Abstract
A biomechanical model-based deformable image registration incorporating specimen-specific changes in material properties is optimized and evaluated for correlating histology of clinical prostatectomy specimens with in vivo MRI. In this methodology, a three-step registration based on biomechanics calculates the transformations between histology and fixed, fixed and fresh, and fresh and in vivo states. A heterogeneous linear elastic material model is constructed based on magnetic resonance elastography (MRE) results. The ex vivo tissue MRE data provide specimen-specific information for the fresh and fixed tissue to account for the changes due to fixation. The accuracy of the algorithm was quantified by calculating the target registration error (TRE) by identifying naturally occurring anatomical points within the prostate in each image. TRE were improved with the deformable registration algorithm compared to rigid registration alone. The qualitative assessment also showed a good alignment between histology and MRI after the proposed deformable registration.
Keywords: Biomechanical models, correlative pathology, deformable registration, finite element model, magnetic resonance elastography
INTRODUCTION
Radiation therapy (RT) is a primary and effective modality to treat and often cure cancer.[1] To deliver this treatment, high resolution diagnostic-quality images (e.g., CT and MR) are necessary for the process of identifying and delineating the tumor and planning the radiation dose. Although, there have been remarkable developments in the imaging technology and dose delivery devices, many studies have shown that inaccuracies in the definition of the tumor boundaries have contributed to most of the post-treatment side effects.[2] The ability to compare and validate imaging with the gold standard of the histopathology is a key approach to overcoming this limitation.[3] Although this comparison is beneficial for advancement of RT, it is challenging due to the substantial changes that occur during the process of histology including gross tissue sectioning, processing, paraffin embedding, and fine sectioning to create histology slices. The main solution to compensate for the changes in the tissue is deformable registration.
Several different approaches to match histology slices with MRI have been investigated and reported in the literature. Solutions can be generally classified based on the choice of nonrigid transformation employed in the algorithm.
One technique is to use free-form deformations (FFD) as the transformation, which was first proposed by Rueckert et al.[4] for the application of breast MRI. Dauguet et al.[5] used the “blockface” or the digital photographs of the sectioned baboon brain slices to assist in the registration of histology to MR. In their approach, blockface images were registered to MRI using mutual information-based FFD. They applied the resulting deformation field to the histology images, which were previously rigidly registered to the blockface images. Alic et al.[6] registered a dense histology volume, reconstructed by up to 100 histology slides of a xenograft tumor developed in a mouse, using a mutual information-based FFD to in vivo MRI via an intermediate ex vivo MRI. These two mentioned works[5,6] are based on intermediate registration and the registration performances were qualitatively shown to be good. However, no quantitative measures of registration accuracy is reported and the performance of their algorithm is highly dependent on the number of histology slices which is limited (around 10) for the case of prostatectomy.
Recently, Chappelow et al.[7] developed an algorithm based on multiattribute combined mutual information (MACMI) to benefit from all the information provided in multiprotocol MR. After finding the corresponding slices between histology and MRI of prostate, a 2D FFD registration by MACMI was applied to histology to match with MRI. Mazaheri et al.[8] used a similar approach for matching MRI and histology of prostate with a simpler optimization objective function to drive the 2D FFD registration. Their objective function was based on volume overlap of the prostate on MRI and histology. Both of these works[7,8] were based on corresponding 2D histology and MR image slices which limits the registration accuracy to be meaningful only in the plane of imaging.
The second group applies thin plate spline (TPS) to transform the images. An average TRE of 0.82 mm and 6% error in volume alignment was achieved by Zhan et al.[9] to match ex vivo MRI and histology in five prostate specimens. The registration was performed using 3D TPS based on boundary landmarks and automatically detected internal landmarks. Although the methodology of this work is promising for histology to ex vivo registration, automatic finding of corresponding landmarks between in vivo and histology is a difficult task that has not been investigated yet. Ward et al.[10] developed an image-based slicing method to accurately section the prostate specimens parallel to in vivo MR imaging plane. They achieved TRE of 0.85 mm by warping the 2D slices of MRI to their corresponding histology slides. However, their registration does not account for any out-of-plane deformation which can occur during gross sectioning and microtome slicing.
