Abstract
Recent advances in research technology have allowed researchers to study bacteria in their natural environment. Dental biofilm forms via an ordered sequence of events, resulting in structured and functionally organized species rich microbial community and modern molecular biological techniques have identified about 1000 different bacterial species in the dental biofilm, twice as many as can be cultured. Sites for biofilm formation include all kinds of surfaces: natural materials above and below ground, metals, plastics, medical implant materials—even plant and body tissue. Wherever you find a combination of moisture, nutrients and a surface, you are likely to find biofilm. The biofilm is used to describe the communities of micro-organisms attached to a surface; such microbes are usually spatially organized into three-dimension structure and are enclosed in matrix of extracellular material derived both from the cells themselves and from the environment. Dental biofilm pathogenicity in the oral cavity is magnified by specific biofilm characteristics and modern molecular biological techniques have identified about 1000 different bacterial species in the dental biofilm, twice as many as can be cultured. Adaptation to a biofilm lifestyle involves regulation of a vast set of genes, and the micro-organisms are thus able to optimize phenotypic properties for the particular environment.
Keywords: Bacteria, biofilm, plaque
INTRODUCTION
It is estimated over 95% of bacteria existing in nature are in biofilms.[1] The slime layer that forms on rocks in streams is a classic example of a biofilm. Biofilms are ubiqui-tous; they form on virtually all surfaces immersed in natural aqueous environments. Biofilms form particu-larly fast in flow systems where a regular nutrient supply is provided to the bacteria. The reason for the existence of the biofilm is that it allows the micro-organisms to stick and to multiply on surfaces. Micro-organisms undergo a wide range of physiological and morphological adaptations in response to environmental changes. In biofilms, different gradients of chemicals, nutrients and oxygen create micro-environments to which micro-organisms must adapt to survive. The perception and processing of chemical information from the environment form a central part of the regulatory control of these adaptive responses. Adaptation to a biofilm lifestyle involves regulation of a vast set of genes, and the micro-organisms are thus able to optimize phenotypic properties for the particular environment. Consequently, biofilm micro-organisms differ phenotypically from their planktonic counterparts. The last 10-15 years have seen emergence of several important new findings and concepts regarding the etiopathogenesis of periodontal disease and this includes the recognition of dental bacterial plaque as a microbial biofilm.[2]
CHARACTERISTICS OF BIOFILM
A biofilm environment confers certain properties to bacteria that are not seen in the nomadic state, a fact that explains the importance of recognizing dental plaque as a biofilm and not as bacteria in the planktonic state.[2] Some of the distinctive properties of biofilms are discussed and explained below.
Structure of biofilm
Biofilms are composed of micro-colonies of bacterial cells (15-20% by volume) that are non-randomly distributed in a shaped matrix or glycocalyx (75-80% volume).[3] The bacteria in the biofilm cluster together to form sessile, mushroom-shaped colonies. Each micro-colony is an independent community with its own customized living environment. Rapid formation of visible layers of micro-organisms due to extensive bacterial growth accompanied by excretion of copious amount of extracellular polymers is typical for biofilms. In the lower levels of most biofilms a dense layer of microbes is bound together in a polysaccharide matrix with other organic and inorganic materials.[4] The successive layer is a loose layer, which is often highly irregular in appearance and may ex-tend into the surrounding medium. The fluid layer bordering the biofilm may have a rather “stationary” sub layer and a fluid layer in motion.[5] There is the presence of voids or water channels between the micro-colonies that were present in these biofilms. The water channels permit the passage of nutrients and other agents throughout the biofilm, acting as primitive “circulatory” system.[3] Nutrients make contact with sessile (attached) micro-colonies by diffusion from the water channel to the micro-colony rather than from the matrix.[4] Each micro-colony is a tiny, independent community containing thousands of compatible bacteria. Different micro-colonies may contain different combinations of bacterial species. Bacteria in the center of a micro-colony may live in a strict anerobic environment, while other bacteria at the edges of the fluid channels may live in an aerobic environment. Thus, the biofilm structure provides a range of customized living environments (with differing pH, nutrient availability and oxygen).[6] Micro-colonies occur in different shapes in biofilms that are governed by shear forces due to passage of fluid over the biofilm. At low shear forces, the colonies are shaped like towers or mushrooms, while at higher shear forces, the colonies are elongated and capable of rapid oscillation.[3]
Exopolysaccharides-the backbone of the biofilm
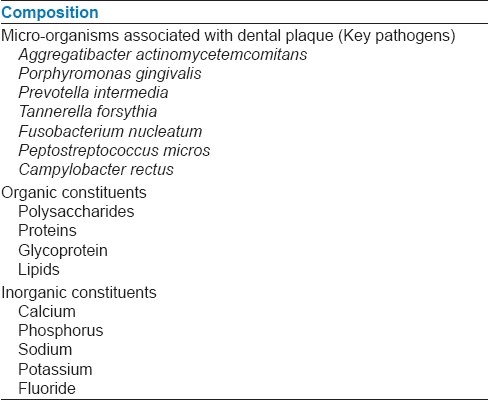
