Abstract
Objective
To examine associations between haplotypes of the serotonin 1B receptor gene and individual differences in anger and hostility.
Methods
Data were analyzed from a study of 361 university students (47% male). Participants were genotyped at 5 polymorphisms in the HTR1B gene (rs11568817, rs130058, rs6296, rs6297, rs13212041), including promoter and 3′UTR polymorphisms with opposite functional effects on gene expression. Participants reported their emotional states across 30 consecutive days for up to four years. Haplotype pairs were constructed statistically and assigned to a level of HTR1B expression based on the presence of the functional polymorphisms.
Results
Six haplotypes accounted for >97% of chromosomes. Three low expression haplotypes contained the 3′UTR variant (rs13212041 A-allele) that enables a microRNA-mediated reduction in expression. One intermediate expression haplotype contained the 3′UTR A-allele paired with the high-activity promoter. Two high expression haplotypes contained the 3′UTR variant (rs13212041 G-allele) that attenuates microRNA-mediated reduction in expression. Men with low expression haplotypes reported greater anger and hostility than men with one or two high expression haplotypes. Diplotype classification accounted for 8.4% of the variance in men’s anger and hostility, primarily due to the 3′UTR polymorphism (rs13212041), but with some contribution of the functional promoter combination (rs11568817, rs130058). Associations with anger and hostility were not found in women.
Conclusions
These findings extend our understanding of the genetic basis of anger and hostility by showing that newly characterized HTR1B haplotypes, particularly those with rs13212041, which modulates microRNA-mediated regulation of HTR1B expression, may have important implications for aggression-related phenotypes among young men.
Keywords: SEROTONIN 1B RECEPTOR, AGGRESSION, ANGER, HOSTILITY, MICRORNA, MIR-96
Introduction
Individual differences in anger and hostility underlie a variety of adverse or risky health conditions including elevated cardiovascular risk (Miller et al., 1996), metabolic and immune dysfunction (Kiecolt-Glaser et al. 2005; Boyle et al., 2007; Goldbacher & Matthews, 2007; Gouin et al., 2008), intimate partner violence (Norlander & Eckhardt, 2005), maintenance of heavy drinking beyond young adulthood (Costanzo et al. 2007), drug dependence (De Moja & Spielberger, 1997) and suicide (Baud, 2005; McGirr & Turecki, 2007). Because emotional style is partly heritable (Bouchard, 2004; Cates et al., 1993; Miles & Carey, 1997), it is important to identify genes that increase risk for this emotional phenotype.
Animal research on aggression may provide insight into the genetic basis of anger and hostility. Aggression is closely related to, but distinct from, the constructs of anger and hostility (Buss & Perry, 1992; Martin, Watson & Wan, 2000). Whereas aggression refers to observable behaviors of an organism involving approach and attempted harm towards others, anger and hostility refer to internal emotional and cognitive experiences of an organism. Feelings of anger and hostility do not always lead to aggression, but they can precipitate the “hotter,” impulsive form of aggression (Berkowitz, 1983). Aggressive behavior is also used as a proxy for an angry/hostile phenotype in studies of cardiovascular risk among higher order primates (e.g., Manuck, Kaplan, & Clarkson, 1983). Taken together, these links suggest that genes that increase susceptibility to aggression may also increase susceptibility to angry and hostile emotions.
In the animal literature, studies suggest a role of the serotonin 1B receptor (HTR1B) in aggressive behavior (for reviews Clark & Neumaier, 2001; Oliver & van Oorshot, 2005; Popova, 2006). For example, mice lacking serotonin 1B receptors behave more aggressively than their wild-type littermates (Saudou et al., 1994) and drugs that increase the activity of the serotonin 1B receptor have anti-aggressive effects on mice placed in paradigms designed to elicit impulsive aggressive behaviors (Miczek & de Almeida, 2001). Given this evidence, researchers have sought to identify human HTR1B variants that might influence either the activity of the serotonin receptor or its level of expression and in turn, the associated aggressive phenotype.