In a recent work by Ceritoglu et al.,[11] large deformation diffeomorphic metric mapping (LDDMM) was used to achieve an average surface distance of 0.4 mm between corresponding areas on histology images and MRI of nine monkey brains. The accuracy of the algorithm was improved by incorporating manually segmented regions of interest. LDDMM guarantees a one-to-one relationship between the registered images. Nonetheless, the applicability of this algorithm is highly influenced by the image intensity profile of the modalities used in registration. The intensity profiles can be affected by histology artifacts (tears, fissures, and missing tissue).
The last general approach is based on biomechanically derived transformations. McNiven et al.[12] used a biomechanical-based registration to match histology images of the prostate with in vivo MRI. Appropriate material property models derived from the literature were used in this initial investigation. The approach was based on rigidly reconstructed histology which resulted in average TRE distribution of 1.3, 1.2, and 1.9 mm in the left/right, anterior/posterior, and superior/inferior directions, respectively.
In this paper, an optimum biomechanical-based deformable registration is proposed to register whole-mount histology images to in vivo MRI of prostate. The mechanical behavior of the prostate 3D model is optimized by assigning specimen-specific material properties obtained using an ex vivo quasi-static magnetic resonance elastography (MRE) technique[13] for prostate specimens. A three-step deformable registration workflow is used to register histology images to in vivo MRI [Figure 1]. The histology slices are also reconstructed based on a 2D deformable registration technique. Compared to the relevant literature mentioned above, the proposed technique is minimally dependent on the number of histology slides and is fully performed in 3D and therefore it can account for out-of-plane deformations. It is primarily based on finding boundary alignment for driving the registration rather than image intensities which might fail due to artifacts and distortion on the images. Analysis of the registration accuracy is performed by calculating TRE to quantify the accuracy measurements and by visual inspection of the registered images. This work is a feasibility study and an initial proof of concept. Further experiments and comparisons with other well-known deformable registration algorithms are on going.
Figure 1.
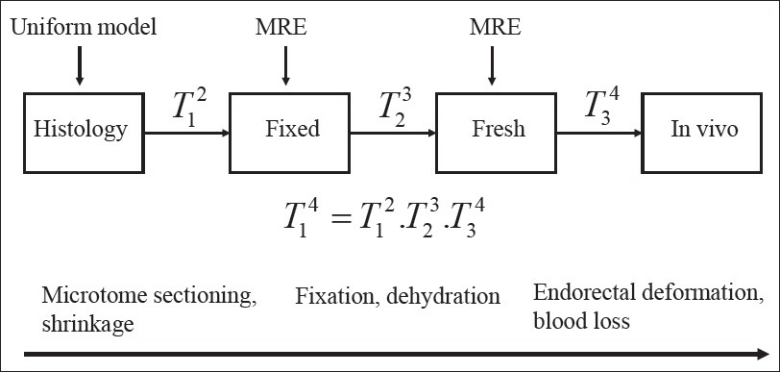
Summary of the deformable registration workflow. The process starts by registering histology to fixed, and then fixed to fresh, and finally fresh to in vivo. For the last two steps MRE-derived material properties are added to the biomechanical modeling
MATERIALS AND METHODS
Image Acquisitions
Seven sets of images are acquired for each of four patients who have undergone radical prostatectomy:
In vivo T2-weighted (T2w) MR images prior to prostatectomy, 0.27×0.27×3 mm mm voxel size (termed “in vivo”).
Ex vivo fresh T2w MR images of the prostate before fixation, 0.3×0.3×0.3 mm voxel size (“fresh”).
Ex vivo fresh MRE of the prostate before fixation, 0.5×0.5×3 mm voxel size (“fresh MRE”).
Ex vivo fixed T2w MR images of the prostate after fixation, 0.3×0.3×0.3 mm voxel size (“fixed”).
Ex vivo fixed MRE of the prostate after fixation, 0.5×0.5×3 mm mm voxel size (“fixed MRE”).
Digital gross photographs of the sectioned slices with 3 mm thickness (“gross”).
Digital images of the whole-mount histology slides with 4 μm thickness (“histology”).
The in vivo MR scans are obtained 3-5 weeks prior to surgery using an endorectal coil, where the imaging plane is set to be perpendicular to the endorectal coil. For whole-specimen fixation the prostatectomy specimens are submersed in 10% neutral-buffered formaldehyde solution for approximately 60 hours to insure complete penetration of fixative. Histology slides are carefully prepared by experienced pathology technicians to minimize damage to the tissue and distortion of the digitized histology images.
