CONSPECTUS
Exploration across the fields of biology, chemical biology, and medicine has led to an increasingly complex, albeit incomplete, view of the interactions that drive life's processes. The ability to monitor and track the movement, activity, and interactions of biomolecules in living cells is an essential part of this investigation. In our laboratory, we have endeavored to develop tools that are capable not only of monitoring protein localization but also reporting on protein structure and function. Central to our efforts is a new strategy, bipartite tetracysteine display, that relies on the specific and high-affinity interaction between a fluorogenic, bis-arsenical small molecule and a unique protein sequence, conformation, or assembly.
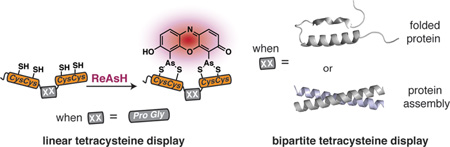
In 1998, a small-molecule analogue of fluorescein with two arsenic atoms, FlAsH, was shown by Tsien and coworkers to fluoresce upon binding to a linear amino acid sequence, Cys-Cys-Arg-Glu-Cys-Cys. Later work demonstrated that substituting Pro-Gly for Arg-Glu optimized both binding and fluorescence yield. Our strategy of bipartite tetracysteine display emanated from the idea that it would be possible to replace the intervening Pro-Gly dipeptide in this sequence with a protein or protein partnership, provided the assembled protein fold successfully reproduced the approximate placement of the two Cys-Cys pairs. In this Account, we describe our recent progress in this area, with an emphasis on the fundamental concepts that underlie the successful use of bis-arsenicals such as FlAsH and the related ReAsH for bipartite display experiments. In particular, we highlight studies that have explored how broadly bipartite tetracysteine display can be employed and that have navigated the conformational boundary conditions favoring success.
To emphasize the utility of these principles, we outline two recently reported applications of bipartite tetracysteine display. The first is a novel, encodable, selective, Src kinase sensor that lacks fluorescent proteins but possesses a fluorescent readout exceeding that of most sensors based on Förster resonance energy transfer (FRET). The second is a unique method, called complex-edited electron microscopy (CE-EM), that facilitates visualization of protein–protein complexes with electron microscopy. Exciting as these applications may be, the continued development of small-molecule tools with improved utility in living cells, let alone in vivo, will demand a more nuanced understanding of the fundamental photophysics that lead to fluorogenicity, as well as creative approaches toward the synthesis and identification of new and orthogonal dye–tag pairs that can be applied facilely in tandem. We describe one example of a dye–sequence tag pair that is chemically distinct from bis-arsenical chemistry. Through further effort, we expect that that bipartite tetracysteine display will find successful use in the study of sophisticated biological questions that are essential to the fields of biochemistry and biology as well as to our progressive understanding of human disease.
Introduction
Exploration across the fields of biology, chemical biology, and medicine has led to an increasingly complex, albeit incomplete view of the interactions that drive life's processes. The ability to monitor and track the movement, activity, and interactions of biomolecules, especially in living cells, is an essential part of this investigation. As chemists, we seek to understand these macromolecular events at a level of resolution that reveals their energetic and structural underpinnings. This objective demands the development of increasingly sophisticated methods to image not just the location of a biomolecule of interest, but also to reveal its structure and /or function. Ongoing research in the field of chemical biology has contributed a number of techniques that accomplish this goal, including Förster resonance energy transfer (FRET),1,2 activity based protein-profiling, 3,4 bimolecular complementation,5,6 and enzyme ligations mediated in trans.7,8 We have chosen to approach this goal using a family of fluorogenic small molecules, bisarsenicals9 or bis-boronic acids, whose fluorescence is dependent on an interaction with a discrete protein sequence, conformation, or assembly. This Account describes our recent progress in this area, with an emphasis on the fundamental concepts that underlie the use of these molecules for live cell experiments. Recent reviews have provided practical details that are useful for laboratories seeking to make use of bis-arsenicals. 10–12 Here, instead, we discuss our own work framed within the context of applying these molecules to explore the structure and function of proteins in the cell. In particular, we highlight the application of bis-arsenicals in a strategy we term bipartite tetracysteine display.
The History of Tetracysteine Display
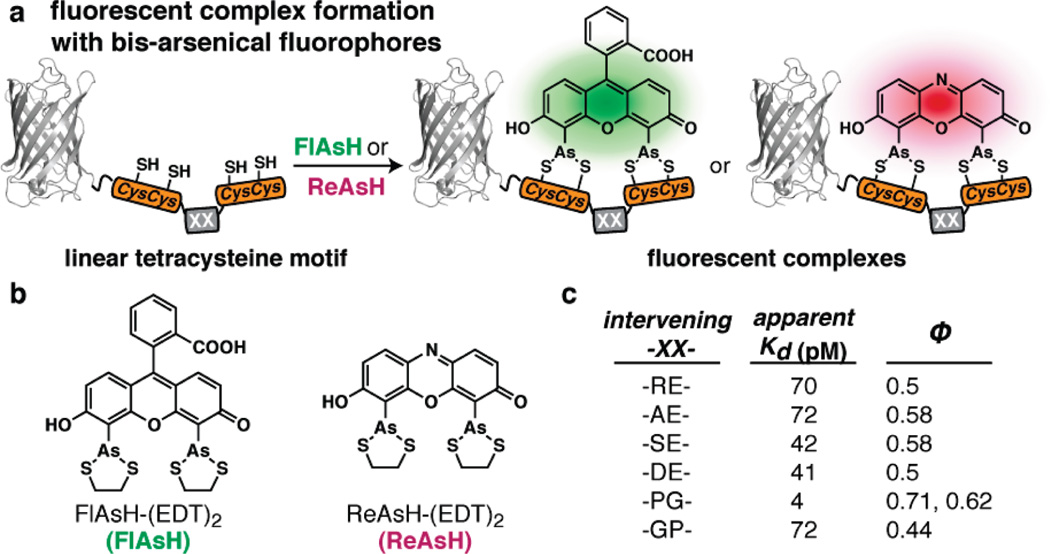
In 1998, Griffin and co-workers reported the synthesis and protein-binding properties of a bis-arsenical analogue of fluorescein, FlAsH (Figure 1).9,13 FlAsH exhibited limited fluorescence when coordinated to simple dithiol ligands such as ethanedithiol (EDT), but became increasingly fluorescent when bound by the four thiols present within the linear sequence CCRECC. When this sequence was encoded at the N- or C-terminus of a protein expressed in mammalian cells, treatment with FlAsH allowed the subcellular location of the protein to be determined using fluorescence microscopy. Since FlAsH is both small and cell permeable and the motif is encodable, their pairing facilitated intracellular protein labeling with temporal control but without many of the disadvantages associated with fluorescent proteins.14 Since this first report, FlAsH and closely related analogues have been used to study the β2-adrenergic receptor,15 β-tubulin,16 the HIV-1 Gag protein,17 GPCR activation,18 and α-synuclein amyloid formation,19 among other interesting and relevant protein targets.20–25
FIGURE 1.
Linear tetracysteine display, as described by Tsien and co-workers.9,13 (a) Cartoon depicting the fluorescent labeling of an engineered protein displaying a linear tetracysteine motif. (b) Chemical structures of FlAsH and ReAsH. (c) The identity of the intervening sequence has a substantial effect on the apparent Kd as well as on the quantum yield (Φ) of the peptide–bis-arsenical complex.