To date, a select group of HTR1B polymorphisms have been examined in relation to human aggressive phenotypes. The most frequently studied HTR1B variant, rs6296 [a synonomous exonic single nucleotide polymorphism (SNP): G861C], has been associated with aggressive behavior in children (Davidge et al., 2004) and antisocial alcoholism (Lappalainen et al., 1998; cf. Kranzler et al., 2002). However these allelic associations with aggressive phenotypes are not consistently found (New et al., 2001). Inconsistent or null associations have also been reported between rs6296 and other phenotypes related to aggression and impulsivity including suicidality (with the 861C allele overrepresented among individuals with personality disorders in New et al., 2001, but not Huang et al., 1999; Nishiguchi et al., 2001; Rujescu et al., 2003; or Zouk et al., 2007); attention deficit hyperactivity disorder (found in Hawi et al., 2002; Quist et al., 2003, Smoller et al., 2006; but not Ickowicz et al., 2007) and substance abuse (Huang et al., 2003). Although the 861C allele was associated with a 20% lower average number of serotonin 1B receptor binding sites in 96 post-mortem brain samples (Huang et al., 1999), there is no evidence that the G861C SNP itself is functional; rather, the high degree of linkage disequilibrium (LD) among previously studied markers in HTR1B (Cigler et al., 2001; Duan et al., 2003; Proudnikov et al., 2006; Zouk et al., 2007) suggests that this marker may be in LD with other functional HTR1B polymorphisms.
The absence of splicing (HTR1B is encoded by a single exon) or common (>2% frequency) amino acid coding polymorphisms suggests that any functional variants would exist in regulatory regions outside the protein coding region. Indeed, two such functional variants have been described. Duan et al. (2003) provided the first report of functional HTR1B polymorphisms. Using in vitro reporter gene expression assays, these investigators showed that two SNPs in the promoter region, −261G paired with the −161 A allele [rs11568817 (T-261G) and rs130058 (A-161T)], increase the binding of transcription factors in the HTR1B promoter producing a 2.3-fold increase in gene transcription in transfected cells. They noted that these promoter SNPs were in significant LD with the commonly examined G861C marker (rs6296). Two groups have examined these promoter SNPs with respect to human phenotypes. Zouk et al. (2007) investigated the T-261G, A-161T promoter polymorphisms, plus other previously investigated SNPs (G861C, A1180G, and the lesser studied C129T), and showed that only the lower expression promoter region −161T allele predicted higher scores on informant reports of the Buss–Durkee Hostility Inventory in a sample of suicide completers. The −161T allele was overrepresented among suicide completers as well (Zouk et al., 2007). Proudnikov et al. (2006) used a hybrid molecular and statistical haplotyping procedure to examine HTR1B haplotype associations with heroin dependence. They observed that haplotypes containing a G-allele at rs6297 (A1180G), a SNP in the proximal region of the HTR1B 3′UTR (Sanders et al., 2001), which has not been demonstrated to have direct functional effects, were protective for heroin dependence (Proudnikov et al., 2006). Interestingly, the protective effect of the G-allele at rs6297 was unrelated to genotype at either the promoter T-261G/A-161T, C129T or G861C alleles, suggesting that it may be linked to other more distal functional polymorphisms.
Recently, evidence for a second functional regulatory variant was reported by Jensen et al. (2008) who characterized a SNP occurring in the distal 3′UTR of HTRIB messenger RNA (rs13212041; A1997G) that disrupts a binding site for the microRNA, miR-96. MicroRNAs are 20-21 nucleotide ribonucleic acids that regulate gene expression by binding to complementary sites on messenger RNA triggering mRNA degradation and/or inhibition of translation (Ambros, 2004; Chendrimada et al., 2007; Baek et al., 2008; Hutvágner & Zamore, 2002; Kiriakidou et al., 2007; Mathonnet et al., 2007; Selbach et al., 2008; Yekta et al., 2004; Wakiyama et al., 2007). Jensen et al. used a luciferase reporter gene assay to show that the rs13212041 polymorphism modulates miR-96 regulation of gene expression. The A-allele mRNA was repressed by miR-96, while the G-allele attenuated this miR-96 regulatory function. Importantly, introduction of a compensatory change in mir96 rescued repression of the G allele, confirming a direct interaction between mir96 and the regulatory element in HTR1B.