Tissue Sectioning Process
To obtain histology slides that are similar to in vivo images, the specimen must be cut at the same angles as the in vivo MR imaging planes. Therefore, the angle of cutting with respect to the resected specimen needs to be estimated. Based on the initial experiments, the most effective angle to best match the sectioned tissue slices with in vivo imaging plane is the rotation angle about left to right axis which is denoted by θ throughout this manuscript. Depending on the availability of gold seeds fiducial markers in the prostate (which were used to guide earlier treatment with external beam radiation therapy), two approaches are proposed to determine θ. If the patient has fiducial markers, then a point-based rigid registration between fiducial points on the fresh and in vivo images is used to obtain θ. Alternatively, fresh images are digitally resampled at several different angles and visually compared to in vivo images to find the best match. To evaluate the accuracy of estimating θ, a metric denoted by Δ is developed. This metric measures the agreement between the relative position of the urethra with respect to the prostate boundary between in vivo MR and fresh ex vivo MR:
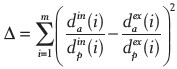
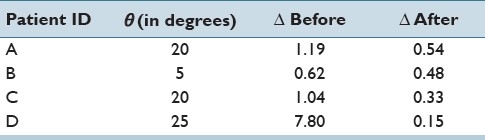
where dain(i) is the vertical distance of the urethra to the anterior part of the prostate boundary in the in vivo MR slice number i, and m is the number of corresponding slices on both in vivo MR and fresh that the urethra is visible. Superscript ex and subscript p represent the ex vivo fresh MR and vertical distance of the urethra to the posterior part of the prostate boundary, respectively. The vertical distances are determined by manual measurements. According to its definition, Δ is unitless. Ideally, Δ should be zero in case where the position of urethra in the ex vivo fresh MR exactly matches with that of the in vivo MR. Therefore, it is expected to have a smaller Δ for the resampled fresh MR with the estimated Δ. Table 1 shows that for all four patients Δ has been well reduced after resampling the fresh image set with the estimated θ. The angles are rounded up to multiples of 5° due to technical precision limit of the device used to cut the specimens. Further investigations on the proposed metric (Δ) and also to determine the accuracy of θ are ongoing.
Table 1.
Estimated sectioning angles denoted with θ (in degrees) and their errors based on the similarity of position of the urethra to the in vivo MR, represented by θ, before and after resampling fresh imageset with the estimated θ. Δ is unitless
Once estimated θ is verified by an expert radiation oncologist, the fixed specimen is sectioned at that angle with 3 mm slice thickness to replicate the thickness of the MR in vivo imaging. Digital photographs of these gross slices are then taken.
3D Reconstruction of Histology
Unlike the MR images which are inherently a 3D image, the whole-mount histology slices must be first reconstructed into a 3D volume to enable deformable registration to the corresponding images. To compensate for the deformations and misalignment between histology slices, each histology slice is first registered to its corresponding gross slice using a 2D deformable registration based on moving least squares (MLS).[14] The MLS algorithm is a landmark-based registration that warps the image using a locally rigid transformation based on the displacement of the control points. These control points are landmarks manually identified on the prostate boundary and urethra on the gross slice and corresponding histology slice. It should be noted that the gross slices are not naturally aligned due to the manual setup of each slice to obtain digital photographs. Therefore, prior to registering histology to gross slices, a 2D rigid registration is performed between gross slices and the resliced fixed MR image set at the estimatedθ. The rationale behind the use of the fixed image set is twofold: first, gross slices are acquired from the fixed tissue by 3 mm sectioning with the estimated θ and second, the magnitude of deformation to the tissue is minimum compared to other states (i.e., fresh and in vivo). The only source of deformation between the fixed MRI and gross slices is serial sectioning, which has been empirically observed to be small. Second, the fixed image set is already aligned since the images are obtained from the whole specimen. The urethra, the prostate boundary, and other anatomical features were used to guide the manual 2D rigid registration between gross slices and the resliced fixed image set with 3 mm slice thickness. The entire process is illustrated in Figure 2 part (a) shows the 2D rigid registration between gross slices and the resliced fixed image, while part (b) displays the 2D deformable registration based on MLS to obtain the warped histology slides. Finally, the prostate boundary is contoured on each warped histology slice using MIPAV (NIH, Bethesda, MD, USA). These contours are then converted into a 3D triangular mesh using an implementation of the marching cube algorithm. The 3D triangular mesh is then used in the biomechanical registration.
Figure 2.