The initial report describing FlAsH proposed that the bis-arsenical recognized a motif consisting of four cysteine side chains spanning a single α-helical turn. This design feature harkens back to classic work of Ruan et al.26 and Ghadiri and Choi27 who reported 20 years ago that metal ions could coordinate appropriately spaced side chains displayed on an α-helix. With respect to FlAsH, it was discovered subsequently that even seemingly minor changes to the original tetracysteine motif led to major changes in the stability and quantum yield of the resulting bis-arsenical–peptide complex (Figure 1c). The number and identity of residues separating the two Cys-Cys pairs were most critical; the brightest and highest affinity complexes were formed with an intervening sequence composed of the dipeptide Pro-Gly.28 Substitution, addition, or deletion of even a single amino acid to the intervening sequence decreased affinity or brightness or both. Notably, Pro-Gly is not preferred in an α-helix and instead favors the formation of a β-hairpin containing a type II β-turn;29 indeed, subsequent NMR studies confirmed that the Pro-Gly β-hairpin places the two cysteine pairs into close proximity with a geometry suitable for bis-arsenical binding.30 Interpreted practically, these structure–activity studies identified a robust and predictable tool that could be, and has been, implemented generally by cell biologists to locate and track discrete proteins.31
There is, however, a more conceptual interpretation of these structure–activity relationships: that the primary function of the Pro-Gly intervening sequence is to optimize the spacing and relative orientation of the two Cys-Cys pairs. High affinity results from an increase in effective molarity; brightness results from a favored geometry that minimizes quenching (vide infra). This interpretation suggests that it should be possible to replace the Pro-Gly intervening sequence with a much longer sequence, perhaps even a full-length protein, in which the Cys-Cys pairs are remote in primary sequence but are brought into proximity by the native protein fold. A corrollary of this idea is that Pro-Gly could even be substituted by an oligomeric protein assembly, as long as the Cys-Cys pairs were arranged suitably in the associated state. If the simple Pro-Gly intervening sequence could be replaced in this way, then the utility of bis-arsenical labeling could be expanded to report on changes in protein conformation or association by linking the binding affinity and concomitant fluorescent signal of the dye to protein tertiary and quaternary structure. This conceptualization led to the development of bipartite tetracysteine display.
Conformationally Sensitive Tetracysteine Motifs
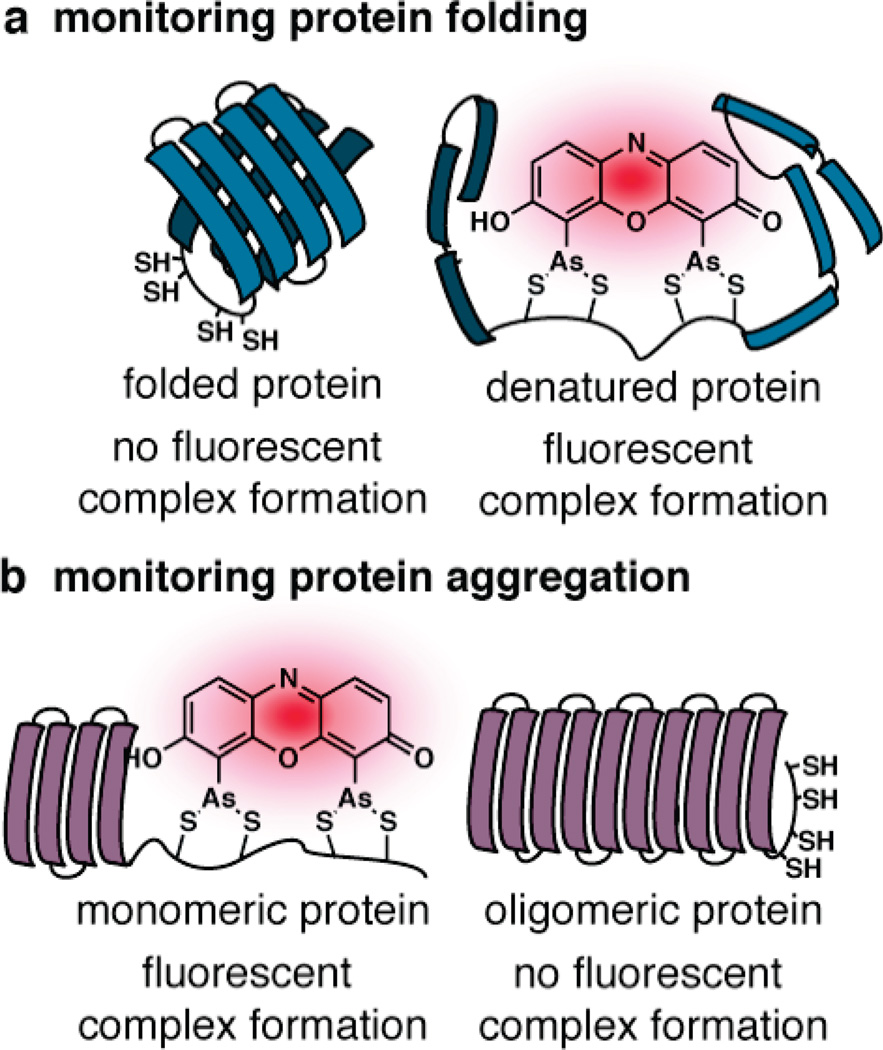
The idea that the fluorescence of a tetracysteine-coordinated bis-arsenical dye could be linked to global protein structure was confirmed through the inadvertent discovery that certain linear tetracysteine motifs could be used themselves to monitor protein conformational changes. Such behavior was first described by Ignatova and Gierasch for the cellular retinoic acid binding protein 1 (CRABP 1), containing a linear tetracysteine motif embedded within an internal loop.32 It was anticipated that FlAsH would bind to the engineered CRABP 1 only when the protein was properly folded. Surprisingly, FlAsH fluorescence was observed only when it was unfolded, either through mutation or addition of denaturants (Figure 2a). Although unexpected, this observation is noteworthy because it could, in theory, allow FlAsH binding to be applied to monitor the effects of cellular chaperones on protein structure and stability. Similar strategies have been applied to study proteins involved in mis-folding diseases such as Alzheimer's and Huntington's disease (Figure 2b).33–36 In some cases, the tetracysteine motif is labeled by FlAsH only when the protein is monomeric, whereas in others it is only labeled when the protein is misfolded or oligomeric. These reports support the view that the fluorescence of a bis-arsenical–protein complex is sensitive to local structure and/or environment.
FIGURE 2.
Monitoring conformational changes using traditional tetracysteine display. Cartoon depicting the use of ReAsH to (a) monitor changes in protein folding and (b) monitor changes in protein assembly.
Beyond Linear Labeling Strategies: Bipartite Tetracysteine Display
Our entry into this field began with a simple question: would it be possible to replace the Pro-Gly intervening sequence that is used typically in bis-arsenical labeling experiments with an appropriately substituted protein or protein partnership? We hypothesized that proteins engineered in this way would bind the fluorophore with high affinity only when the optimal geometry of the tetracysteine ligand was enforced by the protein's native conformation. Additionally, if the bis-arsenical binding site were to become distorted by virtue of mutation or interaction, then the binding affinity and brightness would diminish. Binding of the bis-arsenical molecule in this way would be evocative of natural metal binding sites, which are often constructed from side chains that are isolated in primary sequence but adjacent when the protein is properly folded.37,38 Using this strategy, which we called bipartite tetracysteine display, it would be possible to distinguish the proteins' conformation or association state through fluorescent labeling.
