Abstract
Histopathological images are an important resource for clinical diagnosis and biomedical research. From an image understanding point of view, the automatic annotation of these images is a challenging problem. This paper presents a new method for automatic histopathological image annotation based on three complementary strategies, first, a part-based image representation, called the bag of features, which takes advantage of the natural redundancy of histopathological images for capturing the fundamental patterns of biological structures, second, a latent topic model, based on non-negative matrix factorization, which captures the high-level visual patterns hidden in the image, and, third, a probabilistic annotation model that links visual appearance of morphological and architectural features associated to 10 histopathological image annotations. The method was evaluated using 1,604 annotated images of skin tissues, which included normal and pathological architectural and morphological features, obtaining a recall of 74% and a precision of 50%, which improved a baseline annotation method based on support vector machines in a 64% and 24%, respectively.
Keywords: Basal Cell Carcinoma, Histopathology Images, Automatic Annotation, Visual Latent Semantic Analysis, Non-negative Matrix Factorization, Bag of Features
INTRODUCTION
Recent advances in microscopical acquisition technology have allowed to collect huge numbers of histopathological images, an important resource for the diagnosis act as well as for pathologist training.[1] The interest in developing the suitable image technology to address the automatic analysis of this kind of images has rapidly grown over the last years.[2–4] As a consequence, a new research area, called bioimage informatics, has emerged integrating data mining, database visualization, extraction, searching, comparison and management of biomedical visual data.[3,5] This area combines both image analysis and computational techniques to provide powerful tools that facilitate high-throughput/high-content analysis of biological tissues.[5]
Automatic annotation of histopathological images is a very challenging problem. In contrast with natural images, high level annotations are not usually associated to particular objects in the image. In histopathological images, annotations are related to pathological lesions, morphological and architectural features, which encompass a complex mixture of visual patterns that allow to decide about the illness presence. In general, images with the same annotations present a high visual variability, which can be generated by several factors, starting with the inevitable uncertainty coming from the fact that a very complicated 3D biological structure is randomly projected onto a 2D image i.e. the tissue must become a solid structure from which surface cuts of barely 1-5 μm are obtained. For achieving so, tissues are subjected to a series of histological procedures: they are initially fixated with a basic aldehyde, then dehydrated, embedded and finally cut. This chain of events is very susceptible to different kinds of cumulative errors that result in histopathological images with a complex mix of patterns and sub-patterns that only can be interpreted by an expert, even in cluttered biological circumstances. In addition, image capturing parameters such as environment illumination, exposure time, microscope magnification, etc., are a source of image variability. Therefore, the relevant visual pathological patterns highly change their visual appearance according to their spatial location, severity and co-occurrence with others biological structures. Figure 1 shows examples of some histopathological images of skin tissues with different annotations associated with acellular, cellular and architectural features, illustrating the visual variability problem.
Figure 1.
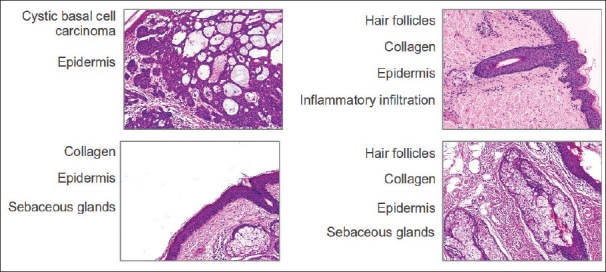
Example of histopathological images globally annotated with multiple annotations (multilabeled images). These images correspond to the test data set used in this work and they have a resolution of 1024 × 768 pixels. Histopathological annotations of morphological and architectural features such as epidermis, collagen, and hair follicles appear in different images illustrating the high-visual variability for the same annotation
Commonly, bioimage analysis methods encompass two main components: a feature extraction and representation process that allows to properly describe the visual image content, which ideally should be robust to the visual variability problem of histopathological annotations, and an interpretable knowledge extraction process, capable of linking low-level visual patterns and high-level annotations. In this paper we propose a novel strategy for automatic annotation of histopathological images, which combines a part-based image representation (bag of features, BOF), a latent topic model (non-negative matrix factorization, NMF) and a probabilistic annotation strategy that allows to connect visual latent topics with high-level annotations. The proposed method provides both a robust automatic annotation method and a coarse location of them inside the images. The proposed method has a remarkable characteristic, it is exclusively trained with images that exhibit only one histopathological annotation. However the resulting annotation model is able to assign multiple histopathological annotations to full microscopical field of views. Therefore, it is not necessary to collect a representative training set that includes images that have different combinations of histopathological annotations. To the best of our knowledge, this is the first work that proposes an automatic annotation algorithm based on a part-based image representation and a probabilistic latent topic model in histopathological images. The proposed approach was evaluated using a set of images of a skin cancer, known as basal cell carcinoma, which contains regions with ten different histopathological annotations, including acellular, cellular, and architectural features (i.e. collagen, sebaceous glands, hair follicles, inflammatory infiltration, eccrine glands, epidermis) and pathological lesions (i.e. nodular basal cell carcinoma, morpheiform basal cell carcinoma, micro-nodular basal cell carcinoma, cystic basal cell carcinoma).