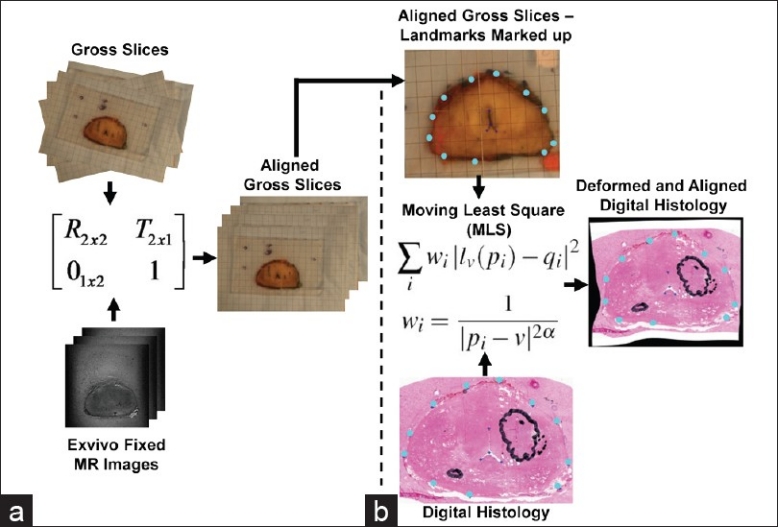
Histology reconstruction. (a) 2D rigid registration between gross slices and their corresponding fixed images. (b) 2D deformable registration based on MLS between histology slides and gross slices
Deformable Registration Workflow
The proposed biomechanical model-based deformable registration workflow based on finite element modeling (FEM) consists of a three-step registration [Figure 1]. A 3D triangular surface mesh of each image set is reconstructed based on the delineated prostate boundary contours. At each registration step, a primary 3D volumetric FEM is registered to a secondary 3D surface mesh. A volumetric mesh is constructed using Hypermesh software (HyperWorks 10.0, Altair Engineering Inc., MI, USA) with tetrahedral elements based on the 3D triangular surface mesh. Appropriate material properties are then assigned to the 3D FEM of the primary based on the MRE results. Boundary conditions are obtained from projection of the primary surface nodes to the surface of the 3D model of the secondary. Next, a commercial finite element analysis (FEA) package (Abaqus, v6.9, Abaqus Inc., Providence, RI, USA) finds the displacement of each node in the 3D tetrahedral mesh of the primary by solving the constitutive equations derived from the boundary conditions and the material models.
In the proposed three-step registration illustrated in Figure 1 different distortions and deformations are resolved by means of biomechanical modeling. First, geometric changes due to microtome sectioning and shrinkage are partially compensated by 2D deformable registration using the MLS technique. However to further increase the accuracy and to account for out-of-plane deformations, a uniform linear elastic 3D FEM of histology is built. This 3D model is then deformed to the fixed state using the biomechanical deformable registration as denoted by T12. Second, changes due to fixative solution and dehydration during the fixation process are modeled in the next step of deformable registration represented as T23. In this step, the result of T12 is deformed to the fresh state. The last step, T34, models the differences occurred between the in vivo and the fresh ex vivo MRI by deforming the result of T23 to in vivo. These deformations are mainly due to the pressure caused by the endorectal coil at the time of in vivo scan and blood loss in the ex vivo imaging because of prostatectomy. Thus, the registration of histology to fixed (T12) is based on a homogenous elastic material model, while the registrations of fixed to fresh (T23) and the fresh to the in vivo (T34) are performed using a heterogeneous specimen-specific material property model.
Development of the Material Model
A quasi-static MRE technique[13] is used to acquire 3D voxel-wise Young's modulus (E) maps of the prostate specimen with linear elasticity assumption in the fresh and the fixed states. Initial validation of this technique using canine prostate showed that the obtained E values were accurately comparable with those measured by elastometer indentations.[13] Prior to correlating the E maps to the FEM, unrealistic values of Young's modulus due to MRI artifacts are detected by setting lower and upper thresholds, i.e., magnetic susceptibility artifacts due to air pockets between the gel could lead to errors in the calculated E maps.[13] The lower threshold for the fresh and the fixed MRE scans varied between 10e4 and 10e-2 KPa for different patients. The frequency of such values is less than 1% on average between patients. This value is chosen based on the fact that 1st percentile was decreased by more than 100% comparing with 0.9th percentile. Therefore, values less than this threshold can be considered as noise. The upper threshold is determined based on a comparison between 99.9th percentile and 99th percentile. If the difference between the two values is more than the 99th percentile value, then the values of 99.9th percentile is chosen as the upper threshold. Upper thresholds for the MRE scans varied between 500 KPa and 1500 KPa for different patients. Values higher than the upper threshold and values lower than the lower threshold are considered too high and too low, respectively. These noisy values are then replaced with the median of their surrounding normal values. Finally, a 3D Gaussian filter with the standard deviation of 4 is applied to the entire volume of the MRE data to further reduce the effect of noise and artifacts (see left part of the Figure 3). A method is proposed to find the element-voxel relation between 3D tetrahedral FEM of each state and its corresponding E maps. After matching the center of geometry of the 3D FEM and that of the prostate MRE data, the center of geometry of each tetrahedral element will fall in one of the voxels of the 3D E maps. Therefore, each element is assigned to its corresponding E values measured by the MRE technique as visualized in the right hand side of Figure 3.