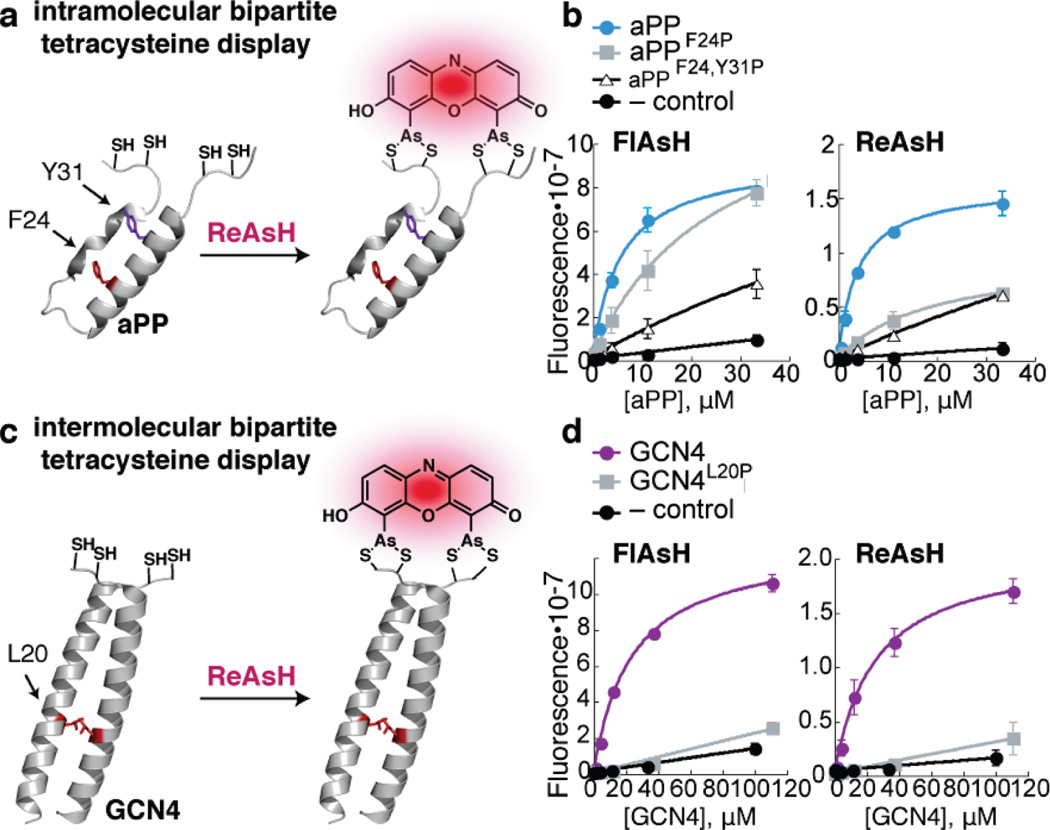
The feasibility of bipartite tetracysteine display was assessed first by Luedtke et al. using four model protein domains: the miniature proteins avian pancreatic polypeptide (aPP) and Zip4, and the leucine zippersGCN4, and Jun. aPP and Zip4 served as models for intramolecular bipartite display, whereas GCN4 and Jun served as models for bipartite display between two independent protein domains.39 Each domain was modified with Cys-Cys pairs that were positioned to recapitulate the requisite binding site only when the domains were folded. In all cases, the bis-arsenical dyes FlAsH and ReAsH bound the folded domains or complexes with apparent dissociation constants ranging from 400 nmto 30 µM even in the presence of excess dithiol competitor (Figure 3). The resulting complexes were fluorescent and, as expected, composed of a 1:1 ratio of dye to folded or associated protein. When the protein fold or assembly was disrupted by even a single point mutation, both affinity and brightness were lost. A subtlety not to be overlooked is that although FlAsH binding increased the thermal stabilities of the destabilized aPP and GCN4 variants (as judged by temperature-dependent circular dichroism), it did not increase the extent of α-helical structure at temperatures below the TM.39
FIGURE 3.
Bipartite tetracysteine display: initial results. (a) Schematic representation of intramolecular bipartite tetracysteine display. (b) Misfolded bipartite tetracysteine variants of aPP (F24P,Y31P) bind FlAsH and ReAsH (25 nM) with diminished affinities relative to the wild-type bipartite tetracysteine constructs. (c) Schematic representation of intermolecular bipartite tetracysteine display. (d) Monomeric bipartite tetracysteine variants of GCN4 (L20P) bind FlAsH and ReAsH (25 nM) with diminished affinities relative to the wild-type bipartite tetracysteine dimer.
These in vitro studies were followed by experiments to confirm that bipartite bis-arsenical labeling was effective in living cells. Despite the purported toxicity associated with bis-arsenical dyes, both linear and bipartite tetracysteine labeling strategies have been implemented successfully in a variety of mammalian cell lines. In our case, fluorescent protein fusions of aPP and GCN4 were prepared; bis-arsenical labeling was observed only when the proteins were in the proper conformation or assembly state. It is often underemphasized that bis-arsenical dyes often show high levels of nonspecific cellular labeling that can be mitigated, though not fully eliminated, by treatment with competing dithiols. Even in the presence of this persistent background, it was possible to measure a fluorescent signal that was more than 400% above background in the case of properly folded and assembled aPP and GCN4, as judged by flow cytometry. As the background signal derives predominantly from cytosolic ligands, further increases in signal to noise are possible when visualizing events at the cell surface through judicious application of total internal reflectance techniques. In summary, the Leudtke et al. work showed that bipartite tetracysteine display was not only feasible but also useful for monitoring protein folding and assembly in live cells.
Binding Site Requirements
Our initial efforts in this area demonstrated that bipartite tetracysteine display could function effectively in the context of small protein domains containing Cys-Cys pairs located near the N- and C-termini of predominantly α-helical motifs. Structural studies of arsenic dithiolates suggest that the ideal geometry of a bis-arsenical binding site is a rhombus with 4–5 Å separating thiols on one axis and 6–8 Å on the other.40 Thus, we anticipated that the affinity of FlAsH and ReAsH for a tetracysteine motif would be greatest if the four thiols complemented this geometry. In the aPP and GCN4 leucine zipper domains studied previously (see above), the thiols in the Cys-Cys pairs were separated by about 7 Å and located in regions near and/or appended to the N- and C-termini, allowing them to adopt the ideal binding geometry (Figure 4a,b). A critical next step was to evaluate whether effective bis-arsenical binding sites could be engineered into regions with less inherent conformational flexibility, including non-α-helical secondary structures and larger, globular domains. In addition to considering simply the relative spacing of the four thiols that comprise the bis-arsenical binding motif, we also questioned the extent to which protein secondary structure and the underlying conformational dynamics might modulate fluorescence. Our goal was to assemble a set of guiding principles that could be implemented by us and by others seeking to use bipartite tetracysteine display across a broad range of systems.
FIGURE 4.
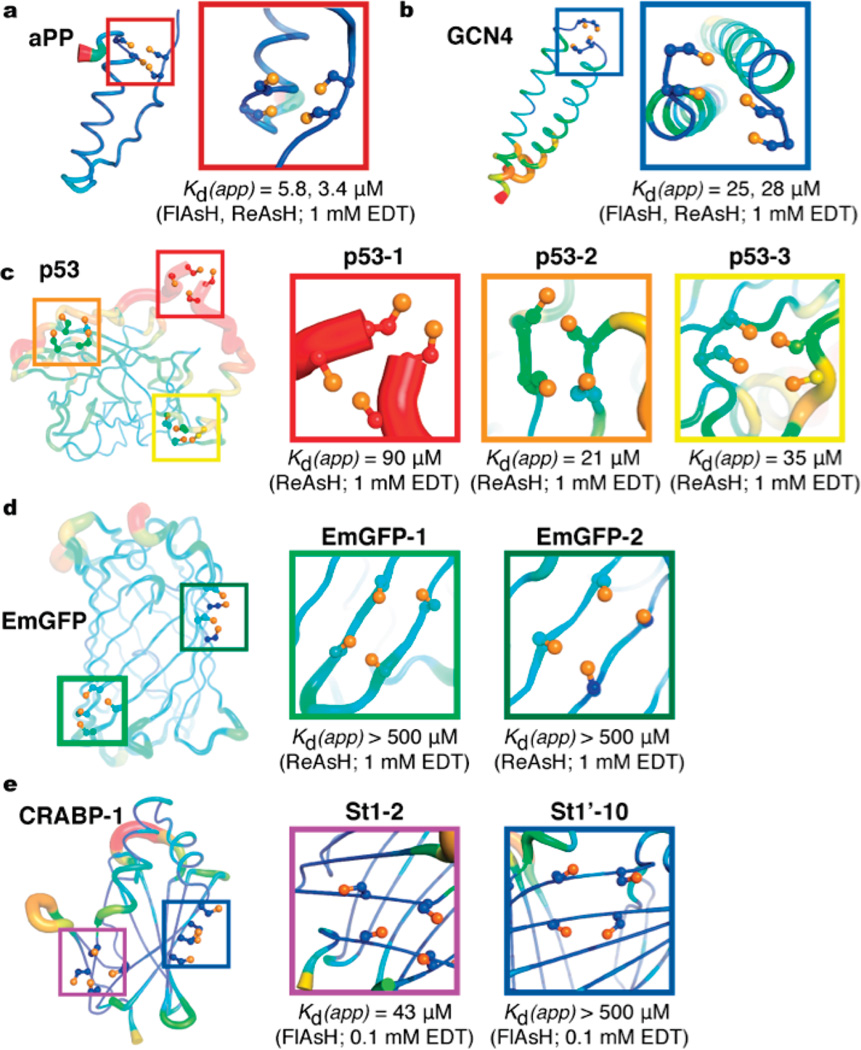
Structural dependence of bipartite tetracysteine binding sites. Crystallographic B-factor putty rendering for bipartite tetracysteine mutants of (a) aPP (PDB 2BF9), (b) GCN4 (PDB 2ZTA), (c) p53 (PDB 1TUP), (d) EmGFP (PDB 1EMA), and (e) CRABP-1 (PDB 1CBI). Close up views of the bis-arsenical binding site are shown to the side, along with the corresponding apparent Kd values.