These molecular differences found in Jensen et al. (2008) suggested that A-allele individuals have reduced HTR1B expression, and greater risk of aggressive behaviors, compared to G-allele individuals. Indeed, Jensen et al. (2008) found that university students with two copies of the microRNA-repressed HTR1B A-allele reported a greater history of aggressive behaviors (e.g., starting fires, damaging property, fighting) than individuals heterozygous or homozygous for the G-allele. Genotype differences were not found in non-aggressive anti-social behaviors (e.g., lying). As yet, it is not known whether this 3′UTR microRNA targeting polymorphism (rs13212041) is in LD with other known polymorphisms (i.e., rs11568817, rs130058, rs6296, rs6297) and whether these polymorphisms act synergistically or, potentially, antagonistically to influence risk for other aggression-related emotional phenotypes such as anger and hostility. For example, it is not known whether the 3′UTR (rs13212041) and 5′-promoter functional alleles (rs11568817, rs130058) are inherited separately or together, and if together, what those inherited combinations are. In addition, linkage with the 3′UTR could explain previously found effects. For example, co-inheritance of the 1180G allele of rs6297 and the 3′UTR G-allele of rs13212041 could explain why rs6297 has been linked to other impulsive disorders (Proudnikov et al., 2006) in the absence of both known functional effects of rs6297 and associations of it with currently known functional SNPs.
In this report, we examined the recently characterized 3′UTR functional SNP (rs13212041; A1997G; Jensen et al., 2008), the two functional promoter region SNPs (rs11568817, rs130058; Duan et al., 2003), and two other commonly studied SNPs, rs6296 and rs6297, and their associations with individual differences in anger and hostility. Because these markers span a relatively short genetic interval of 2,258 base pairs, they are expected to show considerable LD. Thus, we also examined the pattern of LD among markers and identified naturally occurring haplotypes. With this approach, we could determine whether the 5′-promoter and 3′UTR functional alleles are inherited together or separately, as well as their linkage with the G861C and A1180G alleles, which others have examined in relation to human phenotypes. We then classified the haplotypes expected to have lower versus higher levels of gene expression based on prior molecular research showing functional effects of rs11568817, rs130058, and rs13212041. Finally, we tested whether individuals with the predicted lower expression HTR1B diplotypes scored higher on their reports of anger and hostility than those with predicted higher HTR1B expression diplotypes.
Self-reports of anger and hostility were measured using real-time data capture techniques (Stone, Shiffman, Atienza & Nebeling, 2007). Participants completed a daily report of current feelings of anger and hostility for 30 consecutive days, for up to four years. By aggregating multiple emotion state reports over time, we could obtain a highly reliable and stable assessment of anger and hostility over a four-year period of early-adulthood, roughly from ages 18 to 21. This approach also reduced error in the emotion measurement, thereby increasing the statistical power to detect genotype-phenotype associations.
The present study uses the same human sample as in Jensen et al. (2008) but it extends the prior report in three critical ways. First, the present investigation helps to integrate prior literature by genotyping the four most commonly occurring polymorphisms in addition to the one in the 3′UTR and testing their LD. Second, the present investigation uses a haplotype approach to examine the natural pattern of inheritance for these five markers. A haplotype approach provides a biological point of reference for the inheritance of the potentially synergistic or opposing effects of these polymorphisms. Third, the present investigation uses a more extensive phenotype measure. Whereas Jensen et al. (2008) analyzed a single checklist of past aggressive behaviors, the present investigation analyzed real-time reports of anger and hostility measured in a robust and ecologically valid fashion over four years. Moreover, the content of the questions—asking about common everyday emotions (anger and hostility) rather than relatively uncommon aggressive events (setting fires; starting fights; as in Jensen et al.)— may further increase power to detect genetic associations via more refined differentiation among subjects.
