Abstract
Mineralization of bone matrix is an important process in bone formation; thus defects in mineralization have been implicated in bone mineral density (BMD) and bone structure alterations. Three central regulators of phosphate balance, ALPL, ANKH, and ENPP1, are central in the matrix mineralization process; therefore, the genes encoding them are considered important candidates genes for BMD and bone geometry. To test for an association between these three candidate genes and BMD and bone geometry traits, 124 informative single-nucleotide polymorphisms (SNPs) were selected and genotyped in 1513 unrelated subjects from the Framingham offspring cohort. Initial results showed that SNP rs1974201 in the gene ENPP1 was a susceptibility variant associated with several hip geometric indices, with the strongest p value of 3.8 ×10−7 being observed for femoral neck width. A few modest associations were observed between SNPs in or near ALPL and several bone traits, but no association was observed with ANKH. The association signals observed for SNPs around rs1974201 were attenuated after conditional analysis on rs1974201. Transcription factor binding-site prediction revealed that the HOXA7 binding site was present in the reference sequence with the major allele, whereas this potential binding site is lost in the sequence with the minor allele of rs1974201. In conclusion, we found evidence for association of bone geometry variation with an SNP in ENPP1, a gene in the mineralization pathway. The alteration of a binding site of the deregulator of extracellular matrix HOXA7 warrants further investigation.
Keywords: MINERALIZATION, ENPP1, ALPL, ANKH, BONE GEOMETRY, BONE MINERAL DENSITY, ASSOCIATION
Introduction
Osteoporosis is a major public health problem that is associated with disability and mortality and a huge socioeconomic burden owing to its complication of bone fractures. As the world’s population ages, the prevalence rates of osteoporotic fractures increase in parallel with approximately 200 million women worldwide suffering from osteoporosis. Low bone mineral density (BMD) and unfavorable bone geometry are two of the major determinants of bone strength and hence risk factors for osteoporotic fracture. These traits are under strong genetic regulation, with heritability estimates ranging from 0.5 to 0.9 for bone mineral density(1–4) and 0.3 to 0.6 for bone geometry.(5) Identification of genes affecting risk of osteoporosis therefore is important for disease prevention and treatment.
Bone modeling and remodeling are essential in maintaining healthy bone. Bone modeling during skeletal maturation produces a skeleton with a given mineralization and size, whereas bone remodeling removes old bone by osteoclastic bone resorption coupled with bone formation by osteoblasts. Therefore, defects in the bone-formation process affect not only BMD but also bone structure. During bone formation in both bone modeling and remodeling, osteoblasts first undergo proliferation and differentiation, and eventually they produce bone matrix and mediate bone deposition and mineralization. Mineralization of the extracellular matrix of bone is an essential element of bone development, maintenance, and repair. Consequently, the genes encoding for major factors controlling bone mineralization(6) seem to be very promising candidates for association studies with bone-strength phenotypes. In mineralization, hydroxyapatite is formed by crystallization of calcium and inorganic phosphate ions in chondrocyte-and osteoblast-derived matrix vesicles.(7) However, the formation of hydroxyapatite is antagonized by inorganic pyrophosphate (PPi).(8) Therefore, a tight balance between the concentration of inorganic phosphate (Pi) and PPi is essential to maintain proper bone mineralization. Three central regulators mediate this process, namely, alkaline phosphatase liver/bone/kidney (ALPL), ankylosis progressive homologue (mouse) (ANKH), and ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1). ENPP1 modulates physiologic mineralization by generating PPi from trinucleotide phosphate. ALPL hydrolyzes PPi into Pi, whereas ANKH mediates the transport of intracellular and extracellular PPi.(6) Defects in bone mineralization or excess of ectopic ossification have been observed in knockout or mutant mice of alpl,(6,9) ankh,(10) and enpp1.(6,11) We previously reported that polymorphisms in the distal region of the ANKH gene were significantly associated with serum circulating levels of biomarkers involved in bone metabolism, such as osteoprotegerin(12) and parathyroid hormone.(13) We also found previously significant associations between the polymorphisms mapped to a promoter region of ANKH and several skeletal size traits, including body height and length of long bones,(14) suggesting that this gene also may be involved in bone geometry variation. However, the contributions of variants in these genes to BMD and bone geometry variations in humans remain poorly understood. Therefore, an investigation of the effects of polymorphisms located in these genes in another population is important at least in two respects—to test the effect of these genes on bone mass and geometry variations and to verify that this effect is not spurious or population-specific. Here we studied these three mineralization pathway–related candidate genes with osteoporosis-related traits in a well-characterized sample of men and women from the Framingham Osteoporosis Study.
Materials and Methods
Study population
The Framingham Osteoporosis Study is an ancillary study of the population-based Framingham Heart Study (FHS). The 1513 totally unrelated participants in this study are a subset from the Framingham offspring cohort that provided blood samples for DNA and that had bone phenotypes. These subjects were not selected on the basis of any trait but rather based only on having bone phenotypes and DNA for genotyping. The detailed design of the Framingham Osteoporosis Study has been previously described.(15) The study was approved by the Institutional Review Boards for Human Subjects Research of Boston University and Hebrew Rehabilitation Center, and informed consent, including consent for genetic analyses, was obtained from all study participants.