A significant insight into the limitations of bipartite tetracysteine display with respect to secondary structure came from the work of Goodman et al.41 This study evaluated the ability of bipartite tetracysteine display to function in the context of two proteins whose architectures are replete with non-α-helical secondary structure: EmGFP, a classic β-barrel protein containing six parallel and five antiparallel β-strands, and p53, which has a more complex fold containing β-strands, α-helices, and at least six discrete regions of highly variable length that lack defined secondary structure. Goodman et al. explored this structural diversity by preparing Cys-Xaa-Cys pairs in proximal β-strands of EmGFP and Cys-Cys pairs in loops of p53 variants. The affinity of ReAsH for proteins containing these varied binding sites was monitored. Under highly permissive conditions in the absence of competing dithiol, varied ReAsH affinity and fluorescence was observed when the Cys-Xaa-Cys pairs, which better complement the side chain display of a β-sheet, were introduced into two different sets of adjacent β-strands within EmGFP. Virtually no binding (apparent Kd > 500 µM) was detected under stringent conditions containing dithiols at concentrations that approximate those found within a cell. In contrast to the results obtained with EmGFP significant ReAsH affinity and fluorescence was observed when the Cys-Cys pairs were introduced into three different loops within p53, even under stringent, physiological conditions (apparent Kd = 21 µM) (Figure 4c). The challenges associated with introducing a bipartite tetracysteine motif into a β-sheet rich protein were confirmed by Krishnan and Gierasch, who substituted pairs of cysteines (Cys-Xaa-Cys) into CRABP 1.42 It was possible to label these tetracysteine variants with FlAsH (apparent Kd = 43 µM), but only under permissive binding conditions using low concentrations of dithiol competitor and high protein concentrations.
Taken together, these results highlight that many factors contribute to the success of a bipartite tetracysteine display experiment. One consideration is that differences in quantum yield can result from changing the local fluorophore environment. This observation is not surprising considering that linear tetracysteine sequences modified by a discrete flanking sequence (FLNCCPGCCMEP) possess a binding affinity that mirrors the minimal motif, but form complexes with a 6-fold increase in contrast.31 Another key concern is that the distance between the engineered thiols may not match exactly with the distance expected based on structural information. A comparison of the binding sites on EmGFP and p53 indicates that there is a correlation between ReAsH affinity and overall flexibility, which implies that this labeling strategy will function best when the binding site can be engineered within flexible regions of a protein.
Applications of Bipartite Tetracysteine Display
The ability of bipartite tetracysteine display to detect subtle changes in protein conformation or assembly suggested that it would be possible to employ this technique to monitor protein functions that involve complex assembly or induced conformational changes. It was our thought that bipartite tetracysteine display might offer advantages compared to the more commonly used strategies, such as Förster resonance energy transfer (FRET). For instance, methods that monitor these events using changes in FRET efficiency often suffer from low signal-to-noise and the absence of temporal control. By contrast, methods based on bipartite tetracysteine display measure changes in association or conformation directly with a turn-on fluorescent readout. This unique advantage is highlighted in two unrelated and well-characterized systems.
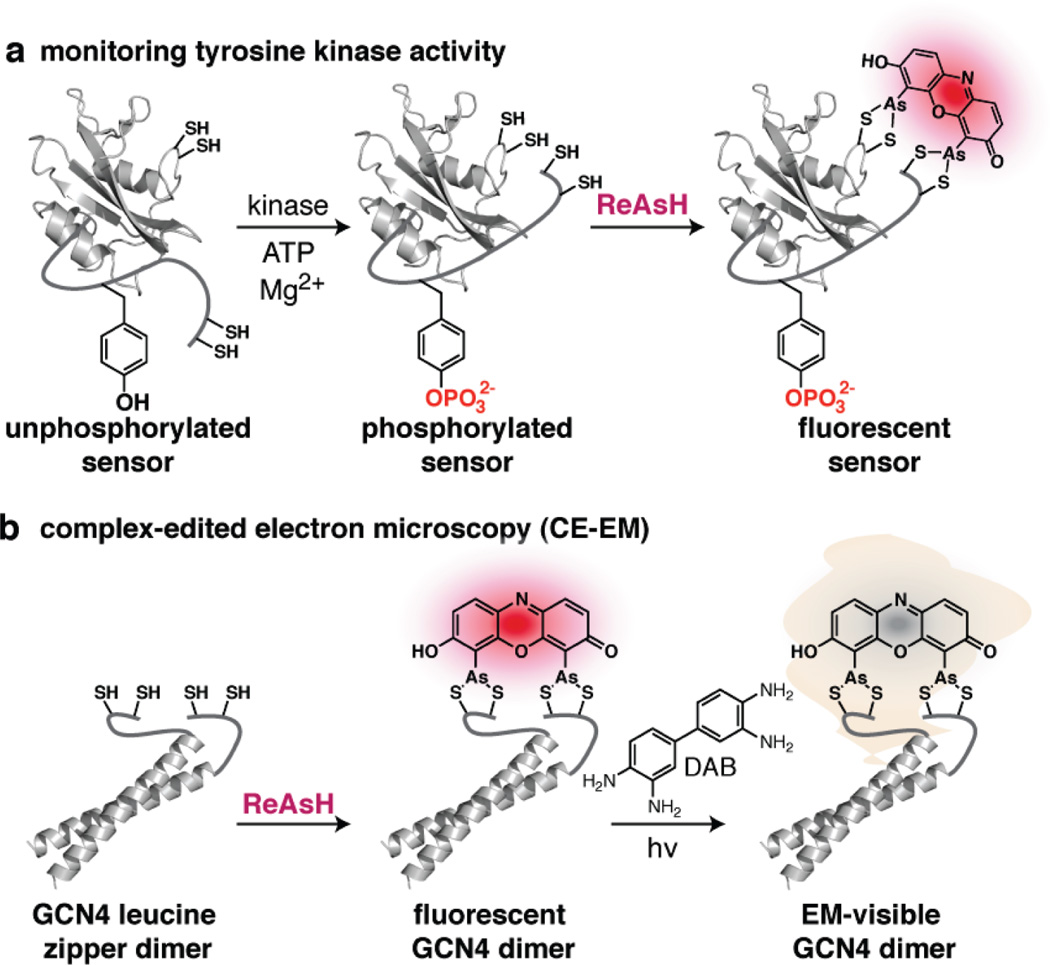
One recently reported application of bipartite tetracysteine display is the development of an encodable sensor for Src family kinase activity.43 The general design described by Ray-Saha and Schepartz is reminiscent of classic FRET-based sensors reported initially by Miyawaki et al. and later by Ting et al., in which a FRET donor (CFP) and a FRET acceptor (YFP) are separated by an engineered protein module composed of a tyrosine-containing Src kinase substrate and a proximal SH2 domain.44,45 Building on the work of Goodman et al., Cys-Cys pairs were introduced into flexible loops at the C-terminus of the Lck SH2 domain and a proximal region of the tyrosine-containing substrate (Figure 5a). The positions of the cysteine pairs were chosen so that a ReAsH binding site would be recapitulated if and only if the phosphorylated tyrosine within the substrate region was nestled into the requisite SH2 binding pocket. Twelve first generation designs were evaluated; one of these, E2, was labeled with ReAsH and fluoresced only upon phosphorylation. The emission of sensor E2 after incubation with ReAsH increased by more than 75% upon phosphorylation, an increase that outperforms most reported FRET-based Src kinase sensors. In addition, E2 appears to be uniquely selective within the Src family, as it acts as a substrate for both Src and Yes, but not Fyn or Lck, all of which are Src A subfamily members. Perhaps the most interesting aspect of sensor E2 is that it responds only to phosphorylation at one of two closely spaced tyrosine (Y762) residues, a feature that is not observed for sensors based on fluorescent protein fusions.
FIGURE 5.
Applications of bipartite tetracysteine display. (a) Schematic representation of a Src kinase sensor. A conformational change that occurs upon phosphorylation by Src kinase can be detected using ReAsH. (b) Schematic representation of complex-edited electron microscopy (CE-EM). Irradiation of the ReAsH complex in the presence of diaminobenzidine (DAB) polymerizes the DAB surrounding each protein complex. The EM signal resulting from subsequent treatment with OsO4 is therefore edited to visualize only those proteins that are closely interacting.
