eli Lilly/ISIS | Phase II | |
| Pateamine A | N/A | Ex vivo cell culture | |
| DMDA-Pat A | N/A | Active in preclinical models | |
| Hippuristanol | N/A | Active in preclinical models | |
| Rocaglamides (Silvestrol) | N/A | Active in preclinical models | |
| Mnk | Cercosporamide | Eli Lilly | Active in preclinical models |
| CGP57380 | Novartis | Ex vivo cell culture |
Figure 1.
Diagram illustrating rapamycin-resistance pathways. (A) Inhibition of mTORC1 by rapalogs leads to dampening of the S6K/IRS feedback loop (in gray box) and increased signaling flux to PI3K and Ras. (B) Increased levels of eIF4E can circumvent effects on cell proliferation and cell survival (in gray box) mediated by inhibition of mTORC1 by rapamycin. See text for details.
Rapamycin resistance can also be imparted by overexpression of eIF4E, which has been identified as a genetic modifier of the rapamycin response (Fig. 1B) (Wendel et al. 2004, 2006; Mills et al. 2008). Given that neither 4E-BP phosphorylation status nor p70S6K activity serve as reliable predictive markers of rapamycin sensitivity in human cancers (Noh et al. 2004; Satheesha et al. 2011), eIF4E expression levels should be assessed as a potential marker to inform on rapamycin resistance (Satheesha et al. 2011). These results suggest that direct inhibitors of eIF4E (and eIF4F) may synergize with rapamycin to diminish resistance.
TOR-Kinase Inhibitors (TOR-KI)
Rapamycin acts through an unusual allosteric mechanism wherein it binds FKBP12 and it is this dimer that then interacts with mTOR (Chen et al. 1995). Binding of FKBP12-rapamycin to mTOR induces a conformational change that specifically weakens the mTOR-Raptor interaction (Kim et al. 2002). In vitro kinase assays with mTORC1 have shown that prolonged incubation with rapamycin is necessary to inhibit 4E-BP1 phosphorylation (Burnett et al. 1998), which differs kinetically from what is observed with p70S6K, where rapamycin rapidly blocks mTORC1 phosphorylation of p70S6K. This is recapitulated in vivo, where rapamycin fully suppresses p70S6K phosphorylation but fails to completely dephosphorylate 4E-BP1 (Choo et al. 2008). The molecular basis responsible for these different phosphorylation kinetics is not well understood but may relate to rapamycin-induced structural changes in mTOR that exert more profound effects on mTOR-p70S6K association than mTOR-4E-BP association, allowing uninterrupted signaling flux in the presence of rapamycin. Consistent with such a model is the finding that raptor binds 4E-BP1 more efficiently than p70S6K (Hara et al. 2002). Furthermore, the FKBP12-rapamycin complex fails to effectively suppress mTORC2 kinase activity, limiting its use as a fully active inhibitor of mTOR (Sarbassov et al. 2004).
These issues prompted development of mTOR kinase inhibitors to block both mTOR complexes. Indeed several novel mTOR inhibitors were identified that specifically compete with ATP for access to the kinase active site and some of these compounds are currently under intensive clinical development (Table 1) (Wander et al. 2011). The TOR-KIs are superior to rapalogs in their ability to suppress growth, proliferation, and protein synthesis (Feldman et al. 2009; Thoreen et al. 2009) and they target mTOR irrespective of its protein-binding partners. One important and unanticipated result from these studies is that at least two TOR-KIs, Torin1 and PP242, more potently inhibit 4E-BP1 phosphorylation than do the rapalogs (Feldman et al. 2009; Thoreen et al. 2009). These results prompted a reexamination of how 4E-BP phosphorylation is regulated by mTOR. It is known that mTORC1 phosphorylates 4E-BP1 in a hierarchical manner, although there is some disagreement in the literature about how this occurs (Gingras et al. 1999a, 2001; Choo et al. 2008). In vitro, mTORC1 can phosphorylate 4E-BP1 on Thr37/46 (Burnett et al. 1998). Yet in some settings, rapamycin fails to block the phosphorylation of these sites in vivo (Choo et al. 2008). In contrast, rapamycin is very effective at allowing dephosphorylation of 4E-BP1 on Ser 65 and Thr 70 in vivo (Gingras et al. 1999a; Choo et al. 2008). The ability of rapamycin to suppress translation is variable between cell lines and correlates with differences in the extent of 4E-BP1 phosphorylation (Beretta et al. 1996; Choo et al. 2008).
In contrast, TOR-KIs rapidly and fully inhibit mTOR-mediated phosphorylation of both 4E-BP1 and p70S6K and notably offer an improvement in their ability to suppress translation initiation (Feldman et al. 2009; Garcia-Martinez et al. 2009; Thoreen et al. 2009; Yu et al. 2010). Fully dephosphorylated 4E-BP1 is much better at inhibiting eIF4E function and translation initiation than its partially dephosphorylated variants (Mothe-Satney et al. 2000). Inhibition of mTORC1 with TOR-KIs suppresses cell proliferation in an mTORC1/4E-BP1/2-dependent manner, whereas cell growth inhibition is mediated by mTORC1/p70S6K (Dowling et al. 2010). Oddly, however, in the absence of 4E-BP1/2, rapamycin can still mediate a partial block of cell proliferation suggesting that rapalogs influence mTOR activity through a more complex mechanism than is currently appreciated (Dowling et al. 2010).
Dual-Specificity PI3K/TOR-KI
A more recent class of mTOR inhibitors that also target PI3K has been characterized and entered into clinical development (Table 1) (Wander et al. 2011). The added advantage of these compounds over rapalogs is that they have the potential to block reactivated PI3K following p70S6K inhibition. One limitation of these inhibitors is specificity. It turns out that some of these drugs have off-target inhibitory effects for related PIKK family members owing to shared structural overlap (Toledo et al. 2011). Although the effectiveness of this class of compounds at inhibiting eIF4F formation remains to be carefully assessed, an interesting link between the PI3K pathway and MYC has recently emerged. Elevated eIF4E and c-MYC levels can lead to PI3K/TOR-KI resistance (Ilic et al. 2011). In a breast cancer setting, MYC amplification leads to PI3K-independent tumor cell survival and resistance to PI3K inhibitors (Liu et al. 2011). A chemical genetics screen further validated c-MYC activation as a means to overcome the proliferative block induced by PI3K/TOR-KIs (Muellner et al. 2011). Elevated MYC and eIF4E levels are expected to increase eIF4F formation independent of mTOR (Jones et al. 1996; Rosenwald 1996; Lin et al. 2008) suggesting that inhibition of translation initiation may be a prominent mechanism by which PI3K/TOR-KIs exert their effects. Taken together, these results suggest that targeting components of the translation apparatus directly under mTOR control would be one avenue to overcome this resistance.