BMD measurements and HSA
BMD (g/cm2) at the L2–L4 vertebral spine (LS BMD) and femoral neck (FN BMD) was measured by dual-energy X-ray absorptiometry (DXA) with a Lunar DPX-L (Lunar Corp., Madison, WI, USA) between 1996 and 2001. An interactive computer program (Hip Structural Analysis, HSA)(16) was used to derive a number of structural variables from the femoral DXA scans. The regions assessed were the narrowest width of the femoral neck (NN), which overlaps or is proximal to the standard Lunar femoral neck region, and the femoral shaft (S)—1.5 times the minimum neck width distal to the intersection of the neck and shaft axes (see ref. (17) for details). HSA provided measures of outer diameter at both femoral regions (NN WIDTH and S WIDTH), as well as neck shaft angle (NSA) and femoral neck length (FNL, defined as the distance from the center of the femoral head to the intersection of neck and shaft axes). These measures have been well described in previous publications.(18,19) In total, six traits are reported here (LS BMD, FN BMD, NN WIDTH, S WIDTH, NSA, and FNL). Coefficients of variation for the different component variables were reported previously for LS BMD (0.9%), FN BMD (1.7%),(20) and the geometric indices, ranging from 3.3% (NN WIDTH) to 9.1% (FNL).(21)
Other characteristics
Variables potentially influencing BMD and hip geometry were obtained at the time of DXA measurement along with overall medical history. Details of these measurements have been published previously.(22) These variables included age, sex, height, weight, and, for women, estrogen use and menopausal status. Weight was measured using a standardized balance-beam scale. Height (without shoes) was measured to the nearest ¼ in using a stadiometer. Tobacco use (ie, cigarette smoking) was assessed at each examination as number of cigarettes per day and used in this study as current, former, or nonsmoker at time of the scan. Men and women were classified into one of two estrogen status categories: (1) premenopausal or postmenopausal women on estrogen-replacement therapy (estrogen-replete) or (2) men or postmenopausal women not on estrogen, where menopause was defined as having no menstrual period for at least 1 year (estrogen-depleted).
DNA extraction, SNP selection, and genotyping
Genomic DNA was extracted from peripheral blood leukocytes using a phenol/chloroform extraction method, and DNA from immortalized lymphoblast cell lines was salt-precipitate extracted (detailed description has been reported previously(23)). For all three candidate genes, SNP tagging was done in a genomic region 20 kb upstream and 10 kb downstream of each gene. For ANKH and ALPL, we selected tag SNPs to account for at least 80% of the common haplotypes,(24) as defined in 2006. Tag SNP selection software TagSNPs (www-rcf.usc.edu/~stram) was applied to the CEPH population based on Phase I HapMap to choose tagging SNPs that were then genotyped in Framingham participants. The TagSNP selection algorithm is based on optimizing the squared correlation between estimates of the number of copies of a particular haplotype h and the true number of copies of haplotype h (Rh2) carried by a subject, averaging over all possible genotype data under an assumption of Hardy-Weinberg equilibrium (HWE).(25) Genotyping for SNPs in ALPL and ANKH was done using ABI Taqman SNP genotyping assays. Strategies in SNP tagging and genotyping methods for ENPP1 have been described previously.(26) In brief, coverage of ENPP1 reached 100% using variants with minor allele frequency (MAF) ≥ 5%, with HWE p ≤.001 and r2 ≥0.70 based on HapMap Phase II (Release 21) CEU population.
For ALPL and ANKH, since the initial tagging SNPs were selected and genotyped based on Phase I HapMap, we improved gene coverage by extracting additional genotypes from 21 and 40 SNPs in and around these genes, respectively, from the Affymetrix 550K Genechip that was used in the Framingham cohorts. The Affymetrix 550K Genechip provides consistently high coverage in different populations. It is comprised of two arrays, and approximately 262,000 and 238,000 SNPs were genotyped by Nsp and Sty arrays, respectively. Gene coverage of ALPL and ANKH eventually reached 74% and 78% of variants whose minor allele frequency is 5% or more with an r2 ≥ 0.70 based on HapMap Phase II (Release 21) CEU population. Finally, with the addition of these SNPs, 34, 50, and 40 SNPs in total were included in ALPL, ANKH, and ENPP1, respectively, with an average intermarker distance of 2.8 kb and capturing at least 84% (r2 threshold of 0.7) of the total number of SNPs in these three candidate genes, according to HapMap Phase II (Release 21) CEU population.
Statistical analysis
The genotyping quality of each SNP was first checked for the call rate, MAF, and HWE using the PLINK software.(27) SNPs with MAF less than 0.05, call rate less than 90%, and HWE p value greater than .001 were excluded from further analysis. Subjects with genotyping rate < 90% also were excluded from further analysis.
The p values, β coefficients, and standard errors were determined using linear regression with an additive model to determine the association of the SNPs with BMD and hip geometry; all analyses were adjusted for age, sex, height, weight, estrogen status, smoking, and alcohol consumption in the linear regression model. Since we observed a strong association between rs1974201 and several hip geometric indices, a conditional analysis also was performed to investigate whether the significant associations in ENPP1 were mainly driven by the effect of rs1974201.
Two approaches were used to correct for multiple testing: (1) 1 million permutations were conducted in PLINK(27) using the command mperm to control for the family-wise error rate, and (2) a Bonferroni correction was applied to correct for 6 traits and 103 markers (618 tests), with the resulting p value significant threshold at 8.1 ×10−5 (because these 6 traits are not totally independent, this correction threshold is considered conservative).
Transcription factor binding site (TFBS) prediction
Because rs1974201 is located in intron 24 of ENPP1, we hypothesized that it may be functional and located in or near an intronic enhancer(28–30); therefore, we investigated the presence of a possible TFBS in the flanking sequence of rs1974201. The potential effect of rs1974201 on TFBS alteration was first predicted using software Alibaba2 (available at www.gene-regulation.com/pub/programs/alibaba2/index.html). The prediction was cross-validated subsequently using an independent TFBS prediction tool (MATINSPECTOR).(31) Matrices used for the TFBS prediction can be filtered by the tissues they are associated with in MATINSPECTOR. Matrix filters of “skeleton” and “bone and bones” were used in the TFBS prediction. Core similarity and matrix similarity were obtained from the program output; in brief, core similarity was defined as the highest conserved positions of the matrix.(31) An optimized matrix similarity was used as the threshold for assessing false-positive rate. When matrix similarity is equal to or larger than the threshold, the number of false-positive matches is minimized.