Dental biofilm is primarily composed of micro-organisms; micro-organisms exists within an intercellular matrix that consists of organic and inorganic materials derived from saliva, gingival crevicular fluid and bacterial products, which have been given in Table 1. Exopolysaccharides (EPS) are produced by the bacteria in the biofilm and are the major components of the biofilm making up 50-95% of the dry weight. They play a major role in maintaining the integrity of the biofilm and as well as preventing desiccation and attack by harmful agents.[4] In addition, they also bind essential nutrients such as cations to create a local rich environment favoring specific micro-organisms. The EPS matrix could also act as a buffer and assist in the retention of extracellular enzymes (and their substrates) enhancing substrate utilization by bacterial cells. One distinguished feature of the oral biofilms is that many of the micro-organisms can both synthesize and degrade the EPSs.[3] The fundamental structural unit of the biofilm is the micro-colony. Proximity of cells within the micro-colony provides an appropriate environment for creation of nutrient gradients, exchange of genes and quorum sensing. Since micro-colonies may be organized mass of multiple species, the cycling of various nutrients through redox reactions can readily occurs in biofilm.
Table 1.
Composition of biofilm
Physiological heterogeneity of biofilm
Bacterial species can exhibit extremely different physiological states in a biofilm even though they are separated by a distance of as little as 10 μm. The use of micro-electrodes has shown that pH can vary quite remarkably over short distances within a biofilm.[3] Measurement of oxygen and other gases has demonstrated that certain micro-colonies that are completely anerobic even though composed of single species and grown in ambient air. Thus, studies to date indicate that the sessile cells growing in mixed biofilms can exist in an almost infinite range of chemical and physical micro-habitants within the microbial communities.[4]
QUORUM SENSING
Some of the unique functions of biofilms are dependent on the ability of the bacteria and micro-colonies within the biofilm to communicate with one another. Quorum sensing or cell density-mediated gene expression in the bacteria involves the regulation of expression of specific genes through the accumulation of signaling compounds that mediate intercellular communication.[3] Quorum-sensing signaling represents a signaling pathway that is activated as a response to cell density.[7] Such systems are found in both Gram-positive and Gram-negative micro-organisms. The stimuli of quorum-sensing systems are signal molecules, called autoinducers.[8] The autoinducers are produced at a basal constant level, and the concentration thus is a function of microbial density. Perception of the signal occurs at a concentration threshold. The term “quorum” is used to describe this kind of signal system, since a certain number of micro-organisms must be present for the signal to be sensed and for the population to respond to the signal. In Gram-positive bacteria the signaling molecules are secreted peptides, whereas in Gram-negative bacteria two different systems of quorum sensing, which use different types of autoinducers, have so far been described. The first system was initially described in Vibrio fischeri as the mechanism that controls the expression of bioluminescence in this micro-organism. Over the past few years, similar systems have also been found in different genera of Gram-negative bacteria, and it has become clear that this system monitors the density of cells by producing acylated homoserine lactones (AHLs), whose structure depends on the bacteria that produce them. In Vibrio harveyi this first system has been called system 1, and hence the autoinducer that controls it is called AI-1. In this case the hydroxybutyryl homoserine lactone is the autoinducer. A second quorum-sensing system has been described in V. harveyi. The structure of the autoinducer for this system, which has been called AI-2, is still unknown, although it has been reported that its synthesis is dependent on the luxS gene.[9] This second system seems to be more widespread among the microbial world than the one that uses AHLs as autoinducers, and homologues for luxS have been identified in a large number of both Gram-positive and Gram-negative micro-organisms. Autoinducer-2 is an umbrella designation that covers a collection of molecules from the spontaneous rearrangement of 4, 5-dihydroxy-2, 3-pentanedione, which is the product of luxS gene. The molecule called autoinducer-2 was a universal signal mediating message among the species in mixed communities. This idea is distinct from the regulation of expression mediated by autoinducer-1, a family of acyl homoserine lactones which regulates gene expression in genetically identical cells. Since bacteria within the biofilms reach a high density, it has been suggested that quorum sensing might play a key role in bacterium-bacterium communication and, therefore, in the formation of biofilms.[9] Quorum sensing may give biofilm their distinct properties. For example, expression of gene for antibiotic resistance at high-cell density may provide protection. Quorum sensing also helps the potential to influence community structure by encouraging the growth of beneficial bacteria (to the biofilm) and discouraging the growth of competitors. It is also possible that physiological properties of bacteria in the community may be altered through quorum sensing.[4] The knowledge of bacterial biofilm property of quorum sensing helps to define the microbial etiology of plaque-mediated diseases and is challenging current practices for diagnosis and treatment.