Therapeutic Strategies Directly Targeting eIF4F
There are several strategies under way to develop drugs that impede eIF4F activity, with efforts targeting different functions: (1) competing for eIF4E binding to the m7GpppN cap structure, (2) uncoupling the eIF4E-eIF4G interaction, (3) targeting eIF4E production, (4) blocking eIF4A activity, and (5) inhibiting eIF4E phosphorylation. All of these have diverse effects on cellular and therapeutic outcomes and all have shown some promising results.
Blocking eIF4E-Cap Interaction
The archetypal inhibitors of eIF4E activity in the field of mRNA translation are undoubtedly the cap analogs (Shatkin et al. 1982; Grudzien-Nogalska et al. 2007b). At the level of translation, these act by outcompeting nascent capped mRNA transcripts for eIF4E binding and preventing the preinitiation ribosomal complex from binding and commencing translation. The original competitors, first described over 30 years ago, were simple m7GDP or m7GTP derivatives. Now more than 75 analogs have been synthesized and extensively tested in vitro, with diverse chemical modifications from simple substitution of the methylated residue to bulkier aromatic groups and remodeling of the nucleotide backbone (Hickey et al. 1977; Adams et al. 1978; Darzynkiewicz et al. 1981, 1987, 1989; Cai et al. 1999; Grudzien-Nogalska et al. 2007a,b; Kowalska et al. 2008; Jemielity et al. 2010; Su et al. 2011). The best compounds that bind to eIF4E are, not surprisingly, those that model the natural mRNA cap residue—minimally, a m7GpppN-like molecule—preserving the contacts between the π-π electron stacking of the eIF4E tryptophan residues (W56 and W102) and the m7G ring, as well the electrostatic attractions between the basic residues of eIF4E (R122, R157, and K162) and the 5′-5′ triphosphate bridge (Marcotrigiano et al. 1997; Niedzwiecka et al. 2002).
These compounds have been invaluable for the biophysical and biochemical study of eIF4E in vitro but their application in vivo has been limited owing to poor cross-membrane transport and low cellular stability arising from both intracellular and extracellular hydrolysis (Wagner et al. 2000; Jemielity et al. 2010). Two recent synthetic routes have been developed to circumvent such limitations, both involving modification of the phosphate backbone with nonnatural functional groups. One study focused primarily on altering the α, β, and γ phosphate groups to prevent hydrolysis by the decapping enzymes DcpS and/or Dcp1/2 (Grudzien-Nogalska et al. 2007a; Kowalska et al. 2008; Su et al. 2011). When attached at the 5′ end of a luciferase-driven construct, a phosphoborate moiety introduced at the β position supported translation efficiencies similar to that of the parent m2 7,2′GpppG compound but imparted greater mRNA stability in vivo (Su et al. 2011)—the latter attributed to a combined effect of protection from decapping by association with eIF4E (Schwartz and Parker 2000) and increased resistance to endogenous Dcp1/2 (and probably DcpS as well) degradation.
Similarly, but with an emphasis toward more therapeutically viable eIF4E cap analog-based inhibitors, Ghosh et al. (2009) have published the synthesis of phosphoramidate derivates of m7GTP. These “pronucleotides” harbor protecting groups that, on removal by endogenous phosphoramidases (histidine triad nucleotide-binding proteins), are converted to active derivatives. These cap analogs were able to inhibit cap-dependent translation in vitro in reticulocyte lysates, as well as in vivo when coinjected with reporter constructs into fertilized zebrafish eggs (Ghosh et al. 2009). Intriguingly, one of these (4Ei-1) was also able to block eIF4E-dependent epithelial-to-mesenchymal (EMT) transition of these eggs. Although promising, their therapeutic relevance remains unclear because both classes of cap derivatives appear unable to cross phospholipid bilayers (requiring either lipofection or ectopic injection), which precludes their use in more clinically relevant assays. Moreover, the use of cap analogs could influence cellular physiology more broadly than just via eIF4E-dependaent protein synthesis: For example, processes that rely on the nuclear cap-binding proteins CBP20 and CBP80, such as NMD, nuclear export, splicing, mRNA 3′-end formation, and miRNA processing, may also be affected (Kim et al. 2008; Maquat et al. 2010). Alternate pharmacological routes may therefore be required to more directly inhibit eIF4E function.
Uncoupling eIF4E-eIF4G Interaction
As mentioned previously, a majority of the driver mutations in cancers up-regulate mTOR signaling, which ultimately promotes the eIF4E-eIF4G interaction. Reverting this dysregulation using nonphosphorylatable 4E-BP constructs often halts tumor cell proliferation and limits the extent of disease progression in mouse models, suggesting that inhibitors designed to prevent the binding of eIF4E to eIF4G would be therapeutically beneficial. Several modified peptides encompassing the eIF4G/4E-BP hydrophobic eIF4E-binding motif coupled to cell-penetrating moieties have been developed and most have proven successful in various model systems with few nonspecific effects (Herbert et al. 2000; Ko et al. 2009; Brown et al. 2011). Most of these peptides bind fairly efficiently to tagged recombinant eIF4E (in the low- to mid- nanomolar range), can inhibit cap-dependent translation in vivo and induce cancer cell lines to undergo apoptosis. In one impressive example, a 4E-BP-like peptide coupled to a gonadotropin-receptor agonist dramatically decreased tumor burden following intraperitoneal (i.p.) injection in an ovarian cell xenograft mouse model with seemingly no detectable toxicity (Ko et al. 2009).
In a search for small molecules that can disrupt the eIF4E-eIF4G interaction, the Wagner group has designed a high-throughput screen. To identify such compounds, chemical libraries were assayed that decreased the fluorescence polarization of a labeled eIF4G-like peptide when bound by recombinant eIF4E (Moerke et al. 2007). The most potent hit, called 4EGI-1, was evaluated further and found to inhibit cap-dependent translation in extracts, down-regulate known eIF4E-dependent transcripts in vivo, and trigger apoptosis in several tumor lines (Moerke et al. 2007). Based on nuclear magnetic resonance (NMR) spectra, 4EGI-1 was shown to directly bind eIF4E. Counterintuitively, 4EGI-1 did not appear to prevent 4E-BP1 from binding to eIF4E but rather promoted the interaction. Whether or not this “gain-of-function” activity contributes to some of the in vitro and in vivo effects observed remains to be tested. 4EGI-1 has biological properties associated with it that appear to be independent of its effects on cap-dependent translation (Fan et al. 2010; Pruvot et al. 2011) and medicinal chemistry efforts are now required to remove these features from 4EGI-1.