Results
Baseline characteristics
Table 1 presents the baseline characteristics of the Framingham offspring subjects. There were 1513 phenotyped participants. In this sample, men and women were of similar age. As expected, male participants were heavier, taller, and in general had larger average BMD and geometric measures than females. Current smokers constituted 11.3% of the men and 13.3% of the women. Of the total 797 women, 372 were either premenopausal or on estrogen-replacement therapy.
Table 1.
Clinical and Demographic Information of the Studied Framingham Offspring Cohort Participants
| Continuous variables | Men (n =716) | Women (n =797) |
|---|---|---|
| Mean SD | Mean SD | |
| Age (years) | 62.0 ±9.1 | 60.6 ± 9.0* |
| Height (cm) | 174.81 ±6.71 | 161.39 ± 6.18* |
| Weight (kg) | 87.67 ±15.55 | 71.71 ± 15.35* |
| Alcohol (oz/week) | 3.56 ±4.72 | 1.74 ± 2.46* |
| LS BMD (g/cm2) | 1.20 ±1.17 | 1.12 ± 0.75 |
| FN BMD (g/cm2) | 0.85 ±1.12 | 0.80 ± 0.87 |
| NSA (degrees) | 127.09 ±18.39 | 126.20 ± 15.31* |
| FNL (cm) | 5.90 ±2.08 | 5.19 ± 1.55* |
| NN width (cm) | 3.78 ±1.58 | 3.25 ± 1.85* |
| S width (cm) | 1.84 ±4.38 | 2.21 ± 3.63* |
| Categorical variables | n | % | n | % |
|---|---|---|---|---|
| Smoking =never | 214 | 30.3 | 279 | 35.4† |
| Smoking =former | 413 | 58.4 | 405 | 51.3† |
| Smoking =current | 80 | 11.3 | 105 | 13.3† |
| Estrogen statusa =low | NA | NA | 416 | 52.8 |
| Estrogen status =high | NA | NA | 372 | 47.2 |
Estrogen status: “Low” means postmenopausal women; “high” means either premenopausal or women on estrogen.
Indicates significant difference at p <.01 between males and females using t test.
Indicates significant difference at p <0.01 between males and females using chi-square test
Genotyping frequencies, linkage disequilibrium (LD), and single-marker association
Genotype data were available for 124 SNPs. The genotyping frequencies and allele information for these 124 SNPs are summarized in Supplemental Table 1. One SNP with a call rate of less than 0.9 and MAF greater than 0.05 and 20 additional SNPs with MAF less than 0.05 were filtered out, leaving 103 SNPs in the final analysis. Coverage of ALPL, ANKH, and ENPP1 reached 74%, 74%, and 100%, respectively. The LD pattern of these 103 SNPs in the three genes is presented in Supplemental Fig. 1 A–C.
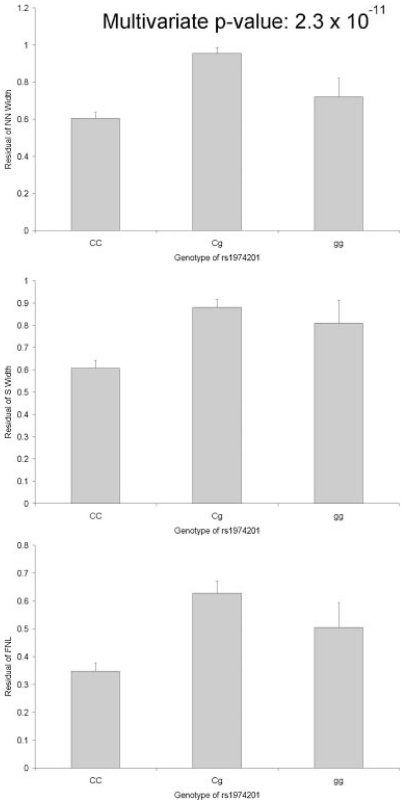
Results of single-marker associations with a nominal p value of less than .05 are summarized in Table 2. All association results (including insignificant results) for each SNP with BMD and hip geometric indices are provided in Supplemental Tables 2 and 3, respectively. Although there were several nominally significant findings for SNPs in the three genes of interest, only SNP rs1974201 in ENPP1 remained statistically significant after Bonferroni correction for multiple hypothesis testing (corrected for 6 traits and 103 markers; Bonferroni-corrected p values of .039, .01, and .00023 for FNL, S WIDTH and NN WIDTH, respectively) or permutation approach (1 million permutations; empirical p values of .005, .001, and 3.8 ×10−5 for FNL, S WIDTH, and NN WIDTH, respectively). By evaluating the estimated means and standard errors of each genotype of rs1974201, we observed that the genetic model deviated from additivity (Fig. 1A–C) despite the fact that a large standard error was observed in the homozygous recessive genotype. Therefore, we repeated the association analysis of rs1974201 with the three traits using a dominant model. Interestingly, improved association signals were observed for rs1974201, with all three hip geometric indices with nominal p values of 1.3 ×10−6, 2.5 ×10−6, and 8.5 ×10−11 being observed for FNL, S WIDTH, and NN WIDTH, respectively (Table 3). The estimated effect sizes (β coefficients) of rs1974201 were 0.177, 0.080, and 0.133 for FNL, S WIDTH, and NN WIDTH, respectively. Multivariate analysis using all three bone geometry traits as dependent variables resulted in a stronger association with a p value of 2.3 ×10−11. In addition, previous studies suggested sex-specific association on bone traits.(4,32,33) However, no significant sex-specific signal was revealed when subgroup analysis was performed on either men or women alone because the associations were significant in both genders (data not shown).