The paper is organized as follows: section 2 describes the proposed method based on BOF and NMF. Section 3 presents the experimental evaluation performed using a basal-cell carcinoma data set and the preliminary results obtained for automatic annotation compared with a classical model of Support Vector Machines (SVM). Finally the conclusions are presented in Section 4.
AUTOMATIC ANNOTATION OF HISTOPATHOLOGICAL IMAGES USING NMF
The proposed method for automatic annotation of histopathological images is depicted by the Figure 2. This approach comprises two main stages: i) training, and, ii) prediction. In the former stage, a probabilistic model that is able to automatically generate multiple annotations for new images (multi-label images) is generated from a set of images globally annotated with only one histopathological annotation (mono-label images). In this stage, a training set of images is represented by two matrices F and L, which codify the distribution of the visual information and the annotations of the images contained in the set, respectively. Note that, L will be a sparse matrix with 1 in the annotation assigned to each image and 0 in the other cases. The visual information is represented using a bag-of-features approach.[6] Therefore, F corresponds to a matrix of visual words versus images. To obtain an image representation using latent topics, the matrix F is factorized in two matrices (W and H), using a NMF model that allows to find the probability distribution of visual latent topic models (H) in the images. Finally, the visual latent topics are linked to the annotations distribution (L) using a probabilistic model that generates a matrix (C) with the latent representation of annotations. The prediction stage also starts with the bag of features representation of non-annotated images using the same visual codebook constructed in the training stage. A new image is projected to the latent topic space, given by W, to generate the vector Hnew. Finally, the above vector (Hnew) and the latent representation of annotations (C) are multiplied to obtain the vectors Lnew that indicates the probability that the new image has each histopathological annotation. The new image is finally annotated with the corresponding histopathological annotation associated with one of the morphological features or pathological lesions with the highest probability by a binarization process.
Figure 2.
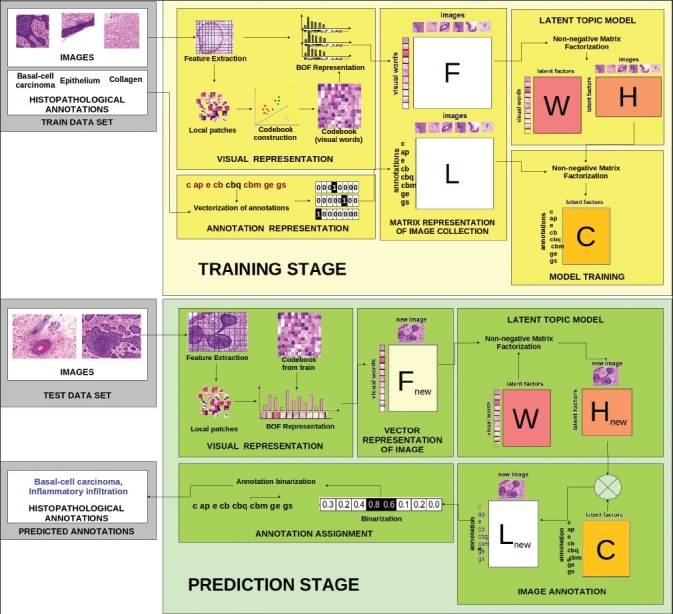
Overview of the proposed method for automatic annotation of histopathological images based on non-negative matrix factorization
The details of the bag of features representation of images, latent topic model, and automatic-annotation process in training and prediction stages, using a probabilistic interpretation of non-negative matrix factorization, are introduced in the following Subsections.