We used a similar strategy in which the eIF4E-eIF4G interaction was monitored using time-resolved fluorescence resonance energy transfer and identified several eIF4E-eIF4G inhibitors, among which the most potent were 4E1RCat and 4E2RCat (Cencic et al. 2011a,b). 4E1RCat blocked cap-dependent but not hepatitis C virus (HCV) internal ribosome entry site (IRES) -dependent translation of a bicistronic dual-luciferase reporter mRNA with an IC50 of ∼25 µM. Like 4EGI-1, 4E1RCat can prevent the association of eIF4G with m7GTP-Sepharose-bound eIF4E but, unlike 4EGI-1, it also blocked 4E-BP1 binding with nearly the same efficacy (Cencic et al. 2011b). 4E1RCat was also pharmacologically active in cells and in mice, where it decreased the rate of overall protein synthesis by ∼30%. Most strikingly, 4E1RCat was able to improve the response to chemotherapy in the Eμ-myc mouse lymphoma model and thus prolong tumor-free survival. This synergy was probably owing to 4E1RCat’s ability to lower Mcl-1 levels, an effect that it shares in common with other inhibitors of translation in vivo.
Targeting eIF4E Levels with Antisense Oligonucleotides (ASOs)
Early proof of principle using antisense-based approaches showed the feasibility of knocking down eIF4E levels to limit the tumorigenic potential of transformed cells, although these experiments often suffered from lack of specificity controls or used short-lived biomolecules (De Benedetti et al. 1991; Rinker-Schaeffer et al. 1993; DeFatta et al. 2000). Second generation ASOs against eIF4E have largely overcome such pitfalls by incorporating extensive phosphate backbone modifications imparting improved nuclease resistance and tissue stability to allow for effective systemic therapeutic delivery. Second generation ASOs targeting eIF4E have been made and results are encouraging (Graff et al. 2007). Despite decreasing overall protein synthesis only minimally (<20%), inhibition of known eIF4E-specific progrowth and prosurvival genes decreased in a dose-dependent manner, halting proliferation and eliciting apoptosis. Impressively, administration of the most potent eIF4E ASO (4E-ASO4, >80% reduction in eIF4E expression) showed pronounced in vivo activity in breast and prostate xenograft models, blunting tumor onset and growth and restricting endothelial cell tube formation, suggesting that eIF4E may also be a possible antiangiogenic target. Thus, targeting eIF4E expression directly might prove to be more sensitive and selective toward tumors through specific down-regulation of oncogenes under eIF4F translational control and afford a much wider therapeutic index and control.
Inhibiting eIF4A Helicase Activity
Another way to inhibit eIF4F-dependent translation initiation is to target eIF4A, the key enzymatic component of the complex. Although both eIF4A’s helicase and ATPase functions are required for efficient ribosome loading and scanning, the consequences of eIF4A enzymatic activity in the mechanism of translation initiation is yet to be precisely defined. It has been suggested that eIF4A is required for unwinding 5′-proximal secondary structure to facilitate 40S ribosome recruitment; that eIF4A actively participates in the preinitiation complex scanning of the 5′UTR; and/or that hydrolysis of ATP may simply be necessary to rearrange protein–protein or protein–mRNA interactions (Rogers et al. 2002; Kapp and Lorsch 2004).
Three small-molecule inhibitors, pateamine A (Pat A), hippuristanol, and silvestrol (and related rocaglamide family members) have been characterized extensively as inhibitors of eIF4A and are currently being explored as potential chemotherapies. All of these compounds were originally discovered and purified from natural sources and all were initially characterized as having strong tumor cell-line-specific cytotoxic properties even though their shared molecular target was then unknown (Higa et al. 1981; Gonzalez et al. 2001; Hood et al. 2001; Hwang et al. 2004). It was fortuitous that all three were identified in a screen designed to identify compounds that could differentially distinguish cap-driven versus IRES-driven translation in Krebs II ascites cell extracts (Novac et al. 2004; Bordeleau et al. 2005, 2006a, 2008). More specifically, they were all able to inhibit the expression of the first cap-dependent luciferase cistron (as part of a dual-luciferase bicistronic capped mRNA transcript) but not the eIF4A-independent HCV-IRES-driven downstream cistron, which provided the first clue that all three were specifically blocking an eIF4A-dependent process. NMR (Lindqvist et al. 2008), affinity selection using immobilized compounds (Bordeleau et al. 2005), and in vitro assays using purified eIF4A (Bordeleau et al. 2008) showed that each was able to bind and affect eIF4A activity directly. Even though all three compounds share a common target, they each have distinct mechanisms of action.
Hippuristanol is an allosteric inhibitor of eIF4A: it prevents both free eIF4 (eIF4Af) and eIF4F complex bound eIF4A (eIF4Ac) from binding RNA, which in turn inhibits eIF4A helicase and ATPase activities (Bordeleau et al. 2006a). Consequently, ribosome loading on exogenous transcripts is prevented as eIF4F activity is greatly reduced. Pat A and silvestrol, on the other hand, promote the binding of recombinant eIF4A to RNA and stimulate by several fold the ATP hydrolysis activity of eIF4A (Bordeleau et al. 2005, 2006b, 2008; Low et al. 2005, 2007). Although perhaps perplexing at first glance, because both are as efficient in blocking ribosome loading as hippuristanol, it seems that either Pat A or silvestrol can only exert this effect on eIF4Af, effectively limiting the amount of eIF4Ac, by sequestering eIF4Af nonspecifically on cellular RNAs (Bordeleau et al. 2005). Intriguingly, Pat A and hippuristanol bind to different regions on eIF4A (the binding of silvestrol has not yet been determined). Hippuristanol appears to make specific contacts in the carboxy-terminal domain (CTD) of eIF4A (Lindqvist et al. 2008), and whereas the exact binding site of Pat A is not well defined, Pat A does not appear to interact with the CTD of eIF4A (M Oberer, J Pelletier, and G Wager, unpubl.) and mutational analysis of eIF4A indicates that it requires the amino-terminal ATP-binding domain of eIF4A for its activity (Low et al. 2005). Similarly, a recently characterized bacterial toxin, BPSL1549, from Burkholderia pseudomallei was shown to deamidate Gln339 of eIF4A, a conserved residue in motifs V and VI within a loop flanking eIF4A’s ATP- and RNA-binding sites (Cruz-Migoni et al. 2011). In fact, this maps precisely to the same region whose residues experienced the strongest chemical shifts in NMR on addition of hippuristanol. The Gln339 modification appeared to transform endogenous eIF4A into a dominant-negative mutant, robustly blocking cellular protein synthesis while inhibiting eIF4A helicase activity and preventing eIF4A recycling (Cruz-Migoni et al. 2011).
Hippuristanol suffers from poor solubility and low potency and thus in vivo studies are scarce, although novel synthetic routes will hopefully resolve these issues (Li et al. 2009; Ravindar et al. 2010, 2011). In one study, hippuristanol was able to selectively inhibit the proliferation of HTLV-1-infected T-cell leukemia cells, both in culture as well as when injected into immunodeficient mice (Tsumuraya et al. 2011). Pat A suffers from a different problem in that it seems to inhibit protein synthesis in an irreversible manner, both in vitro and in vivo, which might make Pat A more difficult to develop for further clinical use (Bordeleau et al. 2005). Nevertheless, in one report, a desmethyl desamino analog of Pat A (DMDA-Pat A) displayed single-agent activity against several human cancer cell xenografts in nude mice, although it appears to inhibit DNA synthesis as well, substantially differing in its mechanism of action from its precursor (Kuznetsov et al. 2009).