Table 2.
Results of Single-Marker Association Tests for Osteoporosis-Related Traits (Only Results With Nominal p Values < .05 Are Shown)
| Traits | SNP | Chr | Positiona | Gene | Betab | SE | Nom. pc | Emp. pd | Bon. pe |
|---|---|---|---|---|---|---|---|---|---|
| LS BMD | rs10917003 | 1 | 21718136 | ALPL | .017 | .008 | .039 | .910 | 1 |
| NSA | rs7535497 | 1 | 21713295 | ALPL | −.405 | .195 | .038 | .803 | 1 |
| rs7523855 | 1 | 21713472 | ALPL | −.4 | .196 | .041 | .825 | 1 | |
| FNL | rs10917003 | 1 | 21718136 | ALPL | .058 | .028 | .0379 | .860 | 1 |
| rs1256335 | 1 | 21762972 | ALPL | −.067 | .03 | .02675 | .770 | 1 | |
| rs9493100 | 6 | 132156773 | .097 | .044 | .02873 | .792 | 1 | ||
| rs858345 | 6 | 132205309 | ENPP1 | .058 | .025 | .02032 | .690 | 1 | |
| rs1974201 | 6 | 132252813 | ENPP1 | .119 | .03 | 6.37 ×10−5 | .005 | .039 | |
| rs7754561 | 6 | 132254386 | ENPP1 | .074 | .028 | .009003 | .418 | 1 | |
| NN Width | rs12025623 | 1 | 21765065 | ALPL | −.029 | .015 | .047 | .906 | 1 |
| rs2275370 | 1 | 21773006 | ALPL | .038 | .018 | .029 | .780 | 1 | |
| rs7752279 | 6 | 132154027 | .036 | .014 | .012 | .489 | 1 | ||
| rs9493100 | 6 | 132156773 | .074 | .025 | .003 | .196 | 1 | ||
| rs858342 | 6 | 132202335 | ENPP1 | .032 | .016 | .049 | .925 | 1 | |
| rs858345 | 6 | 132205309 | ENPP1 | .045 | .014 | .002 | .105 | 1 | |
| rs1044498 | 6 | 132214060 | ENPP1 | .046 | .019 | .017 | .641 | 1 | |
| rs7768480 | 6 | 132245124 | ENPP1 | .091 | .025 | 2.24 ×10−4 | .017 | .138 | |
| rs1799774 | 6 | 132245166 | ENPP1 | .059 | .017 | 4.19 ×10−4 | .030 | .259 | |
| rs1974201 | 6 | 132252813 | ENPP1 | .084 | .017 | 3.78 ×10−7 | 3.78 ×10−5 | 2.34 ×10−4 | |
| rs7754561 | 6 | 132254386 | ENPP1 | .046 | .016 | .005 | .286 | 1 | |
| rs7753048 | 6 | 132259379 | .046 | .022 | .038 | .871 | 1 | ||
| rs9373000 | 6 | 132263398 | .045 | .016 | .005 | .281 | 1 | ||
| S Width | rs6658127 | 1 | 21747976 | ALPL | −.03 | .012 | .017 | .614 | 1 |
| rs12025623 | 1 | 21765065 | ALPL | −.038 | .012 | .002 | .123 | 1 | |
| rs10917008 | 1 | 21766893 | ALPL | −.033 | .013 | .011 | .478 | 1 | |
| rs3767142 | 1 | 21774144 | ALPL | −.027 | .012 | .03 | .795 | 1 | |
| rs7752279 | 6 | 132154027 | .028 | .012 | .018 | .637 | 1 | ||
| rs9493100 | 6 | 132156773 | .074 | .021 | 4.18 ×10−4 | .031 | .258 | ||
| rs943004 | 6 | 132182568 | ENPP1 | .059 | .026 | .023 | .711 | 1 | |
| rs9375830 | 6 | 132188212 | ENPP1 | .034 | .015 | .028 | .774 | 1 | |
| rs2021966 | 6 | 132192131 | ENPP1 | −.028 | .012 | .016 | .601 | 1 | |
| rs858345 | 6 | 132205309 | ENPP1 | .033 | .012 | .006 | .289 | 1 | |
| rs1044498 | 6 | 132214060 | ENPP1 | .032 | .016 | .047 | .918 | 1 | |
| rs7768480 | 6 | 132245124 | ENPP1 | .042 | .021 | .043 | .897 | 1 | |
| rs1799774 | 6 | 132245166 | ENPP1 | .039 | .014 | .005 | .268 | 1 | |
| rs1974201 | 6 | 132252813 | ENPP1 | .06 | .014 | 1.53 ×10−5 | .001 | .010 | |
| rs9373000 | 6 | 132263398 | .038 | .013 | .004 | .209 | 1 |
LS BMD =L2-L4 vertebral spine BMD; FN BMD =femoral neck BMD; NSA =neck shaft angle; FNL =femoral neck length; NN width =femoral neck width; S width =femoral neck shaft width.
Physical position was obtained from dbSNP build 129.
Beta: β Coefficient for minor allele.
Nom. p: Nominal p value.
Emp. p: Empirical p value obtained through 1 million permutations.
Bon. p: Bonferroni-corrected p value for 6 traits and 103 SNPs.
Fig. 1.
Estimated means and standard errors of geometric phenotypes for each genotype of rs1974201. 1a, NN WIDTH; 1b, S WIDTH; 1c, FNL.