Attachment of bacteria
The key characteristic of a biofilm is that micro-colonies within the biofilm attach to a solid surface. Thus, adhesion to a surface is the essential first step in the development of biofilm.[4] Many bacterial species possesses surface structures such as fimbriae and fibrils that aid in their attachment to different surfaces. Fimbriae are found in oral bacteria such as Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis. They are long protein filaments, present singly or in the bundles on the surfaces of the cells.[2] The major component is fimbrillin, a highly antigenic protein encoded by fimA in P. gingivalis and flp in A. actinomycetemcomitans. In both bacteria, fimbriae are thought to be important in colonization because the fimbrial-deficient mutants show reduced ability to bind and invade the epithelial cells and fibroblasts. Fimbriae-mediated epithelial invasion stimulates expression of host cell-adhesion molecules such as intercellular adhesion molecule, vascular cell-adhesion molecule, P-selectin and E-selectin, thus inducing a massive leukocyte response at the site. P. gingivalis 0fimbriae also stimulate IL-1β, IL-1α, TNF-α and granulocyte-macrophage colony-stimulating factor, leading to bone resorption.[2] Tannerella forsythia, P. gingivalis, Treponema denticola and other oral bacteria produce proteolytic enzymes often displayed on their cell surfaces. Dentilisin (T. denticola), PrtH (T. forsythia) and RgpA, RgpB and kgp (P. gingivalis) are the best-characterized enzymes in this group. These enzymes have various virulence components, degrade fibrinogen, laminin, fibronectin and several types of collagens.[2]
ANTIBIOTIC RESISTANCE
It has been recognized for considerable period of time that the organisms growing in the biofilms are more resistant to antibiotics than the same species growing in a planktonic (unattached) state.[4] Estimate of 1000-1500 times greater resistance for biofilm-grown cells than the planktonic cells have been suggested, although these estimates have been considered too high by some investigators. Conventionally, the sensitivity of bacteria to antimicrobial agents is determined on cells grown in liquid culture by the measure-ment of the minimum inhibitory concentration or minimum bactericidal concentration. Given the decreased sensitivity of an organism on a surface to antimicrobial agents, it has been argued that it would be more appropriate to determine the biofilm inhibitory concentration and biofilm killing concentration. Thus, it is important to understand the factors leading to antimicrobial resistance in biofilms such as dental plaque. The important mechanism of resistant appears to be the slower growth rate of bacteria in the biofilm, which makes them less susceptible to many but not all antibiotics. It has been shown in many studies that resistance of bacteria to antibiotics, biocides or preservatives are affected by their nutritional status, growth rate, temperature, pH and prior exposure to subeffective concentrations of antimicrobials.[3] Variations in any of these parameters can lead to a varied response to antibiotics with in a biofilm. The slower growing bacteria often over expresses non-specific defense mechanisms including heat-shock proteins, multidrug efflux pumps and demonstrate increased exopolymer synthesis.[4] When eukaryotic and bacterial cells are exposed to environmental stress (e.g., temperature, pH, redox potential), they synthesize stress proteins such as heat-shock proteins. These proteins protect the cells from damaging effects of the environment. Heat-shock proteins such as GroEL, GroES, DnaK and HtpG have been studied in oral bacteria; heat-shock proteins from A. actinomycetemcomitans stimulate the osteoclast activation and epithelial proliferation at low concentrations and are cytotoxic at high doses.[2] The exopolymer matrix of biofilm, although not a significant barrier in itself to the diffusion of antibiotics, does have certain properties that can retard diffusion. For example, strongly charged or chemically highly reactive agents can fail to reach the deeper zones of the biofilm because the biofilm acts as an ion-exchange resin removing such molecules from the solutions. In addition, extracellular enzymes such as β-lactamase, formaldehyde lyase and formaldehyde dehydrogenase may become trapped and concentrated in the extracellular matrix, thus inactivating the susceptible, typically positively charged, hydrophilic antibiotics. Hydrodynamics and turnover rate of the micro-colonies will also impact on antibiotic effectiveness. Alteration of genotype or phenotype of the cells growing within a biofilm matrix is receiving increased attention. Cells growing within the biofilm express genes that are not observed in the same cells growing in planktonic state and they can retain this resistance for some time after being released from the biofilm.