In terms of preclinical mouse models, silvestrol is at present the best-studied eIF4A inhibitor, mostly owing to good pharmacological tolerance and little apparent nonspecific toxicity in animals (Cencic et al. 2009a; Saradhi et al. 2011). Silvestrol has been tested in multiple settings, from cancer cell line xenografts to oncogene-derived genetic mouse models with encouraging results. It has been tested in xenografts of acute lymphoblastic leukemia (697 ALL), prostate (PC-3), and breast (MDA-MBA-231) cancer lines as well as the Eμ-Tcl-1 (CLL) and Eμ-Myc lymphoma mouse models (Bordeleau et al. 2008; Cencic et al. 2009a; Lucas et al. 2009). In almost all of these cases, administration of silvestrol as single-agent chemotherapy elicited apoptosis and caused tumor regression. In one study, it was observed that silvestrol appeared to be far more toxic toward B cells than T cells, an effect that was magnified in blood samples of CLL patients relative to normal healthy individuals, which suggests preferential killing of proliferating leukemic cells by the drug (Lucas et al. 2009). Likewise, silvestrol treatment alone showed nanomolar in vitro potency against Tsc2−/−Eμ-Myc lymphoma cultures, even in cells rendered rapamycin-resistant by overexpression of the PIM2 kinase (Schatz 2011). In contrast to these studies, silvestrol treatment of Pten+/−Eμ-Myc-derived tumors had minimal effect when used alone, perhaps as a result of the lower doses used (Bordeleau et al. 2008). When combined with doxorubicin, however, silvestrol greatly prolonged the tumor-free survival of mice, matching the synergy observed for combination rapamycin treatment (Bordeleau et al. 2008). Moreover, this combination therapy was even effective against eIF4E-driven Eμ-Myc lymphomas, which do not respond to monotherapy or combination chemotherapy treatment (Bordeleau et al. 2008). This is also what is seen for human AML cell lines, where silvestrol synergizes with the standard-of-care drugs daunorubicin and cytarabine (Cencic et al. 2009b). Finally, as with other inhibitors of translation, silvestrol’s ability to chemosensitize tumors correlates with its ability to inhibit synthesis of Mcl-1 protein. Further improvement of silvestrol’s pharmacology is becoming more feasible as new synthetic routes are helping uncover which structural features determine the activity of rocaglamides (El Sous et al. 2007; Gerard et al. 2007; Roche et al. 2010).
Preventing eIF4E Phosphorylation through Mnk Inhibition
Unlike the 4E-BP/mTOR axis of eIF4E regulation, the molecular and biological consequences of direct phosphorylation of eIF4E have only recently begun to be understood. eIF4E is phosphorylated on a single residue, Ser 209, by two kinases: the MAP kinaselike interacting kinases (Mnk) 1/2 (Pyronnet et al. 1999), which are both activated by the MAPK growth-induced and p38 stress-induced signaling pathways (Buxade et al. 2008). Exactly how the added phosphate residue alters eIF4E activity is not well understood. Theories ranging from increased “clamping” of eIF4E to the cap to greater stability in the eIF4F complex have thus far yielded few conclusive results. In lower organisms, eIF4E phosphorylation seems vital for organismal development, yet in mouse models loss of eIF4E phosphorylation has no noticeable phenotype (Ueda et al. 2004). What is emerging, however, is that eIF4E phosphorylation appears to be important in cancer—overexpression or knock-in of an S209A eIF4E mutant profoundly blunts tumor formation (Topisirovic et al. 2004; Wendel et al. 2007; Furic et al. 2010), which, in some circumstances, appears to rely on phospho-eIF4E’s ability to regulate Mcl-1 expression levels (Wendel et al. 2007). This suggests that small-molecule inhibitors of the Mnk kinases might yield novel anti-cancer agents. Recently, a novel Mnk inhibitor, cercosporamide, was identified from a screen of 300,000 compounds using an in vitro Mnk1 kinase reaction (Konicek et al. 2011). Cercosporamide is more selective for Mnk2 than Mnk1 (IC50 = 0.011 µM versus IC50 = 0.116 µM), also potently inhibits Jak3 (IC50 = 0.031 µM), and is orally bioavailable, acting on tissues (e.g., liver) within 30 min after administration (Konicek et al. 2011). The compound was found to inhibit eIF4E phosphorylation in a dose-dependent manner, block cellular proliferation, and engage apoptosis (Konicek et al. 2010). In a xenograft colon (HCT116) and B16 lung metastasis model, treatment reduced tumor progression and incidence only modestly, probably reflecting the rebound in eIF4E phosphorylation seen after >4 h treatment in mouse tissues. Defining which cancers depend on eIF4E phosphorylation for tumor maintenance will be key in determining whether Mnk1/2 inhibition will become a therapeutically viable option.
Strategies Targeting Ternary Complex Formation
A second key point of regulation of translation initiation is through phosphorylation of eIF2α, an event that limits the availability of the ternary complex, (eIF2•GTP•tRNAiMet), integral for proper start codon selection and the formation of a competent 43S ribosomal scanning complex. eIF2α can be phosphorylated by four different intracellular kinases (PERK, PKR, GCN2, and HRI), which respond to a variety of cellular stress conditions (e.g., amino acid starvation, viral infection, oxidative stress, excess unfolded proteins, heme deficiency) to limit the protein synthetic load in a cell. Given the fact that cancer cells continuously grow, divide, and catabolize, they are generally under tremendous stress to maintain appropriate protein homeostatic load, a complex balancing act between protein biosynthesis, folding, translocation, assembly/disassembly, and clearance. Thus, relative to normal cells, tumors have constitutively higher levels of eIF2α phosphorylation (Rosenwald et al. 2003, 2008). Compounds designed to exploit this discrepancy between malignant and normal cells by tipping the scales of eIF2α phosphorylation could yield therapeutic benefit (and, arguably, may already form part of the mechanism of action of the approved drug bortezomib). Classic reagents used to induce the phosphorylation of eIF2α in cell culture are often too toxic for animal use (e.g., tunicamycin or thapsigargin) and thus finding novel compounds that can hinder ternary complex formation through alternative means is necessary. In a recent screen aimed to ferret out such chemicals, a group of novel activators of HRI was discovered (Chen et al. 2011b). These similarly structured N,N′-diarylurea compounds, the most potent being BTdCPU, were able to specifically activate eIF2α phosphorylation and downstream stress-associated pathways in a variety of conditions. Importantly, all appeared to cause eIF2α phosphorylation through direct activation of HRI and not through nonspecific oxidative damage, the usual route to HRI activation. Moreover, BTdCPU inhibited the growth of tumor cell lines, both in culture and in xenografts in nude mice, an effect that correlated with the induction of eIF2α phosphorylation. BTdCPU appeared to be well tolerated in vivo with little apparent weight loss and surprisingly minimal impact on the hematopoietic system. Whether BTdCPU will develop into an effective anti-cancer agent will depend crucially on the levels of expression of HRI in a given tumor cell. Still, its relatively potent in vivo activity means that it will prove a valuable tool in elucidating the impact of eIF2α phosphorylation and ternary complex formation on both normal cellular physiology and in various disease settings. It will also be interesting to see whether more direct and specific inhibitors of ternary complex formation (like NSC119889 [Robert et al. 2006]) will be able to target a more diverse set of tumor cell types.