Table 3.
Association of rs1974201 With FNL, NN Width, and S Width Using Dominant Model
| SNP | Traits | Beta | SE | Nom. pa | Emp. pb | Bon. pc |
|---|---|---|---|---|---|---|
| rs1974201 | FNL | .177 | .036 | 1.3 ×10−6 | 1.2 ×10−4 | .001 |
| NN width | .133 | .020 | 8.5 ×10−11 | 1.0 ×10−6 | 5.3 ×10−8 | |
| S width | .080 | .017 | 2.5 ×10−6 | 2.0 ×10−4 | .002 |
Nom. p: Nominal p value.
Emp. p: Empirical p value obtained through 1 million permutations.
Bon. p: Bonferroni-corrected p value for 6 traits and 103 SNPs.
Conditional analysis
The other significant association signals in ENPP1 all were attenuated after adjustment for the effect of the index SNP rs1974201 (Supplemental Fig. 2), except for the SNP rs943004, which showed a more significant association with S WIDTH ( p =.009 in the conditional analysis versus p =.02 in the unconditional analysis).
TFBS prediction
The first prediction using Alibaba2 showed that the HOXA7 binding site was present in the sequence featuring the major allele G but absent in the sequence featuring the minor allele C. Using MATINSPECTOR, we observed the presence of home-odomain transcription factors in the sequence just next to the position of rs1974201 (ttttgacttttTAATaata: underlined sequence indicates the matrix sequence, bolded nucleotides indicate the core sequence). The core and matrix similarity were 1 and 0.975, respectively, which exceeds the optimized matrix similarity threshold of 0.95, indicating a low possibility of false-positive detection.
Discussion
This association study directly examined three candidate genes involved in the bone mineralization pathway and revealed a possible role for ENPP1 in bone geometry variation.(34) This approach has several advantages. First, it has a solid biologic background in the selection process of candidate genes, which may help in interpretation of results. Second, focused testing of several informative SNPs in these few genes reduces the multiple-testing problem.
This study identified the genetic variant rs1974201 in ENPP1 as a potentially causal variant for hip geometry variation in the Framingham offspring cohort. The strong association signal observed in this study is unlikely to be a spurious finding because the association p value between rs1974201 and NN WIDTH remained significant after either Bonferroni correction for 618 tests or with 1 million permutations and was consistently associated with three bone geometry traits (FNL, S WIDTH, and NN WIDTH). Based on conditional analysis, the association signals observed with the SNPs around ENPP1 rs1974201 were attenuated, suggesting that these association signals stemmed from the main effect of rs1974201, whereas signals from surrounding SNPs are likely driven by LD with rs1974201. Further, our TFBS prediction revealed a potential alteration of the HOXA7 binding site; HOXA7’s role as a deregulator of extracellular matrix formation is a potential explanation for the strong association observed in this study. Therefore, these findings merit further investigation of the ENPP1 effect on hip geometry and suggest that ENPP1 may be a novel potential target gene in osteoporosis risk prediction.
ENPP1 is an important regulator of PPi production in matrix vesicles and osteoblasts. After the cessation of longitudinal growth, bone remodeling continues on the endosteal surfaces. Osteoclasts resorb a volume of bone, leaving a focal resorptive cavity. After a delay, osteoblasts fill the cavity with a volume of new bone that undergoes rapid primary and then slower secondary mineralization.(35) In mice, enpp1 is required to prevent excessive calcification in several skeletal sites, such as the calvarium and vertebrae. Therefore, enpp1 null mice show hypermineralization abnormalities in these two sites. On the other hand, enpp1 deficiency also causes hypomineralization of long bones, suggesting that enpp1 affects mineralization of the skeleton in a site-specific manner.(36) Interestingly, our finding also is in agreement with a most recent study using a genome-wide N-ethyl-N-nitrosourea (ENU)-induced mutagenesis screen that showed that in mice, a novel mutation C397S in enpp1 was associated with low bone mass, crystal-related arthropathy, and vascular calcification.(37) In that study, low bone mass was observed only with cortical bone in femoral metaphysis and diaphysis, suggesting that enpp1 may be more important in cortical bone metabolism. Although no association was observed between ENPP1 polymorphisms and BMD variation in this study, it may be due to the difference in genetic effect of a common polymorphism versus missense mutation in the pyropho-sphatase/phosphodiesterase domain because it was the latter that significantly affected protein stability of enpp1.(37) Moreover, overexpression of ENPP1 resulted in insulin resistance and defective adipocyte maturation from mesenchymal stem cells.(38) Given the fact that the mesenchymal stem cell is the common ancestor of osteoblast and adipocyte, whether this gene also may affect osteoblast differentiation remains to be elucidated.
Although we examined the association of three candidate genes in the mineralization pathway, it is not surprising that ENPP1 showed the strongest association. First, based on a previous in vivo study, the degree of defects in mineralization of enpp1 knockout mice has been shown to be more severe than that in ankh mutant mice; second, enpp1, but not ankh, is present and is functional in matrix vesicles.(6) In humans, arterial calcification has been observed widely in subjects carrying mutations in the ENPP1 gene.(39–41) Interestingly, we(42) and other groups(43,44) also have reported that arterial calcification is associated with bone loss, osteoporosis, and fracture in both older men and women. Moreover, an association has been reported between several polymorphisms in ENPP1 and radiographic hand osteoarthritis.(45) Thus ENPP1 may be a functional gene that affects both fracture and ectopic ossification pleiotropically, especially in aged subjects who commonly have both increased arterial calcification and risk of fracture (as also was demonstrated in a recent study in mice(37)).