Figure 3.
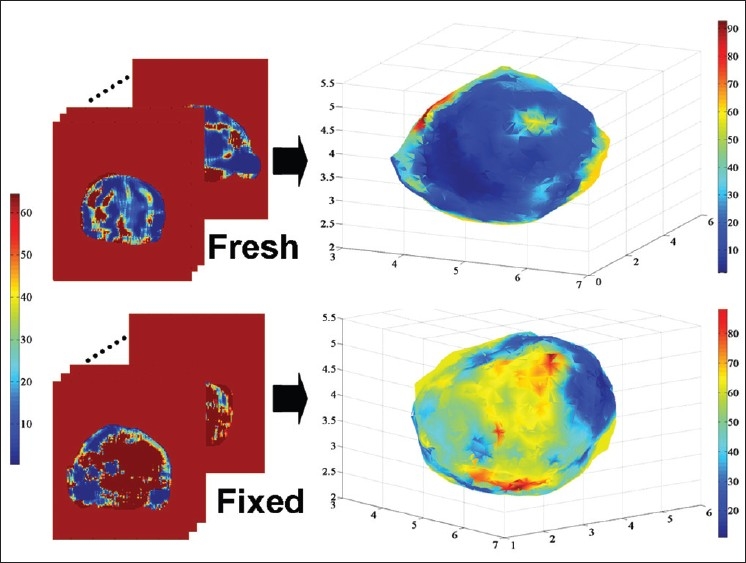
Material model construction based on MRE data. Each element of the model is assigned an E value from the MRE data. The top and the bottom left image sets are the MRE fresh and fixed scans, respectively. The top and the bottom right images display the reconstructed FEM after assigning the E values from MRE. The axes on the right side images represent the position in centimeter. Color maps show the E values in KPa. In the fresh-state E-maps variations in E were noted for diseased tissue and across normal anatomy. Fixation increases Young modulus nonuniformly and as an approximate function of distance from the tissue edge
Registration Evaluation
Target registration error (TRE) was used to quantitatively evaluate the deformable registration performance over rigid registration.[15] For Tij, TRE is defined as the Euclidean distance between the naturally occurring anatomical points identified on image set j and the transformed position of their corresponding points on image set i
TRE=║Tji(Pi)–Pj║,
where Pi and Pj are the corresponding points on imageset i and imageset j, respectively. In this work, rigid and deformable transformations obtained from the registrations are used to calculate TRE. It is important to note that none of the registration algorithms (deformable or rigid) rely on these naturally occurring anatomical points. Therefore, TRE presented in this work provides completely unbiased results. More particularly, Tij is not found based on the corresponding points (i.e., Pi and Pj). In fact, the deformable transformation is found based on the constitutive equations derived from the boundary conditions and the material models.
Distances between surfaces of the histology and fixed 3D meshes were also used to compare rigid and deformable registration performance more clearly for histology to fixed registration step. More specifically, minimum distances of the nodes on the surface of the registered histology mesh to the surface of the fixed mesh (represented by triangular elements) were calculated.