FUTURE PERSPECTIVES
As the development of compounds targeting protein synthesis begins to transition away from preclinical models, the need for assays and biomarkers to reliably gauge drug efficacy and patient stratification becomes most pressing. Unlike the PI3K/TOR-KI inhibitors, where recent work on new pathway-based phosphoprofiling methodologies has proven quite successful in translation into the clinic (Andersen et al. 2010), biomarker readouts for eIF4F activity are less obvious. Polymerase chain reaction (PCR) -based methodologies coupled to next-generation sequencing are sensitive enough to measure polysome-bound mRNAs and may enable one to correlate drug treatment to tumor response on small biopsy specimens while informing on the relative tumorigenic contribution of cap-dependent versus cap-independent translation. In terms of patient stratification, clinical testing will be required to determine whether aberrant PI3K/mTOR activation in tumors can guide treatment modality involving translational inhibitors, particularly in cases of relapse, where increased eIF4F activity has been associated with chemoresistance.
We also anticipate continued screening for novel drug leads that can influence other translation initiation processes and factors, such as (1) AUG selection, which potentially can alter the spectrum of therapeutically targeted mRNAs (Takacs et al. 2011); (2) translation reinitiation, which appears to contribute to the translation of a significant number of mRNA transcripts containing upstream open reading frames (ORFs), as recent deep sequencing data of ribosome-protected mRNA fragments suggests (Ingolia et al. 2011); (3) eIF3, a multisubunit complex that associates with the mTOR/raptor, eIF4G, and eIF4A (Korneeva et al. 2000; Holz et al. 2005) and has been implicated in the transformation process (Marchetti et al. 1995; Zhang et al. 2007, 2008); and (4) eIF4B, an essential factor whose regulation is under p70S6K regulation (Raught et al. 2004; Shahbazian et al. 2006). Mammalian genetic screens using short hairpin RNA (shRNA) libraries will further tease out the unique oncogene and non-oncogene exploitable dependencies of mRNA translation factors and regulators (Solimini et al. 2007). Ultimately, these screens will need to be accomplished in more clinically relevant in vivo model systems that better replicate the natural tumor microenvironment and non-cell-autonomous interactions. Looking back at the evolution of this area of translation over the last 10 years and the remarkable progress that has been made, we are excited about the years to come and the potential that these inhibitors hold for both clinical use and in gaining further insight into the fundamental mechanisms of translational control.
ACKNOWLEDGMENTS
We sincerely apologize to those authors whose work is not cited herein owing to space constraints. Work in the authors’ laboratory on this topic is supported by grants from the Canadian Institutes of Health Research and Canadian Cancer Society Research Institute.
Footnotes
Editors: Nahum Sonenberg, Michael B. Mathews, and John W.B. Hershey
Additional Perspectives on Synthesis and Translational Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Adams BL, Morgan M, Muthukrishnan S, Hecht SM, Shatkin AJ 1978. The effect of “cap” analogs on reovirus mRNA binding to wheat germ ribosomes. Evidence for enhancement of ribosomal binding via a preferred cap conformation. J Biol Chem 253: 2589–2595 [PubMed] [Google Scholar]
- Andersen JN, Sathyanarayanan S, Di Bacco A, Chi A, Zhang T, Chen AH, Dolinski B, Kraus M, Roberts B, Arthur W, et al. 2010. Pathway-based identification of biomarkers for targeted therapeutics: Personalized oncology with PI3K pathway inhibitors. Sci Transl Med 2: 43ra55. [DOI] [PubMed] [Google Scholar]
- Babendure JR, Babendure JL, Ding JH, Tsien RY 2006. Control of mammalian translation by mRNA structure near caps. RNA 12: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J 15: 658–664 [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, Pelletier J 2005. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc Natl Acad Sci 102: 10460–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J 2006a. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2: 213–220 [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, Pelletier J 2006b. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem Biol 13: 1287–1295 [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA Jr, et al. 2008. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest 118: 2651–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Lim JJ, Leonard T, Lim HC, Chia CS, Verma CS, Lane DP 2011. Stabilizing the eIF4G1 α-helix increases its binding affinity with eIF4E: Implications for peptidomimetic design strategies. J Mol Biol 405: 736–753 [DOI] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci 95: 1432–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxade M, Parra-Palau JL, Proud CG 2008. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front Biosci 13: 5359–5373 [DOI] [PubMed] [Google Scholar]
- Cai A, Jankowska-Anyszka M, Centers A, Chlebicka L, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads RE 1999. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry 38: 8538–8547 [DOI] [PubMed] [Google Scholar]
- Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, et al. 2008. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest 118: 3065–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, et al. 2009a. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS ONE 4: e5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Carrier M, Trnkus A, Porco JA Jr, Minden M, Pelletier J 2009b. Synergistic effect of inhibiting translation initiation in combination with cytotoxic agents in acute myelogenous leukemia cells. Leuk Res 34: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, Dingledine R, Fu H, Vajda S, Talbot PJ, et al. 2011a. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J Virol 85: 6381–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Hall DR, Robert F, Du Y, Min J, Li L, Qui M, Lewis I, Kurtkaya S, Dingledine R, et al. 2011b. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci 108: 1046–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers HF 2001a. Antimicrobial agents: Protein synthesis inhibitors and miscellaneous antibacterial agents. McGraw-Hill, New York [Google Scholar]
- Chambers HF 2001b. Antimicrobial agents: The aminoglycosides. McGraw-Hill, New York [Google Scholar]