To our knowledge, there have been no previous studies evaluating the association between osteoporosis-related traits and the ENPP1 gene, although variants in this gene have been studied extensively for association with diabetes-related traits,(46) including several studied by us, such as rs1044498 (K121Q), rs1799774 (delT), and rs7754561. Three, two, and one of these SNPs were nominally associated with NN WIDTH, S WIDTH, and FNL in our study, respectively. We note that the variant most frequently associated with type 2 diabetes, rs1044498/K121Q, is not in LD with our index SNP rs1974201 (r2 =0.02 in Framingham). On the other hand, rs1974201, which showed the strongest association signal in our study, also manifested the strongest association with type 2 diabetic end-stage renal disease (with the minor allele corresponding to a higher risk of disease, p =.0005),(47) suggesting that this SNP may be a functional variant with a pleiotropic effect in different diseases.
New bioinformatics algorithms are being used to improve the predictive specificity of TFBS, whereas a number of the predictions have been validated successfully using functional studies.(48,49) Using the in silico TFBS prediction tools, an alteration of a potential transcription factor binding site of HOXA7 was predicted for rs1974201. Absence of predicted intronic enhancer in the sequence bearing the minor allele may be associated with inefficient gene transcription and hence enhanced bone mineralization because ENPP1 is a deregulator of bone mineralization. Interestingly, similar to ENPP1, HOXA7 is a deregulator of extracellular matrix formation in monocytes(50) and is able to dimerize with PBX1,(51) a product of a novel gene involved in osteoblastogenesis that we have identified recently.(34) Although HOXA7 is involved in extracellular matrix formation in monocytes, it may not be generalizable to bone cells. Therefore, functional studies are required to confirm the prediction and interaction between ENPP1 and HOXA7, especially in mineralized tissue.
The other candidate gene in the current study, ALPL, is a well-known early osteogenic marker in osteoblast differentiation and as such is an excellent candidate for BMD and bone geometry variation. However, we observed only a few modest associations of ALPL with BMD and hip geometry variation, and all became insignificant after correction for multiple testing; this is in agreement with a previous study that did not reveal any significant associations between SNPs in ALPL and BMD variation.(52)
There are several limitations to this study. First, although SNP coverage of ENPP1 was excellent, reaching at least 100% based on an r2 of 0.7 of SNPs with MAF greater than 0.05, our study covered only 74% of all SNPs present in ANKH and ALPL based on the HapMap Phase II (Release 21) CEU population. This may limit the power to detect causal and intermediate-or low-frequency variant(s) that may exist in these genes. Second, the effect size of rs1974201 observed in this study may be an overestimate owing to the “winner’s curse” such that future replications and meta-analyses are important to estimate the true effect size and evidence of association.
In conclusion, this study of 1,513 unrelated white subjects identified rs1974201 in the ENPP1 gene as a potentially causal variant in bone geometry determination. Our study also underscores the importance of having good coverage to be able to identify susceptibility variants in disease gene mapping. Moreover, we also suggest that we should not overlook the importance of bone geometry in future association studies because some genes may be important for bone geometry determination rather than in governing BMD. Future replication and/or functional studies of rs1974201 are needed to validate our findings and to elucidate the underlying molecular mechanism of how this SNP or gene influences bone geometry. Ultimately, the identification of significant variants in a variety of genes contributing to bone density and geometry will contribute to future studies of genetic predisposition to fracture. Novel pathways identified by these studies may be leveraged for future drug development.
Supplementary Data
Acknowledgments
This work was supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (Contract No. N01-HC-25195), an American Diabetes Association Career Development Award, and NIDDK K24 DK080140 (JBM), NHLBI 1U01 HL066582, and grants from the National Institute on Arthritis Musculoskeletal and Skin Diseases, and the National Institute on Aging Grants R01 AR/AG 41398 (DPK) and R01 AR 050066 and AR 057118 (DK). The postdoctoral fellowship of CLC is generously supported by the Men’s Associates of Hebrew SeniorLife. JCF is supported by a Massachusetts General Hospital Physician Scientist Development Award and a Doris Duke Charitable Foundation Clinical Scientist Development Award.