Bag of Features Representation
The visual representation of histopathological images is obtained as a bag of features (BOF).[6] A model inspired by the fact that the visual system perceives an object by integrating its constituent parts.[7,8] Therefore, this representation is basically a histogram of small parts, called visual words, which are defined by a clustering analysis of small patches extracted from an image collection. The general BOF representation approach comprises three main stages: feature detection and description, codebook or visual vocabulary construction, and BOF image representation.[9]
Figure 3 depicts the BOF setup used in this work. The hypothesis underneath the proposed representation is that all biological structures are also represented by a probabilistic model that describe the distribution (histogram occurrence) of quantized small microstructures described by the visual patches. In the first step, the local feature detection consists in extracting small square patches that will be used for describing the whole visual image content. Herein, we extract these patches from a regular image partition of 8 × 8 pixels without overlapping, which corresponds to the minimum resolution that a visual pattern require for covering biological structures such as cell nuclei. On the other hand, taking into account that from a pathologist point of view, visual identification of biological microstructures is based on the stain variations, we use the discrete cosine transform (DCT) coefficients of the RGB color components for describing each patch, because this local feature has been used to effectively describe this kind of variations in small regions.[10,11] Local region descriptor results into a single feature vector of 192 dimensions by the concatenation of the three color component descriptors.[12–14] The second step, the codebook construction, is performed using a k-means clustering algorithm over a sample of patches from the training image set. The number of clusters, k, corresponds to the codebook size. The centroids found with this clustering are the visual words of the codebook and the visual representation of them can be obtained applying the inverse DCT. In this paper the codebook size was set to 700, which is a good value according to,[14] where a systematic experimentation was performed on similar kind of images (histology and histopathology) using this visual features. Finally, each image is represented by a k-bin histogram. This is accomplished by associating the feature vector, describing each patch in the regular grid, to the closest visual word in the codebook. Then, the histogram is generated with each bin counting the number of patches in the image assigned to the corresponding visual word.
Figure 3.
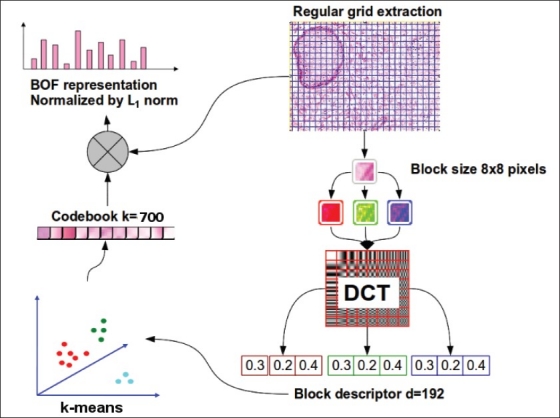
Bag of feature setup used for representing histopathology images. In this work the local features extraction is performed using regular grid extraction and each patch of 8 × 8 pixels is represented by the first coefficients of a discrete cosine transform applied to each color component (RGB) independently, the visual codebook is built using k-means with k = 700, and finally each image is represented by a histogram of 700 bins normalized with L1 norm
Visual Latent Topic Analysis
The BOF representation of an image collection could be seen as a term-vs-document matrix, F ∈ Rn × m, where rows correspond to visual words and columns to images. Each element Fij indicates the frequency of the i-th visual word in the j-th image. The goal of latent topic analysis is to find a set of latent factors that explain the visual content of each image as a mixture of different probability distributions of visual words.
NMF is a well known matrix decomposition approach that approximates a matrix F ∈ Rn × m as a product of two simpler non-negative matrix factors W ∈ Rn × k and H ∈ Rm × k as follows:
F = WHT (1)
with W containing a set of k latent factors that are linearly combined to represent the images in F using the coefficients in H.