- Chen J, Zheng XF, Brown EJ, Schreiber SL 1995. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci 92: 4947–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Guo L, Chen Y, Jiang Y, Wierda WG, Plunkett W 2011a. Homoharringtonine reduced Mcl-1 expression and induced apoptosis in chronic lymphocytic leukemia. Blood 117: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ozel D, Qiao Y, Harbinski F, Chen L, Denoyelle S, He X, Zvereva N, Supko JG, Chorev M, et al. 2011b. Chemical genetics identify eIF2α kinase heme-regulated inhibitor as an anticancer target. Nat Chem Biol 7: 610–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J 2008. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci 105: 17414–17419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Migoni A, Hautbergue GM, Artymiuk PJ, Baker PJ, Bokori-Brown M, Chang CT, Dickman MJ, Essex-Lopresti A, Harding SV, Mahadi NM, et al. 2011. A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A. Science 334: 821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz E, Antosiewicz J, Ekiel I, Morgan MA, Tahara SM, Shatkin AJ 1981. Methyl esterification of m7G5′p reversibly blocks its activity as an analog of eukaryotic mRNA 5′-caps. J Mol Biol 153: 451–458 [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz E, Ekiel I, Lassota P, Tahara SM 1987. Inhibition of eukaryotic translation by analogues of messenger RNA 5′-cap: Chemical and biological consequences of 5′-phosphate modifications of 7-methylguanosine 5′-monophosphate. Biochemistry 26: 4372–4380 [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz E, Stepinski J, Ekiel I, Goyer C, Sonenberg N, Temeriusz A, Jin Y, Sijuwade T, Haber D, Tahara SM 1989. Inhibition of eukaryotic translation by nucleoside 5′-monophosphate analogues of mRNA 5′-cap: Changes in N7 substituent affect analogue activity. Biochemistry 28: 4771–4778 [DOI] [PubMed] [Google Scholar]
- De Benedetti A, Joshi-Barve S, Rinker-Schaeffer C, Rhoads RE 1991. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol Cell Biol 11: 5435–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFatta RJ, Nathan CA, De Benedetti A 2000. Antisense RNA to eIF4E suppresses oncogenic properties of a head and neck squamous cell carcinoma cell line. Laryngoscope 110: 928–933 [DOI] [PubMed] [Google Scholar]
- *.Dobson C, Christodoulou J, Cabrita L 2012. Cold Spring Harb Persepct Biol 10.1101/cshperspect.a012260 [DOI] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M 2006. S6K1- and βTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314: 467–471 [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. 2010. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328: 1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumez H, Gallardo E, Culine S, Galceran JC, Schoffski P, Droz JP, Extremera S, Szyldergemajn S, Flechon A 2009. Phase II study of biweekly plitidepsin as second-line therapy for advanced or metastatic transitional cell carcinoma of the urothelium. Mar Drugs 7: 451–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R, Milburn SC, Hershey JW 1987. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem 262: 380–388 [PubMed] [Google Scholar]
- El Sous M, Khoo ML, Holloway G, Owen D, Scammells PJ, Rizzacasa MA 2007. Total synthesis of (−)-episilvestrol and (−)-silvestrol. Angew Chem Int Ed Engl 46: 7835–7838 [DOI] [PubMed] [Google Scholar]
- Fan S, Li Y, Yue P, Khuri FR, Sun SY 2010. The eIF4E/eIF4G interaction inhibitor 4EGI-1 augments TRAIL-mediated apoptosis through c-FLIP down-regulation and DR5 induction independent of inhibition of cap-dependent protein translation. Neoplasia 12: 346–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM 2009. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol 7: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. 2010. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci 107: 14134–14139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR 2009. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem J 421: 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard B, Cencic R, Pelletier J, Porco JA Jr 2007. Enantioselective synthesis of the complex rocaglate (−)-silvestrol. Angew Chem Int Ed Engl 46: 7831–7834 [DOI] [PubMed] [Google Scholar]
- Ghosh B, Benyumov AO, Ghosh P, Jia Y, Avdulov S, Dahlberg PS, Peterson M, Smith K, Polunovsky VA, Bitterman PB, et al. 2009. Nontoxic chemical interdiction of the epithelial-to-mesenchymal transition by targeting cap-dependent translation. ACS Chem Biol 4: 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N 1999a. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev 13: 1422–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N 1999b. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 15: 2852–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Barral MA, Rodriguez J, Jimenez C 2001. New cytotoxic steroids from the gorgonian Isis hippuris. Structure-activity studies. Tetrahedron 57: 3487–3497 [Google Scholar]
- Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, et al. 2007. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest 117: 2638–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudzien-Nogalska E, Jemielity J, Kowalska J, Darzynkiewicz E, Rhoads RE 2007a. Phosphorothioate cap analogs stabilize mRNA and increase translational efficiency in mammalian cells. RNA 13: 1745–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudzien-Nogalska E, Stepinski J, Jemielity J, Zuberek J, Stolarski R, Rhoads RE, Darzynkiewicz E 2007b. Synthesis of anti-reverse cap analogs (ARCAs) and their applications in mRNA translation and stability. Methods Enzymol 431: 203–227 [DOI] [PubMed] [Google Scholar]
- Gurel G, Blaha G, Moore PB, Steitz TA 2009. U2504 determines the species specificity of the A-site cleft antibiotics: The structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J Mol Biol 389: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110: 177–189 [DOI] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Lamb RF 2005. Restraining PI3K: mTOR signalling goes back to the membrane. Trends Biochem Sci 30: 35–42 [DOI] [PubMed] [Google Scholar]
- Herbert TP, Fahraeus R, Prescott A, Lane DP, Proud CG 2000. Rapid induction of apoptosis mediated by peptides that bind initiation factor eIF4E. Curr Biol 10: 793–796 [DOI] [PubMed] [Google Scholar]
- Hickey ED, Weber LA, Baglioni C, Kim CH, Sarma RH 1977. A relation between inhibition of protein synthesis and conformation of 5′-phosphorylated 7-methylguanosine derivatives. J Mol Biol 109: 173–183 [DOI] [PubMed] [Google Scholar]
- Hidalgo M, Rowinsky EK 2000. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene 19: 6680–6686 [DOI] [PubMed] [Google Scholar]
- Higa T, Tanaka J, Yasumasa T, Hiroyuki K 1981. Hippuristanols, cytotoxic polyoxygenated steroids from the gorgonian Isis hippuris. Chem Lett 11: 1647–1650 [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J 2005. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123: 569–580 [DOI] [PubMed] [Google Scholar]
- Hood KA, West LM, Northcote PT, Berridge MV, Miller JH 2001. Induction of apoptosis by the marine sponge (Mycale) metabolites, mycalamide A and pateamine. Apoptosis 6: 207–219 [DOI] [PubMed] [Google Scholar]
- Hwang BY, Su BN, Chai H, Mi Q, Kardono LB, Afriastini JJ, Riswan S, Santarsiero BD, Mesecar AD, Wild R, et al. 2004. Silvestrol and episilvestrol, potential anticancer rocaglate derivatives from Aglaia silvestris. J Org Chem 69: 3350–3358 [DOI] [PubMed] [Google Scholar]
- Ilic N, Utermark T, Widlund HR, Roberts TM 2011. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci 108: E699–E708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity J, Kowalska J, Rydzik AM, Darzynkiewicz E 2010. Synthetic mRNA cap analogs with a modified triphosphate bridge—Synthesis, applications and prospects. New J Chem 34: 829–844 [Google Scholar]