RESULTS
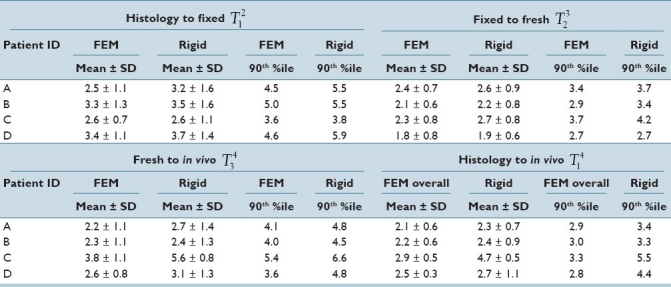
The analysis has been completed on four patients. Table 2 represents mean, standard deviation, and the 90th percentile that shows the value that 90% of the TRE values fall below for each intermediate deformable and rigid registration step and also for the overall deformable and rigid registrations. For each step, different sets of points were chosen, although some of the points were used in multiple steps. This is due to the fact that these naturally occurring anatomical points cannot be found consistently on each image set because of differences in imaging modalities or the specimen condition. Therefore, the overall accuracy is not necessarily computable based on TRE of intermediate steps. The average number of points for all the steps and the patients was 30. From Table 2, it is noticeable that the proposed deformable registration has achieved better accuracy compared to rigid registration. However, the registration accuracy obtained using rigid registration shows that the extent of deformations in the patients analyzed to date is small. Therefore, large differences between the performances of the proposed deformable registration and rigid registration are not expected. Although the proposed deformable registration average performance is modestly better than rigid, error improvements illustrated by a decrease of up to 1.3 mm in the 90th percentile between deformable and rigid registration could potentially have a clinical influence on the correlation of MRI and histology. It is also worth mentioning that because all the registration methods in this paper are performed in 3D, the error values are influenced by the voxel size of the images, which is 0.27 × 0.27 × 3 mm in the in vivo images.
Table 2.
TRE (in mm) for all rigid and deformable registrations steps and the overall. The first column shows the patient ID. FEM denotes the proposed deformable registration using a MRE model. The last two columns of each step represent the value that 90% of TRE values fall below. The average number of naturally occurring anatomical points for all the patients is 30
It is important to note that in the proposed deformable registration workflow which consists of three steps, the goal is to incrementally refine the deformations by obtaining measured biomechanical properties based on two ex vivo MR scans: fresh MRE immediately after the surgery and fixed MRE after approximately 60 hours of fixation. Therefore, the possibility of error accumulation due to several registration steps must be considered with the trade-off of providing the biomechanical modeling deformable registration with more information about the mechanical properties of the specimen measured with MRE.
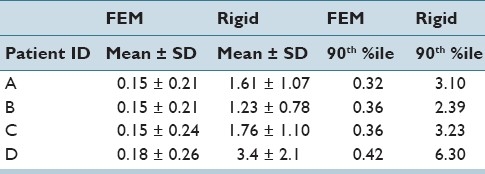
Table 2 also shows that in the histology to fixed registration step the performance of both rigid and deformable registrations is comparable. It should be noted that TRE is a local measure of accuracy and increasing the number of points where the TRE is calculated can help to ensure that the TRE adequately represents the accuracy of local deformation occurring between the prostate images. Therefore, in addition to TRE to demonstrate the effectiveness of deformable registration over rigid registration, the distance between the 3D surfaces of the prostate between histology and fixed with rigid and deformable registration is also calculated. Table 3 shows that with rigid registration the 90th percentile of the distances reaches up to 6.3 mm (for patient D) whereas the 90th percentile value is limited to 0.42 mm (for patient D) with the proposed deformable registration algorithm. Such large surface distances obtained with rigid registration can cause significant issues when comparing histology and fixed images in cases where a small tumor is located close to the boundary of the prostate.
Table 3.
Average, standard deviation, 90th percentile value of the distance between the histology and fixed surfaces (in mm) for step 1 (T21) after rigid and deformable (denoted by FEM) registrations
TRE distribution in three anatomical directions for the overall histology to in vivo registration resulted in 1.2 ± 0.4 mm (mean ± std) in left-right (LR), 0.9 ± 0.4 mm in anterior-posterior (AP), and 1.4 ± 0.5 mm in superior-inferior (SI) directions using the proposed deformable registration compared to 1.4 0.2 mm (LR), 1.6 ± 0.6 mm (AP), and 1.6 ± 1.4 mm (SI) obtained using rigid registration. Compared to published results by Zhan et al.[9] with average TRE of 0.82 mm between histology and ex vivo MR images, the TRE results of histology to fixed registration step (T12) in this work are larger with the average of 2.9 mm. This difference can be explained by the fact that Zhan et al.[9] used 1.5 mm thick histology slices which was further reduced to 0.75 mm by interpolation. It is understandable to achieve submillimeter errors with the reduction of slice thickness from 3 mm to 0.75 mm. Additionally, visual comparison between the ex vivo MR images indicates that Zhan et al.[9] obtained images with more distinguishable features. One of the future goals of this work is to improve the ex vivo MR quality by optimizing the MRI acquisition parameters for prostate specimen. Ward et al.[10] achieved TRE of 0.85 mm with a 2D TPS registration between histology and in vivo MR based on leave-one-out cross-validation experiment using the fiducial points that guide the TPS registration. Since Ward et al.[10] obtained histology slices with approximately perpendicular to SI direction cuts, the overall TRE distribution of 1.2 mm in LR and 0.9 mm in AP achieved in this work is comparable with their reported TRE of 0.85.