The solution to NMF involves iterative optimization techniques using a cost function that describes the “closeness” of WHT to F. Lee and Seung[15] proposed two different cost functions: Euclidean distance and Kullback Leibler (KL) divergence. In this work we use the last one because of its probabilistic interpretation.[16] The optimization problem based on KL divergence is defined as follows:
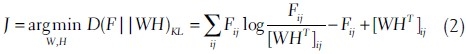
An equivalence between NMF and probabilistic latent semantic indexing (PLSI) was reported by Ding et al.[16] PLSI has a strong statistical foundation that models documents as a mixture of term probabilities conditioned on a latent random variable.[17] The parameters of the model are estimated by a likelihood maximization process based on expectation maximization algorithm. The mixture calculated by PLSI induces a factorization of the original term-document matrix: P(wi, dj) = Σrk = 1P(wi|zk)P(dj|zk)P(zk), if F is normalized according to Fij ← Fij/ΣijFij, it can be interpreted as the joint probability p(wi, dj) = Fij. Ding et al. showed that the factorizations produced by NMF and PLSI are equivalent,[16] with W containing the visual-word-latent-factor conditional probabilities, p(wi|zk), and H the image-latent-factor joint probability, P(dj|zk) P(zk) = P(dj,zk).
In conclusion, NMF generates a model of the image collection that explains the occurrence of visual words in images by a mixture of probability distributions conditioned on a small set of latent factors. These latent factors can be interpreted as general visual patterns. Additionally, each latent factor can be associated to a cluster of images,[18] the centroid of the cluster given by the columns of W and the assignment of images to clusters given by the rows of H, where the values can be interpreted as soft image-cluster membership functions.
Probabilistic Annotation Model
Following the probabilistic model described in the previous subsection, the annotation task can be seen as the process of calculating the annotation-vs-image conditional probabilities, p(lc|dnew), where lc is the c-th annotation and dnew corresponds to the new unannotated image. This is done by extending the latent topic model of the previous subsection with information from the annotations of the training images. This information is represented in a annotation-vs-image matrix, L ∈ Rc × m. The first step is to assign histopathological annotation to each one of the visual latent topics, i.e., to calculate the conditional probability p(lc|zk). This is accomplished by applying NMF to the L matrix as follows:
L = CHT (3)
where H is the same matrix obtained from the visual latent topic factorization, which is kept fixed during the optimization process. After an appropriate normalization and according to the discussion of previous subsection, C contains the annotation-vs-latent-topic conditional probabilities p(lc|zk). The second step is to project the new image to the visual latent space, this is done by applying NMF to solve:
Fnew = WHTnew (4)
where Fnew is the BOF representation of the new image, W is the same matrix obtained from the visual latent topic analysis and is kept fixed during the optimization process. After an appropriate normalization, HnewT contains the joint probabilities p(dnew, zt). Finally, the conditional probability p(lc|dnew) is calculated using Bayes rule and law of total probability as follows:
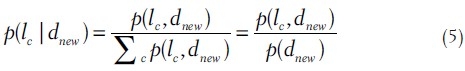
where p(lc, dnew) is the factorized joint probability
![]()
assuming that lc and dnew are independent given zk. It is easy to see that Equation 6 corresponds to the following matrix multiplication:
Lnew = CHTnew (7)
According to the above discussion we propose a straightforward method for automatic annotation of images based on NMF (A2NMF) that consists in two stages (training and prediction) described in Algorithms 1 and 2.
Algorithm 1.
Training stage for automatic annotation of images using NMF

Algorithm 2.