Footnotes
Disclosures
All the authors state that they have no conflicts of interest.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Ralston SH. Genetic determinants of osteoporosis. Curr Opin Rheumatol. 2005;17:475–479. doi: 10.1097/01.bor.0000166385.62851.92. [DOI] [PubMed] [Google Scholar]
- 2.Livshits G, Deng HW, Nguyen TV, Yakovenko K, Recker RR, Eisman JA. Genetics of bone mineral density: evidence for a major pleiotropic effect from an intercontinental study. J Bone Miner Res. 2004;19:914–923. doi: 10.1359/JBMR.040132. [DOI] [PubMed] [Google Scholar]
- 3.Eisman JA. Genetics of osteoporosis. Endocr Rev. 1999;20:788–804. doi: 10.1210/edrv.20.6.0384. [DOI] [PubMed] [Google Scholar]
- 4.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demissie S, Dupuis J, Cupples LA, Beck TJ, Kiel DP, Karasik D. Proximal hip geometry is linked to several chromosomal regions: genome-wide linkage results from the Framingham Osteoporosis Study. Bone. 2007;40:743–750. doi: 10.1016/j.bone.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harmey D, Hessle L, Narisawa S, Johnson KA, Terkeltaub R, Millan JL. Concerted regulation of inorganic pyrophosphate and osteopontin by akp2, enpp1, and ank: an integrated model of the pathogenesis of mineralization disorders. Am J Pathol. 2004;164:1199–209. doi: 10.1016/S0002-9440(10)63208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boskey AL, Boyan BD, Schwartz Z. Matrix vesicles promote mineralization in a gelatin gel. Calcif Tissue Int. 1997;60:309–15. doi: 10.1007/s002239900234. [DOI] [PubMed] [Google Scholar]
- 8.Johnson K, Moffa A, Chen Y, Pritzker K, Goding J, Terkeltaub R. Matrix vesicle plasma cell membrane glycoprotein-1 regulates mineralization by murine osteoblastic MC3T3 cells. J Bone Miner Res. 1999;14:883–92. doi: 10.1359/jbmr.1999.14.6.883. [DOI] [PubMed] [Google Scholar]
- 9.Waymire KG, Mahuren JD, Jaje JM, Guilarte TR, Coburn SP, MacGregor GR. Mice lacking tissue non-specific alkaline phosphatase die from seizures due to defective metabolism of vitamin B-6. Nat Genet. 1995;11:45–51. doi: 10.1038/ng0995-45. [DOI] [PubMed] [Google Scholar]
- 10.Ho AM, Johnson MD, Kingsley DM. Role of the mouse ank gene in control of tissue calcification and arthritis. Science. 2000;289:265–70. doi: 10.1126/science.289.5477.265. [DOI] [PubMed] [Google Scholar]
- 11.Okawa A, Nakamura I, Goto S, Moriya H, Nakamura Y, Ikegawa S. Mutation in Npps in a mouse model of ossification of the posterior longitudinal ligament of the spine. Nat Genet. 1998;19:271–3. doi: 10.1038/956. [DOI] [PubMed] [Google Scholar]
- 12.Vistoropsky Y, Malkin I, Kobyliansky E, Livshits G. Osteoprotegerin plasma levels are strongly associated with polymorphisms in human homologue of the mouse progressive ankylosis (ANKH) gene. Ann Hum Genet. 2007;71( Pt 3):302–7. doi: 10.1111/j.1469-1809.2006.00331.x. [DOI] [PubMed] [Google Scholar]
- 13.Vistoropsky Y, Keter M, Malkin I, Trofimov S, Kobyliansky E, Livshits G. Contribution of the putative genetic factors and ANKH gene polymorphisms to variation of circulating calciotropic molecules. PTH and BGP. Hum Mol Genet. 2007;16:1233–40. doi: 10.1093/hmg/ddm071. [DOI] [PubMed] [Google Scholar]
- 14.Malkin I, Ermakov S, Kobyliansky E, Livshits G. Strong association between polymorphisms in ANKH locus and skeletal size traits. Hum Genet. 2006;120:42–51. doi: 10.1007/s00439-006-0173-6. [DOI] [PubMed] [Google Scholar]
- 15.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families: the Framing-ham offspring study. Am J Epidemiol. 1979;110:281–90. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 16.Beck TJ, Ruff CB, Warden KE, Scott WW, Jr, Rao GU. Predicting femoral neck strength from bone mineral data: a structural approach. Invest Radiol. 1990;25:6–18. doi: 10.1097/00004424-199001000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Karasik D, Cupples LA, Hannan MT, Kiel DP. Genome screen for a combined bone phenotype using principal component analysis: the Framingham study. Bone. 2004;34:547–56. doi: 10.1016/j.bone.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 18.Greenspan SL, Beck TJ, Resnick NM, Bhattacharya R, Parker RA. Effect of hormone replacement, alendronate, or combination therapy on hip structural geometry: a 3-year, double-blind, placebo-controlled clinical trial. J Bone Miner Res. 2005;20:1525–32. doi: 10.1359/JBMR.050508. [DOI] [PubMed] [Google Scholar]
- 19.Rivadeneira F, van Meurs JB, Kant J, et al. Estrogen receptor beta (ESR2) polymorphisms in interaction with estrogen receptor alpha (ESR1) and insulin-like growth factor I (IGF1) variants influence the risk of fracture in postmenopausal women. J Bone Miner Res. 2006;21:1443–56. doi: 10.1359/jbmr.060605. [DOI] [PubMed] [Google Scholar]
- 20.Hannan MT, Felson DT, Dawson-Hughes B, et al. Risk factors for longitudinal bone loss in elderly men and women: the Framingham Osteoporosis Study. J Bone Miner Res. 2000;15:710–20. doi: 10.1359/jbmr.2000.15.4.710. [DOI] [PubMed] [Google Scholar]
- 21.Khoo BC, Beck TJ, Qiao QH, et al. In vivo short-term precision of hip structure analysis variables in comparison with bone mineral density using paired dual-energy X-ray absorptiometry scans from multi-center clinical trials. Bone. 2005;37:112–21. doi: 10.1016/j.bone.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Karasik D, Myers RH, Cupples LA, et al. Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: the Framingham Study. J Bone Miner Res. 2002;17:1718–27. doi: 10.1359/jbmr.2002.17.9.1718. [DOI] [PubMed] [Google Scholar]