- Jones RM, Branda J, Johnston KA, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt EV 1996. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol 16: 4754–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian HM, Talpaz M, Santini V, Murgo A, Cheson B, O’Brien SM 2001. Homoharringtonine: History, current research, and future direction. Cancer 92: 1591–1605 [DOI] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR 2004. The molecular mechanics of eukaryotic translation. Annu Rev Biochem 73: 657–704 [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175 [DOI] [PubMed] [Google Scholar]
- Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH 2008. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol 49: 1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TD, Frick M, le Coutre P 2011. Omacetaxine mepesuccinate for the treatment of leukemia. Expert Opin Pharmacother 12: 2381–2392 [DOI] [PubMed] [Google Scholar]
- Ko SY, Guo H, Barengo N, Naora H 2009. Inhibition of ovarian cancer growth by a tumor-targeting peptide that binds eukaryotic translation initiation factor 4E. Clin Cancer Res 15: 4336–4347 [DOI] [PubMed] [Google Scholar]
- Konicek BW, Stephens JR, McNulty AM, Robichaud N, Peery RB, Dumstorf CA, Dowless MS, Iversen P, Parsons SH, Ellis KE, et al. 2011. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res 71: 1849–1857 [DOI] [PubMed] [Google Scholar]
- Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE 2000. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J Biol Chem 275: 41369–41376 [DOI] [PubMed] [Google Scholar]
- Kowalska J, Lewdorowicz M, Zuberek J, Grudzien-Nogalska E, Bojarska E, Stepinski J, Rhoads RE, Darzynkiewicz E, Davis RE, Jemielity J 2008. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA 14: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73: 585–596 [DOI] [PubMed] [Google Scholar]
- Kuznetsov G, Xu Q, Rudolph-Owen L, Tendyke K, Liu J, Towle M, Zhao N, Marsh J, Agoulnik S, Twine N, et al. 2009. Potent in vitro and in vivo anticancer activities of des-methyl, des-amino pateamine A, a synthetic analogue of marine natural product pateamine A. Mol Cancer Ther 8: 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson TG, Ray BK, Dodds JT, Grifo JA, Abramson RD, Merrick WC, Betsch DF, Weith HL, Thach RE 1986. Influence of 5′ proximal secondary structure on the translational efficiency of eukaryotic mRNAs and on their interaction with initiation factors. J Biol Chem 261: 13979–13989 [PubMed] [Google Scholar]
- Lawson TG, Cladaras MH, Ray BK, Lee KA, Abramson RD, Merrick WC, Thach RE 1988. Discriminatory interaction of purified eukaryotic initiation factors 4F plus 4A with the 5′ ends of reovirus messenger RNAs. J Biol Chem 263: 7266–7276 [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345: 544–547 [DOI] [PubMed] [Google Scholar]
- Li W, Dang Y, Liu JO, Yu B 2009. Expeditious synthesis of hippuristanol and congeners with potent antiproliferative activities. Chemistry 15: 10356–10359 [DOI] [PubMed] [Google Scholar]
- Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J 2008. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res 68: 5326–5334 [DOI] [PubMed] [Google Scholar]
- Lindqvist L, Oberer M, Reibarkh M, Cencic R, Bordeleau ME, Vogt E, Marintchev A, Tanaka J, Fagotto F, Altmann M, et al. 2008. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS ONE 3: e1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, Yang J, Semaan DJ, Chen C, Fox EA, et al. 2011. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med 17: 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lorsch J, Dever T, Hinnebusch A, Green R 2012. Cold Spring Harb Persepct Biol 10.1101/cshperspect.a011544 [DOI] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO 2005. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20: 709–722 [DOI] [PubMed] [Google Scholar]
- Low WK, Dang Y, Bhat S, Romo D, Liu JO 2007. Substrate-dependent targeting of eukaryotic translation initiation factor 4A by Pateamine A: Negation of domain-linker regulation of activity. Chem Biol 14: 715–727 [DOI] [PubMed] [Google Scholar]
- Lucas DM, Edwards RB, Lozanski G, West DA, Shin JD, Vargo MA, Davis ME, Rozewski DM, Johnson AJ, Su BN, et al. 2009. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood 113: 4656–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Hwang J, Sato H, Tang Y 2010. CBP80-promoted mRNP rearrangements during the pioneer round of translation, nonsense-mediated mRNA decay, and thereafter. Cold Spring Harb Symp Quant Biol 75: 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R 1995. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol 69: 1932–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961 [DOI] [PubMed] [Google Scholar]
- Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT, et al. 2008. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci 105: 10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. 2007. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128: 257–267 [DOI] [PubMed] [Google Scholar]
- Mothe-Satney I, Yang D, Fadden P, Haystead TA, Lawrence JC Jr 2000. Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol Cell Biol 20: 3558–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellner MK, Uras IZ, Gapp BV, Kerzendorfer C, Smida M, Lechtermann H, Craig-Mueller N, Colinge J, Duernberger G, Nijman SM 2011. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat Chem Biol 7: 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, et al. 2002. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol 319: 615–635 [DOI] [PubMed] [Google Scholar]
- Noh WC, Mondesire WH, Peng J, Jian W, Zhang H, Dong J, Mills GB, Hung MC, Meric-Bernstam F 2004. Determinants of rapamycin sensitivity in breast cancer cells. Clin Cancer Res 10: 1013–1023 [DOI] [PubMed] [Google Scholar]
- Novac O, Guenier AS, Pelletier J 2004. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res 32: 902–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al. 2006. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 66: 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Peltz SW 2007. Therapeutic opportunities in translation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Pelletier J, Sonenberg N 1985a. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40: 515–526 [DOI] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N 1985b. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: Effect of mRNA 5′ secondary structure. Mol Cell Biol 5: 3222–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S 1977. Inhibitors of protein synthesis. Academic Press, New York [Google Scholar]
- Pruvot B, Jacquel A, Droin N, Auberger P, Bouscary D, Tamburini J, Muller M, Fontenay M, Chluba J, Solary E 2011. Leukemic cell xenograft in zebrafish embryo for investigating drug efficacy. Haematologica 96: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J 18: 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW 2004. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23: 1761–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindar K, Reddy MS, Lindqvist L, Pelletier J, Deslongchamps P 2010. Efficient synthetic approach to potent antiproliferative agent hippuristanol via Hg(II)-catalyzed spiroketalization. Org Lett 12: 4420–4423 [DOI] [PubMed] [Google Scholar]