Prediction stage for automatic annotation of images using NMF

EXPERIMENTAL EVALUATION
Basal Cell Carcinoma Data Set
The proposed method was evaluated on a histopathological image data set, which was annotated by an expert, identifying the presence of architectural or morphological features, and pathological lesions inside each image. Images correspond to field of views with a 10X magnification, extracted from Hematoxilyn-eosin (H&E) stained skin tissues diagnosed with different types of basal cell carcinoma. These images contain a particular richness in architectural and morphological features, i.e., characteristic arrangements of cells, surrounded by several combinations of epithelial and connective tissues, also found in many other pathologies.[19]
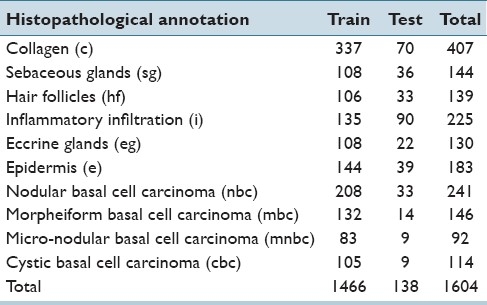
The entire image set, composed of 655 digital images, was randomly divided into training (80%) and test (20%) sets. Square subimages that contained single histopathological annotations were manually cropped from the training image set. Although, there is no typical size for those annotations, because of their large intrinsic variability, subimage size was estimated as an average value of a set of regular regions that the pathologist marked as containing a single histopathological annotation i.e. square subimages of 300 × 300 pixels. A total of 1, 466 training subimages were finally obtained, each containing a single annotation among the ten possibilities. On the other hand, the test set was composed of 138 images of 1024 × 768 pixels, which, in general, are annotated as containing more than one histopathological annotation. Latter, images were globally annotated, i.e., the actual location of these annotations was not provided, which makes the task of automatic annotation even more challenging. The data set distribution by histopathological annotation is detailed in Table 1.
Table 1.
Data set distribution per histopathological annotation for training and test
In order to reduce the visual image variability, a color normalization strategy, based on the transfer of the statistical properties of the stain contributions, was firstly applied.[20] Examples of some morphological features and a pathological lesion (collagen, epidermis, hair follicles and cystic basal cell carcinoma) are shown in Figure 4, in which the large appearance variability exposed by them can be appreciated. For example in the same figure Epidermis refers to outer layer of skin which comprises stratified squamous epithelium, i.e. several layers with different morphology of cells. However, typically a whole digital image of histopathology have one or more visual patterns associated with different morphological and architectural features of tissues like the images shown in Figure 1, which belong to the test image set. These images have in some located regions particular patterns, e.g. epidermis or hair follicles, whereas others are sparsely distributed without a well defined spatial location, e.g. collagen.
Figure 4.
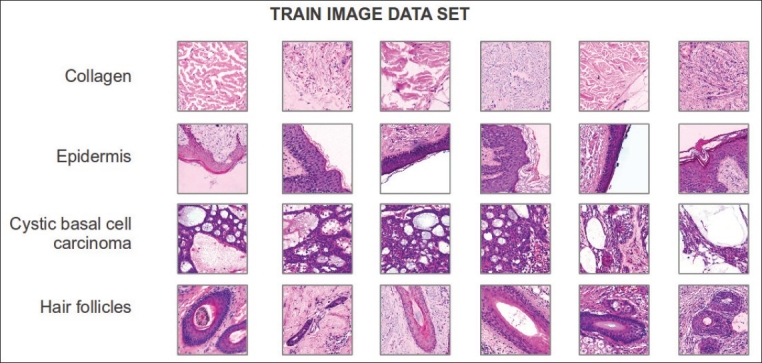
Examples of training images with the corresponding histopathological annotations. These images have a resolution of 300 × 300 pixels and exhibit only one annotation per image
Performance evaluation
The performance of the proposed automatic annotation method was evaluated using standard measures such as precision, recall, accuracy and f-measure which are defined as follows:
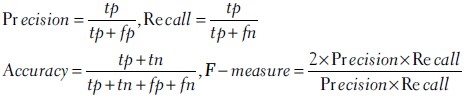
where tp is the number of correctly predicted annotations, fp is the number of wrong predicted annotations, tn is the number of correctly omitted annotations, and fn is the number of missed annotations.
Note that these measures are not evaluated independently by class. This is a better way to evaluate the performance in an automatic annotation task where images simultaneously exhibit multiple annotations.