Another application of bipartite tetracysteine display has been in the field of electron microscopy (EM). It has been known for three decades that certain dyes, undergo photoconversion to generate singlet oxygen (1O2) upon irradiation. Indeed, 1O2 generation (and its resulting reactivity) is a primary source of fluorescence quenching. However, if 1O2 is generated in the presence of diaminobenzidine, it promotes the conversion of diaminobenzidene into an insoluble polymer that becomes electron dense upon treatment with OsO4. These electron dense regions are visible as dark spots in EM images. It has been shown previously that recombinant proteins carrying a linear tetracysteine tag and labeled with ReAsH can be detected at the very high (~5 nm) resolution possible using electron microscopy following the scheme outlined above.46 Using a strategy called complex-edited electron microscopy (CE-EM) described by Dexter and Schepartz, the EM signal that results from ReAsH photoconversion is edited to visualize only those proteins that are closely interacting (Figure 5b). CE-EM was demonstrated with a GCN4 leucine zipper that was engineered to contain the requisite tetracysteine motif only when assembled into a parallel dimer. When the dimer was properly formed, it was possible to observe an EM signal after labeling with ReAsH. As expected, this signal was not observed for GCN4 point mutants that formed significantly less stable dimers. Thus, CE-EM facilitated the selective visualization of protein–protein complexes by electron microscopy. One important feature of this technique is that copies of the protein that are not in complex are not detected. Additionally, the initial labeling steps can occur in live cells.47
The Physical Organic Chemistry of Bis-Arsenical Fluorescence
The mechanistic basis for the pro-fluorescent behavior of FlAsH and ReAsH is poorly understood. Two hypotheses have been proposed to explain why these bis-arsenicals become fluorescent when bound to a tetracysteine motif. One suggests that coordination of a bis-arsenical with a protein-embedded tetracysteine motif hinders rotation about the As–aryl bond and prevents efficient quenching of the excited state via photoinduced electron transfer (PeT).9 The second proposal is that a decrease in ring strain upon ligand coordination eliminates fluorescence quenching.48 It is useful to consider these proposals within the context of current understanding about how PeT influences the fluorescence of well-characterized turn-on sensors such as fluorescein.
A large body of work suggests that the fluorescence of fluorescein and its derivatives are controlled by PeT.49–51 Starting with the observation that the absorbance spectrum of fluorescein's xanthene core was unaffected by changes in the electronic structure of the benzoic acid substituent, Nagano and co-workers proposed that the xanthene and benzoic acid portion of fluorescein are orthogonal and independent in the ground state. Upon excitation, the expected fluorescence output may be PeT quenched via electron donation from the benzoic acid moiety into the xanthene core. As the energy of the benzoic acid HOMO is lowered, for example, upon substitution with electron-withdrawing groups, fluorescence quenching is lost due to the unfavorable energy match of the frontier orbitals.
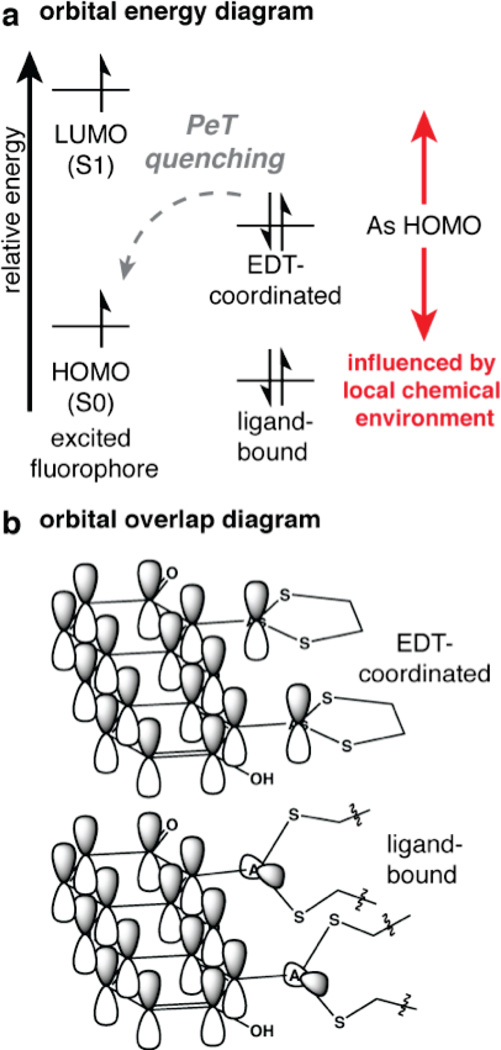
In the context of FlAsH and ReAsH, one can identify two factors that would be required for the As dithiolate to quench the fluorescence of the xanthene or resorufin core: The relative energies of the As HOMO and the fluorophore HOMO must be appropriately matched (Figure 6a), as is the case for fluorescein, and the orbitals must align to allow for sufficient overlap, as is required by orbital theory (Figure 6b). Both prerequisites must be met in the free (EDT-coordinated) state, and one or both must be lost in the ligand-bound complex in order for the bis-arsenical dye to behave fluorgenically. It may be that hindered rotation about the As–aryl bond decreases overlap while holding the As HOMO energy relatively constant, resulting in fluorescence turn-on when bound to the tetracysteine ligand (the “hindered rotation” hypothesis). It may also be that decreased ring strain in the bound state maintains orbital overlap, but decreases the energy of the As HOMO thus eliminating PeT (the “ring strain” hypothesis). It is likely that both mechanisms, and perhaps others, are at play; there is little doubt that a better understanding of the molecular details that control bis-arsenical fluorescence quenching will lead to the synthesis of molecules with improved fluorescence properties.
FIGURE 6.
Two possible mechanisms for photoinduced electron transfer (PeT) quenching in bis-arsenical fluorophores. (a) Orbital energy diagram depicting a PeT quenching mechanism for bis-arsenical fluorophores. (b) Simplified orbital overlap diagram representing a quenched fluorophore (top) in which the As orbitals are in conjugation with the fluorophore π-system, or an unquenched fluorophore (bottom) in which the As orbitals are orthogonal to the π-system.
The Development of Orthogonal Motifs
There are three ways to improve the utility of a bis-arsenical-encodable tag pair: alter the dye photophysics to minimize background, alter the tag to maximize affinity and brightness, or alter the dye structure to facilitate modification. All three approaches have been evaluated since the first report of linear tetracysteine display. New bis-arsenicals have been reported that possess uniquely tuned fluoresence properties,28,50,52 diminished background labeling,53 additional affinity handles,33,54,55 and convenient sites for further functionalization.56 In addition, new tag sequences that alter bis-arsenical binding affinity and kinetics,48 and increase complex brightness57,58 have also been described.
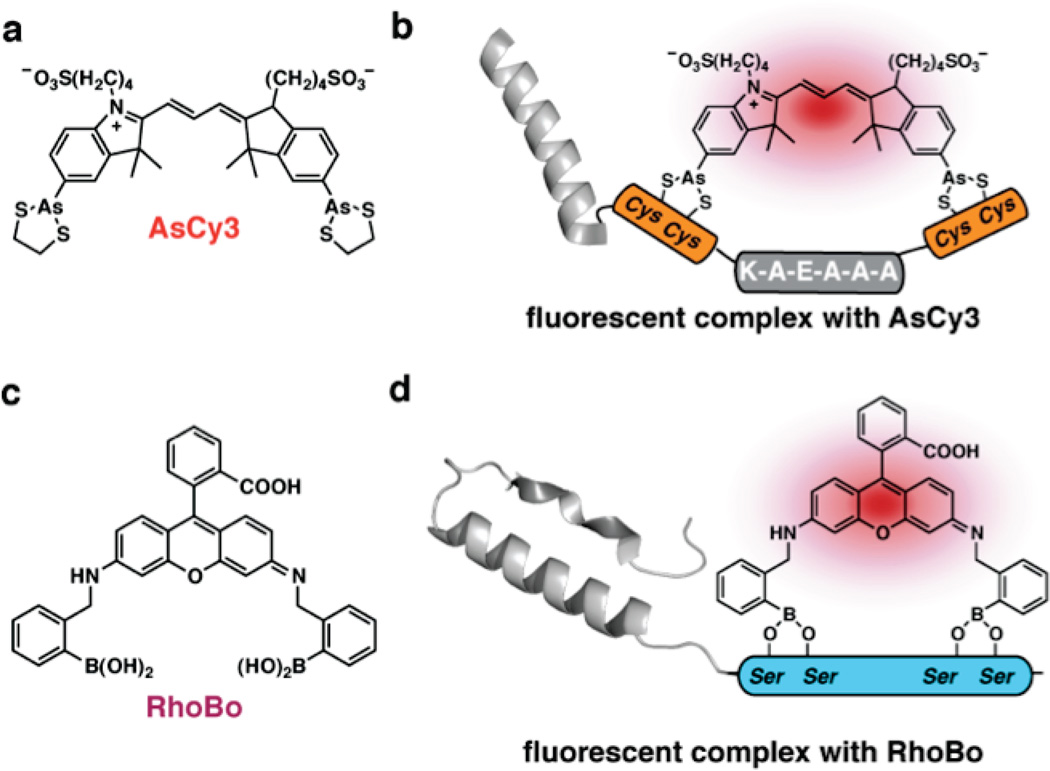
Although the modifications described above offer improvements, they are still limited by the inherent photophysics of the xanthene and resorufin chromphores. A new bis-arsenical that lacks this limitation has been reported by Cao et al.62 This bis-arsenical cyanine dye, AsCy3, possesses both a higher extinction coefficient and higher quantum yield than ReAsH; together these features translate into significantly enhanced brightness (Figure 7a). AsCy3 is the only bis-arsenical dye yet reported that could be appropriate for super-resolution fluorescence microscopy, although that application has yet to be achieved experimentally. The application of cyanine-based bis-arsenicals to super-resolution microscopy may demand new synthetic efforts to incorporate arsenic groups into asymmetric cyanine dyes that are further sulfonated, as these modifications improve photophysics and solubility.59–61
FIGURE 7.