- Ravindar K, Reddy MS, Lindqvist L, Pelletier J, Deslongchamps P 2011. Synthesis of the antiproliferative agent hippuristanol and its analogues via Suarez cyclizations and Hg(II)-catalyzed spiroketalizations. J Org Chem 76: 1269–1284 [DOI] [PubMed] [Google Scholar]
- Rinker-Schaeffer CW, Graff JR, De Benedetti A, Zimmer SG, Rhoads RE 1993. Decreasing the level of translation initiation factor 4E with antisense RNA causes reversal of ras-mediated transformation and tumorigenesis of cloned rat embryo fibroblasts. Int J Cancer 55: 841–847 [DOI] [PubMed] [Google Scholar]
- Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR, et al. 2006. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNAiMet ternary complex availability. Mol Biol Cell 17: 4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Carrier M, Rawe S, Chen S, Lowe S, Pelletier J 2009. Altering chemosensitivity by modulating translation elongation. PLoS ONE 4: e5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche SP, Cencic R, Pelletier J, Porco JA Jr 2010. Biomimetic photocycloaddition of 3-hydroxyflavones: Synthesis and evaluation of rocaglate derivatives as inhibitors of eukaryotic translation. Angew Chem Int Ed Engl 49: 6533–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW Jr, Komar AA, Merrick WC 2002. eIF4A: The godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 72: 307–331 [DOI] [PubMed] [Google Scholar]
- Rosenwald IB 1996. Upregulated expression of the genes encoding translation initiation factors eIF-4E and eIF-2α in transformed cells. Cancer Lett 102: 113–123 [DOI] [PubMed] [Google Scholar]
- Rosenwald IB, Wang S, Savas L, Woda B, Pullman J 2003. Expression of translation initiation factor eIF-2α is increased in benign and malignant melanocytic and colonic epithelial neoplasms. Cancer 98: 1080–1088 [DOI] [PubMed] [Google Scholar]
- Rosenwald IB, Koifman L, Savas L, Chen JJ, Woda BA, Kadin ME 2008. Expression of the translation initiation factors eIF-4E and eIF-2* is frequently increased in neoplastic cells of Hodgkin lymphoma. Hum Pathol 39: 910–916 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP 2004. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med 10: 484–486 [DOI] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH 1994. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78: 35–43 [DOI] [PubMed] [Google Scholar]
- Saradhi UV, Gupta SV, Chiu M, Wang J, Ling Y, Liu Z, Newman DJ, Covey JM, Kinghorn AD, Marcucci G, et al. 2011. Characterization of silvestrol pharmacokinetics in mice using liquid chromatography-tandem mass spectrometry. AAPS J 13: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14: 1296–1302 [DOI] [PubMed] [Google Scholar]
- Satheesha S, Cookson VJ, Coleman LJ, Ingram N, Madhok B, Hanby AM, Suleman CA, Sabine VS, Macaskill EJ, Bartlett JM, et al. 2011. Response to mTOR inhibition: Activity of eIF4E predicts sensitivity in cell lines and acquired changes in eIF4E regulation in breast cancer. Mol Cancer 10: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, Shi W, Zhang Z, Rajasekhar VK, Pagano NC, et al. 2011. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med 208: 1799–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol 20: 7933–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM 2010. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 40: 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N 2006. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J 25: 2781–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ, Darzynkiewicz E, Furuichi Y, Kroath H, Morgan MA, Tahara SM, Yamakawa M 1982. 5′-Terminal caps, cap-binding proteins and eukaryotic mRNA function. Biochem Soc Symp 47: 129–143 [PubMed] [Google Scholar]
- Silvera D, Formenti SC, Schneider RJ 2010. Translational control in cancer. Nat Rev Cancer 10: 254–266 [DOI] [PubMed] [Google Scholar]
- Solimini NL, Luo J, Elledge SJ 2007. Non-oncogene addiction and the stress phenotype of cancer cells. Cell 130: 986–988 [DOI] [PubMed] [Google Scholar]
- Su W, Slepenkov S, Grudzien-Nogalska E, Kowalska J, Kulis M, Zuberek J, Lukaszewicz M, Darzynkiewicz E, Jemielity J, Rhoads RE 2011. Translation, stability, and resistance to decapping of mRNAs containing caps substituted in the triphosphate chain with BH3, Se, and NH. RNA 17: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs JE, Neary TB, Ingolia NT, Saini AK, Martin-Marcos P, Pelletier J, Hinnebusch AG, Lorsch JR 2011. Identification of compounds that decrease the fidelity of start codon recognition by the eukaryotic translational machinery. RNA 17: 439–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Faussat AM, Majdak P, Marzac C, Dubrulle S, Marjanovic Z, Legrand O, Marie JP 2006. Semisynthetic homoharringtonine induces apoptosis via inhibition of protein synthesis and triggers rapid myeloid cell leukemia-1 down-regulation in myeloid leukemia cells. Mol Cancer Ther 5: 723–731 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O 2011. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol 18: 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Ruiz-Gutierrez M, Borden KL 2004. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res 64: 8639–8642 [DOI] [PubMed] [Google Scholar]
- Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J, Mori N 2011. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem Pharmacol 81: 713–722 [DOI] [PubMed] [Google Scholar]
- Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R 2004. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol 24: 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez D 1979. Inhibitors of protein biosynthesis. Mol Biol Biochem Biophys 30: 1–312 [DOI] [PubMed] [Google Scholar]
- Wagner CR, Iyer VV, McIntee EJ 2000. Pronucleotides: Toward the in vivo delivery of antiviral and anticancer nucleotides. Med Res Rev 20: 417–451 [DOI] [PubMed] [Google Scholar]
- Wander SA, Hennessy BT, Slingerland JM 2011. Next-generation mTOR inhibitors in clinical oncology: How pathway complexity informs therapeutic strategy. J Clin Invest 121: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW 2004. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428: 332–337 [DOI] [PubMed] [Google Scholar]
- Wendel HG, Malina A, Zhao Z, Zender L, Kogan SC, Cordon-Cardo C, Pelletier J, Lowe SW 2006. Determinants of sensitivity and resistance to rapamycin-chemotherapy drug combinations in vivo. Cancer Res 66: 7639–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, et al. 2007. Dissecting eIF4E action in tumorigenesis. Genes Dev 21: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Shi C, Toral-Barza L, Lucas J, Shor B, Kim JE, Zhang WG, Mahoney R, Gaydos C, Tardio L, et al. 2010. Beyond rapalog therapy: Preclinical pharmacology and antitumor activity of WYE-125132, an ATP-competitive and specific inhibitor of mTORC1 and mTORC2. Cancer Res 70: 621–631 [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC 2008. PI3K pathway alterations in cancer: Variations on a theme. Oncogene 27: 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pan X, Hershey JW 2007. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem 282: 5790–5800 [DOI] [PubMed] [Google Scholar]
- Zhang L, Smit-McBride Z, Pan X, Rheinhardt J, Hershey JW 2008. An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3. J Biol Chem 283: 24047–24060 [DOI] [PMC free article] [PubMed] [Google Scholar]