Gene transfer
The high density of bacterial cells in a biofilm also facilitates the exchange of genetic information among the cells of the same species and across species and even genera. Conjugation, transformation and transduction have been shown to occur more easily in a biofilm. Biofilm-associated bacteria communicate with each other by way of horizontal gene transfer.[2] Horizontal gene transfer among bacteria is recognized as a major contributor in the molecular evolution of many bacterial genomes.[10] In addition, horizontal gene transfer is responsible for the seemingly uncontrollable spread of antibiotic resistance gene among bacteria in the natural and nosocomial environments. The oral cavity is believed to be an excellent environment in which horizontal gene transfer can occur, as a result of the close and stable proximity of the majority of the bacteria present in dental plaque and the availability of exogenous DNA passing through the oral cavity.
Transformation
Transformation is defined as the uptake and maintenance of DNA. Competence is the physiological state in which the cells can take up DNA. Some oral bacteria, including members of genus Streptococcus, Neisseria and Actinobacillus are naturally competent and have specialized systems for DNA uptake. Transformation has no requirement for live donor cells because the DNA released upon cell death is the principal source of transforming DNA.[10]
Transduction
Transduction is a process where bacterial DNA is packaged into phage heads. When the phage infects a suitable host it injects this bacterial DNA, instead of phage DNA, into the new host. One of the main barriers to the activity of bacteriophage in the oral biofilm is the access to the cells within the extracellular polymeric substances secreted by cells themselves when growing as a biofilm.[10]
Conjugation
Conjugation is the polar transfer of genetic material through direct cell-to-cell contact and is mediated by a variety of specialized genetic elements, such as conjugative transposons and conjugative plasmids. It requires intimate cell-to-cell contact between the donor and recipient cells; this is mediated by variety of specialized intracellular and surface structures.[10] In general, it seems that there are a number of important factors that could actively promote gene transfer in the oral cavity. Both bacteriophage and naked DNA can survive for an appreciable time in human saliva; this survival is necessary for successful gene transfer by transduction or transformation, respectively.
Biofilm formation and contamination of dental unit water
Dental-unit water systems (DUWS) harbor bacterial biofilms, which may serve as a haven for pathogens. The water obtained from dental units via 3-in-1 syringes, air rotors, and low-speed handpieces may be heavily contaminated with micro-organisms and thus may be a potential source of infection for both practice staff and patients. The range of micro-organisms isolated includes both environmental organisms (e.g., Moraxella spp. and Flavobacterium spp.) and opportunistic and true human pathogens (e.g., Pseudomonas, Legionella pneumophila, Mycobacterium spp., and Staphylococcus spp.). The most common cause of DUW contamination is believed to be the formation and subsequent sloughing off of microbial biofilms from the surfaces of tubing within DUWSs.[11] Thus it depicts that bacterial biofilm in DUWs is a widespread problem, and poses a potentially significant risk of infection to dental staff and patients, particularly those who are medically compromised or immunocompromised.[12]
Current challenges and strategies to control biofilm
The key characteristics of biofilm that could be targets for pathogen management include its behavior as an adhesive mass with viscoelastic properties, its activity as a coordinated multispecies community in which cells communicate via small molecules, and its inflammatory disease potential. Dental plaque biofilm cannot be eliminated. However, the pathogenic nature of the dental plaque biofilm can be reduced by reducing the bioburden (total microbial load and different pathogenic isolates within that dental plaque biofilm) and maintaining a normal flora with appropriate oral hygiene methods that include daily brushing, flossing and rinsing with antimicrobial mouthrinse. This can result in the prevention or management of the associated sequelae, including the development of periodontal diseases and possibly the impact of periodontal diseases on specific systemic disorders.[13] When assessing treatment options, an appreciation of the ecology of the oral cavity will enable the enlightened clinician to take a more holistic approach and consider the nutrition, physiology, host defenses and general well-being of the patient, as these will affect the balance and activity of the resident oral microflora. Future episodes of disease will occur unless the cause of any breakdown in homeostasis is recognized and remedied.[14] The recent explosion in the field of biofilm research has led to exciting progress in the development of new technologies for studying these communities, advanced our understanding of the ecological significance of surface-attached bacteria and provided new insights into the molecular genetic basis of biofilm development. The nature of a biofilm helps explain why periodontal diseases have been so difficult to prevent and treat.