Figure 4 illustrates a visualization of a corresponding slice between all the image sets after each registration step in the proposed workflow. Figure 4a–d represent the in vivo, fresh, fixed, and histology of a corresponding slice. Figure 4e displays the checkerboard visualization of the in vivo and deformed histology. The checkerboard visualization of the deformed fresh and in vivo is shown in Figure 4f. The alignment between deformed fixed and fresh images is visualized in Figure 4g. Finally, Figure 4h represents a satisfying alignment between deformed histology and the fixed ex vivo MR. It should be noted that the small black region under the urethra on the in vivo, fresh, and fixed image sets shows one of the gold seeds, and it corresponds to the white region under the urethra on the histology since the actual seed was removed during the sectioning process. Most of the corresponding slices between each image sets have achieved a similar qualitative performance after the proposed deformable registration which indicates an overall good quality of registration.
Figure 4.
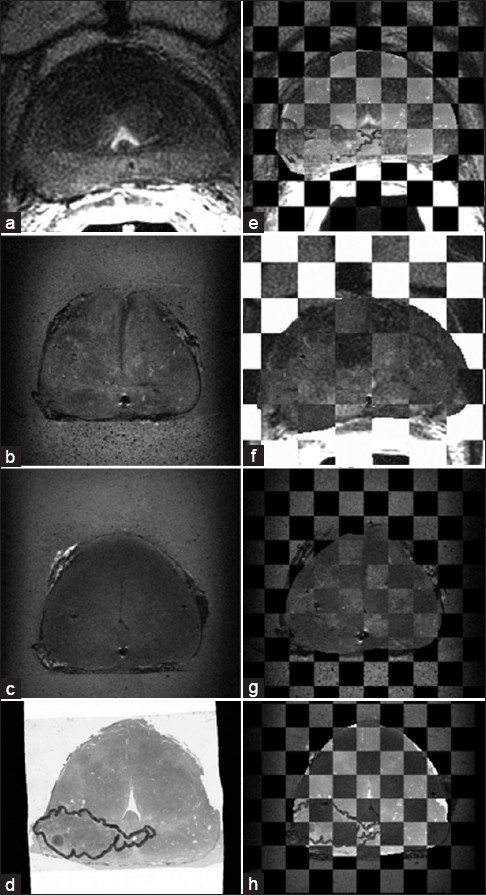
Checkerboard visualization for different registration steps in the proposed workflow. Figures (a)-(d) show the corresponding slice on in vivo, fresh, fixed, and the histology, respectively, (e) is the checkerboard display of the deformed histology overlaid on the in vivo. (f) shows the deformed fresh image overlaid on the in vivo, (g) represents the fresh image overlaid on the deformed fixed, (h) is the overlay of the deformed histology on the fixed image
CONCLUSION
Histology to MR correlation is challenging due to various deformations in the tissue. In this paper, a proof of concept deformable registration workflow is proposed which is primarily based on biomechanical modeling. The registration workflow is shown to be a promising technique for correlating histological slides of the prostate specimen to in vivo MRI. The addition of two intermediate steps of the prostate specimen (fresh and fixed) including the specimen-specific linear elastic heterogeneous material models built from MRE data provides useful information about the extent of deformations during the fixation process. The validation of the proposed technique using four patients resulted in the average TRE of 2.3 mm, which is in the order of the slice thickness, 3 mm. The current overall level of accuracy can be improved by using higher resolution (i.e., smaller slice thickness) in vivo images. Highly accurate registration of histology to high resolution ex vivo images before and after fixation provides unique information for future clinical investigations.