Development of orthogonal dye–tag pairs. (a) Chemical structure of AsCy3. (b) Cartoon depicting the fluorescent complex that forms when AsCy3 binds to an orthogonal tetracysteine sequence. (c) Chemical structure of RhoBo. (d) Cartoon portraying the fluorescent complex that forms when RhoBo binds to a novel tetraserine sequence.
A final potential advantage of AsCy3 is its reported ability to recognize a sequence tag that is orthogonal to that preferred by FlAsH and ReAsH (Figure 7b).62 The As–As distance in AsCy3 was cited as 14.5 Å in the fully extended, planar conformation. By comparison, the As–As distances in FlAsH and ReAsH are roughly 5 Å. It has been proposed that the As–As distance in AsCy3 complements that of Cys-Cys side chain pairs separated by two turns of an α-helix, and the preferred tetracysteine sequence is CCKAEAACC. AsCy3 fails to fluoresce in the presence of the tetracysteine-containing binding site for FlAsH and ReAsH, and FlAsH and ReAsH fail to fluoresce in the presence of the reported AsCy3 tag (unpublished results). These observations suggest that FlAsH (absorbance λmax = 508 nm; emission λmax = 528 nm) and AsCy3 (absorbance λmax = 560 nm; emission λmax = 576 nm) could be applied in combination to monitor two proteins simultaneously.
In spite of the demonstrated utility of bis-arsenicals such as FlAsH and ReAsH, the fact remains that these reagents can be cytotoxic and are difficult to apply in certain cell lines and cellular locales where thiols are easily oxidized.46,63,64 Several considerations led us to consider boronic acids as an orthogonal alternative to bis-arsenicals. First, it has been long been known that boronic acids can bind molecules containing 1,2 and 1,3 diols.65–67 Second, many fluorogenic bis-boronic acids have been synthesized and their fluorescence and solubility properties have been studied in detail.68 One particular bis-boronic acid rhodamine dye (RhoBo) was identified as a molecule potentially suitable for this application, as it could be prepared via a short, straightforward route that would facilitate analogue synthesis, possesses minimal affinity (mM) for monosaccharides, and forms boronate esters that fluoresce at wavelengths useful for live cell microscopy (emission λmax = 520 nm) (Figure 7c). Moreover, the B–B distance in RhoBo approximates the As–As distance in FlAsH and ReAsH, suggesting that RhoBo might form high affinity complexes with the serine-analogue of the preferred tetracysteine motif. Halo et al. recently reported that RhoBo binds peptides or proteins containing a SSPGSS tag in the nanomolar concentration range, undergoes a 4-fold increase in fluorescence intensity upon binding, and prefers the sequence SSPGSS to a range of monosaccharides by at least >105 (Figure 7d).69 RhoBo fails to form stable fluorescent complexes with the plethora of glycans displayed on the mammalian cell surface, as judged by epifluorescent, confocal, and TIRF microscopy. Furthermore, RhoBo crosses the cell membrane, as judged by the presence of fluorescence within the cell interior. A BLAST search revealed more than 200 proteins within the human proteome that contain a SSPGSS sequence, although it has not been established which of these, if any, function as ligands for RhoBo in live cells. Based on these results, it is hypothesized that RhoBo binds weakly and reversibly to the cell surface, but is sequestered in thermodynamically or kinetically more stable complexes within the cell. Further work will be necessary to identify a linear sequence that competes effectively with the large number of hydroxyl-rich targets found in the mammalian cells.
Conclusion
The development of encodable strategies to track the location and movement of proteins in living cells has transformed fundamental and translational discovery in virtually every arena of the life sciences. In our own lab, we have strived to develop tools that are capable of not only monitoring protein localization but also reporting on structure and function. Central to our efforts is a new strategy, bipartite tetracysteine display, that relies on the specific and high-affinity interaction between a fluorogenic, bis-arsenical small molecule and a unique protein sequence, conformation, or assembly.9,39 In this Account, we discuss the observations that guided the development of bipartite tetracysteine display, and the proof-of-concept experiments that verified feasibility in living cells. We describe studies that explored how broadly bipartite display strategy can be employed, and identify the conformational boundary conditions that favor success. To highlight the utility of these principles, we outline two recently reported applications of bipartite tetracysteine display. The first is a novel, encodable, and selective Src kinase sensor that lacks fluorescent proteins but whose fluorescent readout (even in this first generation design) exceeds that of most FRET-based sensors. The second is a unique method, called complex-edited electron microscopy, which facilitates visualization of protein–protein complexes in living cells using electron microscopy. Exciting as these applications may be, the continued development of small molecule tools with improved utility in cellulo, let alone in vivo, will demand a more nuanced understanding of the fundamental photophysics of fluorogenicity as well as creative approaches toward new, orthogonal, dye-tag pairs that can be applied conveniently in tandem. One example of a dye-sequence tag pair that is chemically orthogonal to bisarsenicals is described. Through further effort, we expect that that bipartite display will find successful use in the study of sophisticated biological questions that are essential to the fields of biochemistry and biology, as well as to our progressive understanding of human disease.
Biographies
Alanna Schepartz is currently the Milton Harris '29 Ph.D Professor of Chemistry and a Professor of Molecular, Cellular, and Developmental Biology at Yale University. She obtained her B.S. in Chemistry at the State University of New York, Albany with Shelton Bank and her Ph.D. in Chemistry at Columbia University with Ronald Breslow. Before beginning her independent career, she was an NIH postdoctoral fellow with Peter Dervan at the California Insitute of Technology. Her research interests focus on the design of molecules, small and large, whose structures and/or functions allow them to address fundamental problems at the interface of chemistry, biology, and medicine.
Rebecca Scheck received her undergraduate degree in Chemistry from Columbia University in 2004, where she performed research in the laboratory of Colin Nuckolls. In 2008, Rebecca earned her doctorate in Chemistry at the University of California, Berkeley under the guidance of Matt Francis. She is currently an NIH postdoctoral fellow working with Alanna Schepartz at Yale University.