CONCLUSIONS
Oral biofilms are very heterogeneous in structure and modern molecular biological techniques have identified about 1000 different bacterial species in the dental biofilm, twice as many as can be cultured.[15] Bacteria in a biofilm have a physiology different from that of planktonic cells and live under nutrient limitation and often in a dormant state, thus a biofilm is organized to maximize energy, spatial arrangements and movement of nutrients and byproducts with advantages which includes a broader habitat range for growth, an enhanced resistance to antimicrobial agents and host defense and an enhanced ability to cause disease. Research on microbial biofilms is proceedings on many dimensions, with specific focus on elucidation of the genes specifically expressed by biofilm-associated organisms, assessment of different control approaches for either preventing or remediating biofilm colonization of medical devices, and development of new methods for evaluating the efficacy of these treatments.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Overman PR. Biofilm: A new view of plaque. J Contempt Dent Pract. 2006;1:1–11. [PubMed] [Google Scholar]
- 2.Tatakis DN, Kumar PS. Etiology and pathogenesis of periodontal disease. Dent Clin Nam. 2005;49:491–516. doi: 10.1016/j.cden.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Socransky SS, Haffajee AD. Dental biofilms: Difficult therapeutics target. Periodontol 2000. 2002;28:12–55. doi: 10.1034/j.1600-0757.2002.280102.x. [DOI] [PubMed] [Google Scholar]
- 4.Lindhe J, editor. 4th ed. Oxford, UK: Blackwell Publishing Company; 1998. Clinical Periodontology and implant dentistry book. [Google Scholar]
- 5.Newman MG, Carranza FA, Takei H, Klokkevold PR, editors. 10th ed. USA: Elsevier Health Sciences; 2006. Carranzas clinical Periodontology book. [Google Scholar]
- 6. [cited in 2010]. Available from: http://www.lww.com .
- 7.Scheie AA, Petersen FC. The Biofilm Concept: Consequences for future prophylaxis of oral disease. Cri Rev Oral Bio Med. 2004;15:4–12. doi: 10.1177/154411130401500102. [DOI] [PubMed] [Google Scholar]
- 8.Frias J, Olle E, Alsina M. Periodontal pathogens produce quorum sensing signal molecules. Infect Immun. 2001;69:3431–4. doi: 10.1128/IAI.69.5.3431-3434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr Communication among oral bacteria. Micro Mole Bio Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts AP, Mullany P. Genetic basis of horizontal gene transfer among oral bacteria. Periodontol 2000. 2006;42:36–46. doi: 10.1111/j.1600-0757.2006.00149.x. [DOI] [PubMed] [Google Scholar]
- 11.Walker JT, Bradshaw DJ, Bennett AM, Fulford MR, Martin MV, Marsh PD. Microbial biofilm formation and contamination of dental unit water system in general dental practice. Appl Environ Microbial. 2000;66:3363–7. doi: 10.1128/aem.66.8.3363-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuttlebee CM, O’Donnell MJ, Keane CT, Russell RJ, Sullivan DJ, Falkiner F, et al. Effective control of dental chair unit waterline biofilm and marked reduction of bacterial contamination of output water using two peroxide-based disinfectants. J Hosp Infect Sci. 2002;52:192–205. doi: 10.1053/jhin.2002.1282. [DOI] [PubMed] [Google Scholar]
- 13.Thomas JG, Nakaishi LA. Managing the complexity of a dynamic biofilm. J Am Dent Assoc. 2006;137:10S–5S. doi: 10.14219/jada.archive.2006.0409. [DOI] [PubMed] [Google Scholar]
- 14.Marsh PD. Dental plaque as biofilm and a microbial community- implication for health and disease. BMC Oral Health. 2006;6:S14. doi: 10.1186/1472-6831-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacob M, Cate T. Biofilms, a new approach to the microbiology of dental plaque. Odontology. 2006;94:1–9. doi: 10.1007/s10266-006-0063-3. [DOI] [PubMed] [Google Scholar]