ACKNOWLEDGMENTS
This research was supported in part by grants from the Ontario Institute for Cancer Research, Terry Fox New Frontiers Program Project in Ultrasound for Cancer Therapy, and the Ontario Ministry of Health and Long Term Care. The views expressed do not necessarily reflect those of the OMOHLTC. Dr. Brock is supported through a Cancer Care Ontario Research Chair in Cancer Imaging.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2011/2/2/10/92035
REFERENCES
- 1.Jaffray DA, Siewerdsen JH, Wong JW, Martinez AA. Flat-panel cone-beam computed to-mography for image-guided radiation therapy. Int J Radiat Oncol Biol Phys. 2002;53:1337–49. doi: 10.1016/s0360-3016(02)02884-5. [DOI] [PubMed] [Google Scholar]
- 2.Jeanneret-Sozzi W, Moeckli R, Valley JF, Zouhair A, Ozsahin EM, Mirimanoff RO. The reasons for discrepancies in target volume delineation. Strahlenther Onkol. 2006;182:450–7. doi: 10.1007/s00066-006-1463-6. [DOI] [PubMed] [Google Scholar]
- 3.Teh BS, Bastasch MD, Wheeler TM, Mai W, Frolov A, Uhl BM, et al. IMRT for prostate cancer: defin-ing target volume based on correlated pathologic volume of disease. Int J Radiat Oncol Biol Phys. 2003;56:184–91. doi: 10.1016/s0360-3016(03)00085-3. [DOI] [PubMed] [Google Scholar]
- 4.Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: Application to breast MR images. IEEE Trans Med Imaging. 2002;18:712–21. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 5.Dauguet J, Delzescaux T, Condé F, Mangin JF, Ayache N, Hantraye P, et al. Three-dimensional reconstruction of stained histological slices and 3D non-linear registration with in vivo MRI for whole baboon brain. J Neurosci Methods. 2007;164:191–204. doi: 10.1016/j.jneumeth.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 6.Alic L, Haeck JC, Klein S, Bol K, Van Tiel ST, Wielopolski PA, et al. Multi-modal image registration: Matching MRI with histology. Proceedings of SPIE. 2010;7626:762603-1–9. [Google Scholar]
- 7.Chappelow J, Bloch BN, Rofsky N, Genega E, Lenkinski R, DeWolf W, et al. Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information. Med Phys. 2011;38:2005–18. doi: 10.1118/1.3560879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazaheri Y, Bokacheva L, Kroon DJ, Akin O, Hricak H, Chamudot D, et al. Semi-automatic deformable registration of prostate MR images to pathological slices. J Magn Reson Imaging. 2010;32:1149–57. doi: 10.1002/jmri.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan Y, Ou Y, Feldman M, Tomaszeweski J, Davatzikos C, Shen D. Registering Histologic and MR Images of Prostate for Image-based Cancer Detection. Acad Radiol. 2007;14:1367–81. doi: 10.1016/j.acra.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward A, Crukley C, McKenzie C, Montreuil J, Gibson E, Gomez J, et al. Registration of in vivo prostate magnetic resonance images to digital histopathology images. Prostate Cancer Imaging. Computer-Aided Diagnosis, Prognosis, and Intervention, Lect Notes Comput Sci. 2010;6367:66–76. [Google Scholar]
- 11.Ceritoglu C, Wang L, Selemon LD, Csernansky JG, Miller MI, Ratnanather JT. Large deformation diffeomorphic metric mapping registration of reconstructed 3D histological section images and in vivo MR images. Front Hum Neurosci. 2010;4:43. doi: 10.3389/fnhum.2010.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNiven A, Moseley J, Langer DL, Haider MA, Brock KK. Preliminary Feasibility Study: Modeling 3D deformations of the prostate from whole-mount histology to in vivo MRI. Med Phys. 2009;36:2567. [Google Scholar]
- 13.McGrath DM, Foltz WD, Brock KK. Measuring the Effect of Formalin Fixation on Ex vivo Tissue Material Properties using High Resolution 3D Quasi-Static MR Elastography at 7 Tesla for Improved Biomechanical Registration of Histopathology, and Correlation with the Effect of Fixation on T1,T2 and ADC. Proc Intl Soc Mag Res Med. 2010;18:2504. [Google Scholar]
- 14.Schaefer S, McPhail T, Warren J. Image deformation using moving least squares. ACM Transactions on Graphics (TOG) 2006;3:533–40. [Google Scholar]
- 15.Besl PJ, McKay ND. A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell. 1992;239:56. [Google Scholar]