REFERENCES
- 1.Jares-Erijman EA, Jovin TM. Imaging molecular interactions in living cells by FRET microscopy. Curr. Opin. Chem. Biol. 2006;10(5):409–416. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 2.Medintz IL, Mattoussi H. Quantum dot-based resonance energy transfer and its growing application in biology. Phys. Chem. Chem. Phys. 2009;11(1):17–45. doi: 10.1039/b813919a. [DOI] [PubMed] [Google Scholar]
- 3.Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer. 2010;10(9):630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heal WP, Dang TH, Tate EW. Activity-based probes: discovering new biology and new drug targets. Chem. Soc. Rev. 2011;40(1):246–257. doi: 10.1039/c0cs00004c. [DOI] [PubMed] [Google Scholar]
- 5.Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem. Soc. Rev. 2009;38(10):2876–2886. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prescher JA, Contag CH. Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr. Opin. Chem. Biol. 2010;14(1):80–89. doi: 10.1016/j.cbpa.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Ting AY, Thyagarajan A. Imaging Activity-Dependent Regulation of Neurexin-Neuroligin Interactions Using trans-Synaptic Enzymatic Biotinylation. Cell. 2010;143(3):456–469. doi: 10.1016/j.cell.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Johnsson K, O'Hare HM, Gautier A. Chemical probes shed light on protein function. Curr. Opin. Struct. Biol. 2007;17(4):488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Griffin B, Adams S, Tsien R. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281(5374):269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 10.Adams SR, Tsien RY. Preparation of the membrane-permeant biarsenicals FlAsH-EDT2 and ReAsH-EDT2 for fluorescent labeling of tetracysteine-tagged proteins. Nat. Protoc. 2008;3(9):1527–1534. doi: 10.1038/nprot.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann C, Gaietta G, Zurn A, Adams SR, Terrillon S, Ellisman MH, Tsien RY, Lohse MJ. Fluorescent labeling of tetracysteine-tagged proteins in intact cells. Nat. Protoc. 2010;5(10):1666–1677. doi: 10.1038/nprot.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pomorski A, Krezel A. Exploration of biarsenical chemistry-challenges in protein research. Chem. Bio. Chem. 2011;12(8):1152–1167. doi: 10.1002/cbic.201100114. [DOI] [PubMed] [Google Scholar]
- 13.Tsien R, Adams S, Griffin B, Kerr R, Li W, Llopis J, Miyawaki A, Schafer W, Zlokarnik G. Imaging protein interactions and gene expression in live cells. Mol. Biol. Cell. 1998;9:2A–2A. [Google Scholar]
- 14.Hinner MJ, Johnsson K. How to obtain labeled proteins and what to do with them. Curr. Opin. Biotechnol. 2010;21(6):766–776. doi: 10.1016/j.copbio.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Nakanishi J, Takarada T, Yunoki S, Kikuchi Y, Maeda M. FRET-based monitoring of conformational change of the beta2 adrenergic receptor in living cells. Biochem. Biophys. Res. Commun. 2006;343(4):1191–1196. doi: 10.1016/j.bbrc.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 16.Andresen M, Schmitz-Salue R, Jakobs S. Short tetracysteine tags to beta-tubulin demonstrate the significance of small labels for live cell imaging. Mol. Biol. Cell. 2004;15(12):5616–5622. doi: 10.1091/mbc.E04-06-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudner L, Nydegger S, Coren LV, Nagashima K, Thali M, Ott DE. Dynamic fluorescent imaging of human immunodeficiency virus type 1 gag in live cells by biarsenical labeling. J. Virol. 2005;79(7):4055–4065. doi: 10.1128/JVI.79.7.4055-4065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann C, Gaietta G, Bunemann M, Adams SR, Oberdorff-Maass S, Behr B, Vilardaga JP, Tsien RY, Ellisman MH, Lohse MJ. A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nat. Methods. 2005;2(3):171–176. doi: 10.1038/nmeth742. [DOI] [PubMed] [Google Scholar]
- 19.Roberti MJ, Bertoncini CW, Klement R, Jares-Erijman EA, Jovin TM. Fluorescence imaging of amyloid formation in living cells by a functional, tetracysteinetagged alpha-synuclein. Nat. Methods. 2007;4(4):345–351. doi: 10.1038/nmeth1026. [DOI] [PubMed] [Google Scholar]
- 20.Dyachok O, Isakov Y, Sagetorp J, Tengholm A. Oscillations of cyclic AMP in hormone-stimulated insulin-secreting beta-cells. Nature. 2006;439(7074):349–352. doi: 10.1038/nature04410. [DOI] [PubMed] [Google Scholar]
- 21.Enninga J, Mounier J, Sansonetti P, Tran Van Nhieu G. Secretion of type III effectors into host cells in real time. Nat. Methods. 2005;2(12):959–965. doi: 10.1038/nmeth804. [DOI] [PubMed] [Google Scholar]
- 22.Panchal RG, Ruthel G, Kenny TA, Kallstrom GH, Lane D, Badie SS, Li L, Bavari S, Aman MJ. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc. Natl. Acad. Sci. U.S.A. 2003;100(26):15936–15941. doi: 10.1073/pnas.2533915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, Huang T, Yang L, Pan J, Zhu S, Yan X. Sensitive and Selective Bacterial Detection Using Tetracysteine-Tagged Phages in Conjunction with Biarsenical Dye. Angew. Chem., Int. Ed. 2011 doi: 10.1002/anie.201100334. DOI: 10.1002/anie.201100334. [DOI] [PubMed] [Google Scholar]
- 24.Spille JH, Zurn A, Hoffmann C, Lohse MJ, Harms GS. Rotational diffusion of the alpha(2a) adrenergic receptor revealed by FlAsH labeling in living cells. Biophys. J. 2011;100(4):1139–1148. doi: 10.1016/j.bpj.2010.08.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pace CJ, Huang Q, Wang F, Palla KS, Fuller AA, Gao J. A FlAsH-Tetracysteine Assay for Quantifying the Association and Orientation of Transmembrane alpha-Helices. Chem. Bio. Chem. 2011;12(7):1018–1022. doi: 10.1002/cbic.201000736. [DOI] [PubMed] [Google Scholar]
- 26.Ruan FQ, Chen YQ, Itoh K, Sasaki T, Hopkins PB. Synthesis of Peptides Containing Unnatural, Metal-Ligating Residues - Aminodiacetic Acid as a Peptide Side-Chain. J. Org. Chem. 1991;56(14):4347–4354. [Google Scholar]
- 27.Ghadiri MR, Choi C. Secondary Structure Nucleation in Peptides - Transition-Metal Ion Stabilized Alpha-Helices. J. Am. Chem. Soc. 1990;112(4):1630–1632. [Google Scholar]
- 28.Adams S, Campbell R, Gross L, Martin B, Walkup G, Yao Y, Llopis J, Tsien R. New biarsenical Ligands and tetracysteine motifs for protein labeling in vitro and in vivo: Synthesis and biological applications. J. Am. Chem. Soc. 2002;124(21):6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 29.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 4th ed. New York: W. H. Freeman and Company; 2005. [Google Scholar]
- 30.Madani F, Lind J, Damberg P, Adams SR, Tsien RY, Graslund AO. Hairpin Structure of a Biarsenical-Tetracysteine Motif Determined by NMR Spectroscopy. J. Am. Chem. Soc. 2009;131(13):4613–4615. doi: 10.1021/ja809315x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin B, Giepmans B, Adams S, Tsien R. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 2005;23(10):1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 32.Ignatova Z, Gierasch LM. Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling. Proc. Natl. Acad. Sci. U.S.A. 2004;101(2):523–528. doi: 10.1073/pnas.0304533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taguchi Y, Shi Z-D, Ruddy B, Dorward DW, Greene L, Baron GS. Specific Biarsenical Labeling of Cell Surface Proteins Allows Fluorescent- and Biotin-tagging of Amyloid Precursor Protein and Prion Proteins. Mol. Biol. Cell. 2009;20(1):233–244. doi: 10.1091/mbc.E08-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaspersic J, Hafner-Bratkovic I, Stephan M, Veranic P, Bencina M, Vorberg I, Jerala R. Tetracysteine-tagged prion protein allows discrimination between the native and converted forms. FEBS J. 2010;277(9):2038–2050. doi: 10.1111/j.1742-4658.2010.07619.x. [DOI] [PubMed] [Google Scholar]
- 35.Hu Y, Su B, Kim C-S, Hernandez M, Rostagno A, Ghiso J, Kim JR. A Strategy for Designing a Peptide Probe for Detection of beta-Amyloid Oligomers. Chem. Bio. Chem. 2010;11(17):2409–2418. doi: 10.1002/cbic.201000435. [DOI] [PubMed] [Google Scholar]