As baseline, a state-of-the-art supervised annotation method based on support vector machines (SVM) was used. We train a one-vs-all SVM model with an RBF kernel for each class, the best parameters were chosen using a 10-fold cross-validation over the training data set. As well the proposed approach, the SVM model uses the same BOF image representation for visual content of the images.
The performance of the proposed and baseline annotation methods was evaluated in both training and testing data sets. When evaluating the performance in the training data set, 20% of the training images are withheld during training and later used to evaluate the generalization performance. The purpose of this two-way evaluation was to contrast the performance of the annotation methods in two scenarios: a simple mono-label annotation task, corresponding to annotate images with the same characteristics as that ones used for training, and the original complex multi-label annotation task.
RESULTS
An important parameter for a latent-topic model is the size of latent space dimension, i.e the number of latent topics required for representing the collection visual content. Therefore, the effect of varying the number of latent topics was assessed on the mono-label annotation task. Figure 5 shows the average performance of the proposed method in the training data set against the number of latent topics (dimension of the latent space). With a small number of latent dimensions, the annotation model has a high recall, but with low precision, accuracy and f-measure. This indicates that the annotation model tends to assign a high number of annotations per image. The situation improves with a higher number of latent dimensions, and all the measures steadily increase beyond 32 dimensions.
Figure 5.
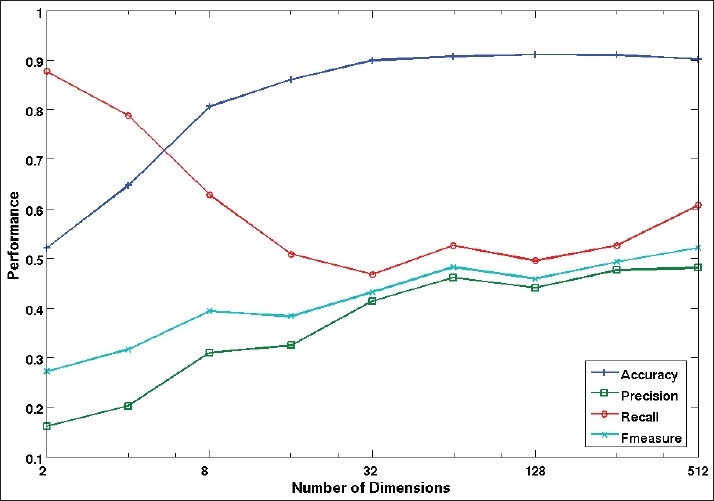
Performance evaluation on training mono-label images by each number of dimensions in the latent space
These results suggest selecting a k value as big as possible. Lee and Seung in[21] suggested a number of latent topics k < nm / (n + m). The reason is that beyond of this value the number of parameters in the factorization, the maximum number of dimensions in the latent space according to the rule (n + m)k, will be greater than the number of values in the original matrix, nm. We decided to use this limit for the experiments, taking into account that n is the codebook size and m is the number of images in training stage. This gives a number of latent topics k = 438 for the mono-label scenario and k = 473 for the multi-label scenario.
The proposed approach was compared against a state-of-the-art SVM model in both mono-label and multi-label scenarios. Table 2 shows the average value for each performance measure, for both scenarios on the respective test set. The results show that the SVM model performs better on the mono-label annotation tasks. The reason could be because the test images are similar to those used in the training stage, i.e. small images containing a unique annotation. However, when test images contain more than one histopathological annotation, the proposed approach takes advantage of the intermediate representation in the latent semantic space, and outperforms the results reported by the SVM learning model.
Table 2.
Average in automatic annotation performance in both experiments with standard performance measures, accuracy (Acc), precision (Pr), recall (Rc), and f-measure (F)
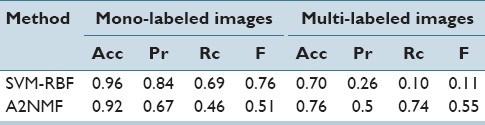
The results suggest that the proposed model is doing a better work characterizing the high visual variability of the different histopathological annotations. A supervised learning model, such as SVM, requires a representative set of training images that exhibit combination of morphological and architectural features of tissues similar to the ones expected in the test set. In contrast, the proposed method initially characterizes the visual variability of the training data set in an unsupervised fashion. Annotations of the training data set are used in a later step to build a probabilistic annotation model that connects latent visual topics with histopathological annotations.