- 23.Cupples LA, Arruda HT, Benjamin EJ, et al. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet. 2007;8( Suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K, Deng M, Chen T, Waterman MS, Sun F. A dynamic programming algorithm for haplotype block partitioning. Proc Natl Acad Sci USA. 2002;99:7335–9. doi: 10.1073/pnas.102186799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bakker PI, Burtt NP, Graham RR, et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006;38:1298–303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- 26.Stolerman ES, Manning AK, McAteer JB, et al. Haplotype structure of the ENPP1 gene and nominal association of the K121Q missense single nucleotide polymorphism with glycemic traits in the Framingham Heart Study. Diabetes. 2008;57:1971–7. doi: 10.2337/db08-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng W, Huang J, Zhang J, Williams T. Identification and analysis of a conserved Tcfap2a intronic enhancer element required for expression in facial and limb bud mesenchyme. Mol Cell Biol. 2008;28:315–25. doi: 10.1128/MCB.01168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wederell ED, Bilenky M, Cullum R, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–64. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–41. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 31.Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–42. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 32.Ioannidis JP, Ng MY, Sham PC, et al. Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. J Bone Miner Res. 2007;22:173–83. doi: 10.1359/jbmr.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ralston SH, Galwey N, MacKay I, et al. Loci for regulation of bone mineral density in men and women identified by genome wide linkage scan: the FAMOS study. Hum Mol Genet. 2005;14:943–51. doi: 10.1093/hmg/ddi088. [DOI] [PubMed] [Google Scholar]
- 34.Cheung CL, Chan BY, Chan V, et al. Pre-B-cell leukemia homeobox 1 (PBX1) shows functional and possible genetic association with bone mineral density variation. Hum Mol Genet. 2009;18:679–87. doi: 10.1093/hmg/ddn397. [DOI] [PubMed] [Google Scholar]
- 35.Kaufman JM, Ostertag A, Saint-Pierre A, et al. Genome-wide linkage screen of bone mineral density (BMD) in European pedigrees ascertained through a male relative with low BMD values: evidence for quantitative trait loci on 17q21-23, 11q12-13, 13q12-14, and 22q11. J Clin Endocrinol Metab. 2008;93:3755–62. doi: 10.1210/jc.2008-0678. [DOI] [PubMed] [Google Scholar]
- 36.Anderson HC, Harmey D, Camacho NP, et al. Sustained osteomalacia of long bones despite major improvement in other hypophosphatasia-related mineral deficits in tissue nonspecific alkaline phosphatase/nucleotide pyrophosphatase phosphodiesterase 1 double-deficient mice. Am J Pathol. 2005;166:1711–20. doi: 10.1016/S0002-9440(10)62481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babij P, Roudier M, Graves T, et al. New Variants in Enpp1 and Ptpn6 genes cause low bone density, crystal-related arthropathy and vascular calcification. J Bone Miner Res. 2009;24:1552–1564. doi: 10.1359/jbmr.090417. [DOI] [PubMed] [Google Scholar]
- 38.Liang J, Fu M, Ciociola E, Chandalia M, Abate N. Role of ENPP1 on adipocyte maturation. PLoS ONE. 2007;2:e882. doi: 10.1371/journal.pone.0000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutsch F, Vaingankar S, Johnson K, et al. PC-1 nucleoside triphosphate pyrophosphohydrolase deficiency in idiopathic infantile arterial calcification. Am J Pathol. 2001;158:543–54. doi: 10.1016/S0002-9440(10)63996-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutsch F, Terkeltaub R. Parallels between arterial and cartilage calcification: what understanding artery calcification can teach us about chondrocalcinosis. Curr Opin Rheumatol. 2003;15:302–10. doi: 10.1097/00002281-200305000-00019. [DOI] [PubMed] [Google Scholar]
- 41.Cheng KS, Chen MR, Ruf N, Lin SP, Rutsch F. Generalized arterial calcification of infancy: different clinical courses in two affected siblings. Am J Med Genet A. 2005;136:210–3. doi: 10.1002/ajmg.a.30800. [DOI] [PubMed] [Google Scholar]
- 42.Szulc P, Kiel DP, Delmas PD. Calcifications in the abdominal aorta predict fractures in men: MINOS study. J Bone Miner Res. 2008;23:95–102. doi: 10.1359/jbmr.070903. [DOI] [PubMed] [Google Scholar]
- 43.O’Neill TW, Silman AJ, Naves Diaz M, Cooper C, Kanis J, Felsenberg D. Influence of hormonal and reproductive factors on the risk of vertebral deformity in European women. European Vertebral Osteoporosis Study Group. Osteoporos Int. 1997;7:72–8. doi: 10.1007/BF01623464. [DOI] [PubMed] [Google Scholar]
- 44.Schulz E, Arfai K, Liu X, Sayre J, Gilsanz V. Aortic calcification and the risk of osteoporosis and fractures. J Clin Endocrinol Metab. 2004;89:4246–53. doi: 10.1210/jc.2003-030964. [DOI] [PubMed] [Google Scholar]
- 45.Suk EK, Malkin I, Dahm S, et al. Association of ENPP1 gene polymorphisms with hand osteoarthritis in a Chuvasha population. Arthritis Res Ther. 2005;7:R1082–90. doi: 10.1186/ar1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyre D, Bouatia-Naji N, Tounian A, et al. Variants of ENPP1 are associated with childhood and adult obesity and increase the risk of glucose intolerance and type 2 diabetes. Nat Genet. 2005;37:863–7. doi: 10.1038/ng1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Y, Jahagirdar BN, Reinhardt RL, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–9. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Tomso DJ, Chorley BN, et al. Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet. 2007;16:1188–200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hata J, Matsuda K, Ninomiya T, et al. Functional SNP in an Sp1-binding site of AGTRL1 gene is associated with susceptibility to brain infarction. Hum Mol Genet. 2007;16:630–9. doi: 10.1093/hmg/ddm005. [DOI] [PubMed] [Google Scholar]
- 50.Leroy P, Berto F, Bourget I, Rossi B. Down-regulation of Hox A7 is required for cell adhesion and migration on fibronectin during early HL-60 monocytic differentiation. J Leukoc Biol. 2004;75:680–8. doi: 10.1189/jlb.0503246. [DOI] [PubMed] [Google Scholar]
- 51.Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, Kamps MP. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci USA. 1997;94:14553–8. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lazary A, Kosa JP, Tobias B, et al. Single nucleotide polymorphisms in new candidate genes are associated with bone mineral density and fracture risk. Eur J Endocrinol. 2008;159:187–96. doi: 10.1530/EJE-08-0021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.