- 36.Ramdzan YM, Nisbet RM, Miller J, Finkbeiner S, Hill AF, Hatters DM. Conformation Sensors that Distinguish Monomeric Proteins from Oligomers in Live Cells. Chem. Biol. 2010;17(4):371–379. doi: 10.1016/j.chembiol.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hecht MH, Richardson JS, Richardson DC, Ogden RC. Denovo Design, Expression, and Characterization of Felix - a 4-Helix Bundle Protein of Native-Like Sequence. Science. 1990;249(4971):884–891. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- 38.Parraga G, Horvath SJ, Eisen A, Taylor WE, Hood L, Young ET, Klevit RE. Zinc-Dependent Structure of a Single-Finger Domain of Yeast Adr1. Science. 1988;241(4872):1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- 39.Luedtke NW, Dexter RJ, Fried DB, Schepartz A. Surveying polypeptide and protein domain conformation and association with FlAsH and ReAsH. Nat. Chem. Biol. 2007;3(12):779–784. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaikh TA, Bakus RC, Parkin S, Atwood DA. Structural characteristics of 2-halo- 1,3,2-dithiarsenic compounds and tris-(pentafluorophenylthio)-arsen. J. Organomet. Chem. 2006;691(9):1825–1833. [Google Scholar]
- 41.Goodman JL, Fried DB, Schepartz A. Bipartite Tetracysteine Display Requires Site Flexibility for ReAsH Coordination. Chem. Bio. Chem. 2009;10(10):1644–1647. doi: 10.1002/cbic.200900207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnan B, Gierasch LM. Cross-Strand Split Tetra-Cys Motifs as Structure Sensors in a beta-Sheet Protein. Chem. Biol. 2008;15(10):1104–1115. doi: 10.1016/j.chembiol.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray-Saha S, Schepartz A. Visualizing Tyrosine Kinase Activity with Bipartite Tetracysteine Display. Chem. Bio. Chem. 2010;11(15):2089–2091. doi: 10.1002/cbic.201000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388(6645):882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 45.Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98(26):15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaietta GM, Giepmans BN, Deerinck TJ, Smith WB, Ngan L, Llopis J, Adams SR, Tsien RY, Ellisman MH. Golgi twins in latemitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 2006;103(47):17777–17782. doi: 10.1073/pnas.0608509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dexter RJ, Schepartz A. Direct Visualization of Protein Association in Living Cells with Complex-Edited Electron Microscopy. Angew. Chem., Int. Ed. 2010;49(43):7952–7954. doi: 10.1002/anie.201003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen B, Cao H, Yan P, Mayer MU, Squier TC. Identification of an orthogonal peptide binding motif for biarsenical multiuse affinity probes. Bioconjugate Chem. 2007;18(4):1259–1265. doi: 10.1021/bc0603900. [DOI] [PubMed] [Google Scholar]
- 49.Miura T, Urano Y, Tanaka K, Nagano T, Ohkubo K, Fukuzumi S. Rational design principle for modulating fluorescence properties of fluorescein-based probes by photo-induced electron transfer. J. Am. Chem. Soc. 2003;125(28):8666–8671. doi: 10.1021/ja035282s. [DOI] [PubMed] [Google Scholar]
- 50.Spagnuolo CC, Massad W, Miskoski S, Menendez GO, Garcia NA, Jares-Erijman EA. Photostability and spectral properties of fluorinated fluoresceins and their biarsenical derivatives: a combined experimental and theoretical study. Photochem. Photobiol. 2009;85(5):1082–1088. doi: 10.1111/j.1751-1097.2009.00565.x. [DOI] [PubMed] [Google Scholar]
- 51.Urano Y, Kamiya M, Kanda K, Ueno T, Hirose K, Nagano T. Evolution of fluorescein as a platform for finely tunable fluorescence probes. J. Am. Chem. Soc. 2005;127(13):4888–4894. doi: 10.1021/ja043919h. [DOI] [PubMed] [Google Scholar]
- 52.Spagnuolo CC, Vermeij RJ, Jares-Erijman EA. Improved photostable FRET-competent biarsenical-tetracysteine probes based on fluorinated fluoresceins. J. Am. Chem. Soc. 2006;128(37):12040–12041. doi: 10.1021/ja063212q. [DOI] [PubMed] [Google Scholar]
- 53.Cao H, Chen B, Squier TC, Mayer MU. CrAsH: a biarsenical multi-use affinity probe with low non-specific fluorescence. Chem. Commun. 2006;24:2601–2603. doi: 10.1039/b602699k. [DOI] [PubMed] [Google Scholar]
- 54.Soh N. Selective chemical labeling of proteins with small fluorescent molecules based on metal-chelation methodology. Sensors-Basel. 2008;8(2):1004–1024. doi: 10.3390/s8021004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tour O, Adams SR, Kerr RA, Meijer RM, Sejnowski TJ, Tsien RW, Tsien RY. Calcium Green FlAsH as a genetically targeted small-molecule calcium indicator. Nat. Chem. Biol. 2007;3(7):423–431. doi: 10.1038/nchembio.2007.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhunia AK, Miller SC. Labeling tetracysteine-tagged proteins with a SplAsH of color: A modular approach to bis-arsenical fluorophores. Chem. Bio. Chem. 2007;8(14):1642–1645. doi: 10.1002/cbic.200700192. [DOI] [PubMed] [Google Scholar]
- 57.Wang T, Yan P, Squier TC, Mayer MU. Prospecting the proteome: Identification of naturally occurring binding motifs for biarsenical probes. Chem. Bio. Chem. 2007;8(16):1937. doi: 10.1002/cbic.200700209. [DOI] [PubMed] [Google Scholar]
- 58.Van Engelenburg SB, Nahreini T, Palmer AE. FACS-Based Selection of Tandem Tetracysteine Peptides with Improved ReAsH Brightness in Live Cells. Chem. Bio. Chem. 2010:489–493. doi: 10.1002/cbic.200900689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levitus M, Ranjit S. Cyanine dyes in biophysical research: the photophysics of polymethine fluorescent dyes in biomolecular environments. Q. Rev. Biophys. 2011;44(1):123–151. doi: 10.1017/S0033583510000247. [DOI] [PubMed] [Google Scholar]
- 60.Yaron D, Silva GL, Ediz V, Armitage BA. Experimental and computational investigation of unsymmetrical cyanine dyes: Understanding torsionally responsive fluorogenic dyes. J. Am. Chem. Soc. 2007;129(17):5710–5718. doi: 10.1021/ja070025z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toutchkine A, Nguyen DV, Hahn KM. Simple one-pot preparation of water-soluble, cysteine-reactive cyanine and merocyanine dyes for biological imaging. Bioconjugate Chem. 2007;18(4):1344–1348. doi: 10.1021/bc060376n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao H, Xiong Y, Wang T, Chen B, Squier TC, Mayer MU. A red Cy3-based biarsenical fluorescent probe targeted to a complementary binding peptide. J. Am. Chem. Soc. 2007;129(28) doi: 10.1021/ja070003c. 8672–. [DOI] [PubMed] [Google Scholar]
- 63.Langhorst MF, Genisyuerek S, Stuermer CAO. Accumulation of FlAsH/Lumio Green in active mitochondria can be reversed by beta-mercaptoethanol for specific staining of tetracysteine-tagged proteins. Histochem. Cell Biol. 2006;125(6):743–747. doi: 10.1007/s00418-005-0136-3. [DOI] [PubMed] [Google Scholar]
- 64.Uttamapinant C, White KA, Baruah H, Thompson S, Fernandez-Suarez M, Puthenveetil S, Ting AY. A fluorophore ligase for site-specific protein labeling inside living cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107(24):10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuivila HG, Keough AH, Soboczenski EJ. Areneboronates from Diols and Polyols. J. Org. Chem. 1954;19(5):780–783. [Google Scholar]
- 66.James TD, Sandanayake KRAS, Shinkai S. Novel Photoinduced Electron-Transfer Sensor for Saccharides Based on the Interaction of Boronic Acid and Amine. J. Chem. Soc. Chem. Commun. 1994;4:477–478. [Google Scholar]
- 67.Cao HS, Heagy MD. Fluorescent chemosensors for carbohydrates: A decade's worth of bright spies for saccharides in review . J. Fluoresc. 2004;14(5):569–584. doi: 10.1023/b:jofl.0000039344.34642.4c. [DOI] [PubMed] [Google Scholar]
- 68.James TD, Shinkai S. Artificial receptors as chemosensors for carbohydrates. Host-Guest Chem. 2002;218:159–200. [Google Scholar]
- 69.Halo TL, Appelbaum J, Hobert EM, Balkin DM, Schepartz A. Selective Recognition of Protein Tetraserine Motifs with a Cell-Permeable, Pro-fluorescent Bis-boronic Acid. J. Am. Chem. Soc. 2009;131(2):438–439. doi: 10.1021/ja807872s. [DOI] [PMC free article] [PubMed] [Google Scholar]