One important characteristic of the proposed method for automatic annotation is its interpretability. Whereas SVM is one of the most powerful models for supervised learning, the generated classifiers are not easily interpretable. The improved interpretability of the proposed method is due to the fact that it is possible to map back the generated labels to particular regions of the image by each morphological and architectural feature. This is accomplished by assigning a histopathological annotation posterior probability to each small image patch. Figure Figure 6 illustrate the concept mapping strategy: a test image is shown in Figure 6a and the corresponding probabilities maps of its patches for each of the ten histopathological annotations are shown in Figure 6b–k. Each of these maps have in the top the real binary membership value of the histopathological annotation (v), the posterior probability of the predicted annotation given the image by the proposed method in Equation 6 (p), anding histopathological annotation according to the step 4 of Algorithm 2 (b).
Figure 6.
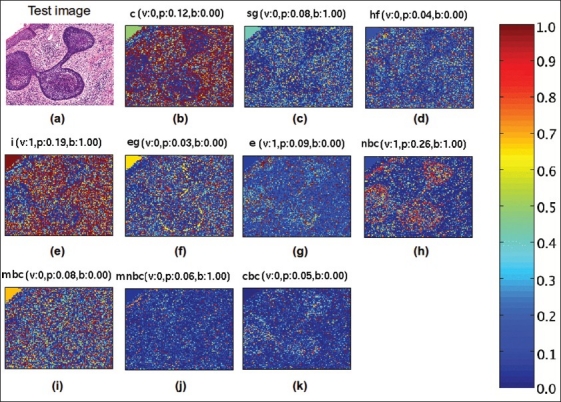
Example of an image from the test data set automatically annotated by the proposed method. The original multilabel image (a) is showed with the salient maps of the patches inside the image according with each one of the 10 histopathological annotations: collagen (b), sebaceous glands (c), hair follicles (d), inflammatory infiltration (e), eccrine glands (f), epidermis (g), nodular basal cell carcinoma (h), morpheiform basal cell carcinoma (i), micro-nodular basal cell carcinoma (j), cystic basal cell carcinoma (k), on the top of each salient image is the real membership of the class (v), the conditional probability estimated by the proposed method (p), and the final concept binarization value (b)
These results are relevant in the biomedical context because the high-variability of architectural and morphological features in healthy and pathological tissues is a common phenomena. In general, it is very difficult to have enough examples of each possible structural arrangement of the morphological features for training a supervised learning model such as the SVM algorithm. This scenario is also a more realistic in biomedical image domain, where regions of interest in the image, which cover an example of biological structures, are commonly annotated by the presence or absence of a given set of histopathological annotations whereas computer-aided diagnosis or image retrieval systems require the annotation of full images.
CONCLUSIONS
This paper presented a novel method for histopathological images annotation with probabilistic support for prediction and spatial location of morphological and architectural features in healthy and pathological tissues. The method was evaluated in a challenging scenario were training images corresponded to small subimages exhibiting only one histopathological annotation, although test images included multiple annotations. The proposed method exhibited an improved performance when compared to a state-of-the-art supervised annotation method. The distinctive characteristic of the proposed method is that it builds an enhanced representation of the visual image collection content in an unsupervised fashion finding latent visual topics, which encode high-level visual patterns.
Histopathological images are particularly challenging to analyze because of their high variability and complex visual structure. The results reported in this paper suggest that latent semantic characterization of the visual structure is a viable alternative to build competitive annotation models for histopathological images.
ACKNOWLEDGMENTS
This work was partially funded by the projects “Automatic Annotation and Retrieval of Radiology Images Using Latent Semantic” number 110152128803 and “Medical Image Retrieval System Based On Multimodal Indexing” number 110152128767, from COLCIENCIAS 521 of 2010.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2011/2/2/4/92031
REFERENCES
- 1.Madabhushi A. Digital pathology image analysis: Opportunities and challenges (Editorial) Imaging Med. 2009;1:7–10. doi: 10.2217/IIM.09.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2006;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 3.Swedlow JR, Goldberg IG, Eliceiri KW. OME consortium. Bioimage informatics for experimental biology. Ann Rev Biophys. 2009;38:327–46. doi: 10.1146/annurev.biophys.050708.133641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kvilekval K, Fedorov D, Obara B, Singh A, Manjunath BS. Bisque: A platform for bioimage analysis and management. Bioinformatics. 2010;26:544–52. doi: 10.1093/bioinformatics/btp699. [DOI] [PubMed] [Google Scholar]
- 5.Peng H. Bioimage informatics: A new area of engineering biology. Bioinformatics. 2008;24:1827–36. doi: 10.1093/bioinformatics/btn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li FF, Perona P. CVPR ‘05: In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05) 2005;2:524–31. [Google Scholar]
- 7.Biederman I. Recognition-by-components: A theory of human image understanding. Psychol Rev. 1987;94:115–47. doi: 10.1037/0033-295X.94.2.115. [DOI] [PubMed] [Google Scholar]
- 8.Olshausen B. Principles of image representation in visual cortex. In: Chalupa LM, Werner JS, editors. The Visual Neurosciences. Cambridge Massachusetts: MIT Press; 2003. pp. 1603–15. [Google Scholar]
- 9.Csurka G, Dance CR, Fan L, Willamowski J, Bray C. Visual categorization with bags of keypoints. Workshop on Statistical Learning in Computer Vision. 2004 [Google Scholar]
- 10.Kamiya Y, Takahashi T, Ide I, Murase H. Lecture Notes in Computer Science. Vol. 5371. New York: Springer Berlin/Heidelberg; 2009. A multimodal constellation model for object category recognition.In MMM ‘09 Proceedings of the 15th International Multimedia Modeling Conference on Advances in Multimedia Modeling; pp. 310–21. [Google Scholar]
- 11.Deselaers T, Ferrari V. Global and Efficient Self-Similarity for Object Classification and Detection.2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’2010) 2010:1633–40. [Google Scholar]
- 12.Cruz-Roa A, Caicedo JC, González FA. Visual mining in histology images using bag of features. 6th International Seminar on Medical Image Processing and Analysis - SIPAIM 2010. 2010 [Google Scholar]
- 13.Díaz G, Romero E. Lecture Notes in Computer Science. Vol. 6419. New York: Springer Berlin / Heidelberg; 2010. Histopathological image classification using stain component features on a pLSA model.CIARP’10 Proceedings of the 15th Iberoamerican congress conference on Progress in pattern recognition, image analysis, computer vision, and applications; pp. 55–62. [Google Scholar]
- 14.Cruz-Roa A, Caicedo JC, González FA. Visual pattern mining in histology image collections using bag of features. Artif Intell Med. 2011;52:91–106. doi: 10.1016/j.artmed.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Lee DD, Seung HS. Algorithms for non-negative matrix Factorization. In: Leen TK, Dietterich TG, Tresp V, editors. Advances in Neural Information Processing Systems. Vol. 13. MIT Press; 2001. pp. 556–62. [Google Scholar]
- 16.Ding C, Li T, Wei P. On the equivalence between non-negative matrix factorization and probabilistic latent semantic indexing. Comput Stat Data Anal. 2008;8:3913–27. [Google Scholar]
- 17.Hofmann T. Unsupervised learning by probabilistic latent semantic analysis. J Mach Learn. 2001;42:177–96. [Google Scholar]
- 18.Ding C, Li T, Jordan MI. Convex and semi-nonnegative matrix factorizations. IEEE Trans Pattern Anal Mach Intell. 2010;32:45–55. doi: 10.1109/TPAMI.2008.277. [DOI] [PubMed] [Google Scholar]
- 19.McGee JO, Isaacson PG, Wright NA. Oxford: Oxford University Press; 1992. Oxford textbook of pathology: Principles of pathology. [Google Scholar]
- 20.Díaz G, Romero E. Micro-structural tissue analysis for automatic histopathological image annotation. Microsc Res Tech. 2011 doi: 10.1002/jemt.21063. doi:101002/jemt21063 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–91. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]