Method
Participants
The sample consisted of 361 university students (168 men; 193 women) from a mid-sized US state university. It was derived from a sample of 575 participants in a four-year longitudinal study who provided DNA samples (N = 416). To avoid population stratification, we limited the analysis sample to individuals of non-Hispanic Caucasian ancestry. This sample included individuals analyzed elsewhere for unrelated hypotheses (Armeli et al., 2008; Covault et al., 2007; Gacek et al., 2008; Gunthert et al., 2007). The average age of sample participants at the start of this four-year study was 18.7 +/ 0.9 yrs with 61% percent freshmen, 32% sophomores, and 7% upper classmen. All participants gave written informed consent under guidelines approved by the Institutional Review Boards at the University of Connecticut (Storrs) and the University of Connecticut Health Center (Farmington).
Measures and Procedure
In each year, participants completed an initial assessment battery one month following the start of either the fall (59% of participants) or spring (41% of participants) semester by accessing a secure website. This on-line assessment included demographic variables and the history of aggression questionnaire reported on previously (Jensen et al., 2008). Approximately two weeks after the initial assessment, participants logged on to a secure website to complete a 5-minute survey between 2:30 and 7:00 p.m. daily for 30 days. The daily survey contained questions about a range of behaviors and experiences, including current emotional states. Using a 5-point scale from 1 (very slightly/not at all) to 5 (extremely), participants rated how “angry” and “hostile” they felt right now, and these responses were averaged within each daily survey for a measure of hostility (α = .76). Participants also rated how “sad” and “dejected” they felt, averaged for sadness (α = .75); “jittery” and “nervous” for anxiety (α = .72); and “cheerful” and “happy” for happiness (α = .88). Participants repeated this same 30-day survey procedure every fall or spring for up to four years.
We initiated DNA collection in year two of the study. At that time, the 535 participants who were thought to be still living in the area were invited to participate in a genetic sub-study, with eligibility to continue in the larger study not contingent on providing a sample for DNA. Of those invited, 416 (77.8 %) consented to provide genetic data, 69 (12.9 %) declined, 34 (6.4%) did not respond and 16 (3.0%) had moved out of the area. After giving informed consent, participants were asked to “swish” Scope™ mouthwash (Procter and Gamble, Cincinnati, Ohio) for 20-30 seconds, and then spit the mouthwash into a collection tube. The 416 participants who consented to provide DNA samples did not differ from the other 119 participants on any of the primary outcome variables, although consenters were more likely to be female (χ2 = 7.74, p < .01), younger (18.7 vs. 19.0 years, p < .05), and to participate in the study longer (3.5 vs. 2.3 years, p < .001).
Genotyping
DNA was extracted from mouthwash-stabilized samples using a commercial DNA isolation protocol (PureGene, Gentra Systems, Minneapolis, MN). The five HTR1B polymorphisms were genotyped using PCR-based TaqMan® allelic discrimination assays. A 10μl PCR reaction was prepared in 1x ABI TaqMan Universal master mix (Applied Biosystems Inc., Foster City, CA) containing 100nM of each Fam and Vic-labeled allele specific TaqMan® MGB probes, and 600nM of each forward and reverse primer (Table I). For the promoter SNPs, rs115688177 and rs130058, primers and probes were as described by Proudnikov et al. (2006). MGB-probes and primers for the remaining 3 SNPs were designed using Primer Express v3.0 software (Applied Biosystems Inc.). The polymerase chain reaction was performed with the following cycling parameters: 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. End point FAM and VIC fluorescence levels were captured using an ABI 7500 Sequence Detection System (Applied Biosystems Inc.) and genotype calls were made based on the level of fluorescence signal. The genotyping assay was repeated for 13% of samples with no discrepancies in genotype calls. Finally, we carried out molecular 2-SNP analysis of the T-261G and A-161T polymorphisms using the A-161T TaqMan probe and downstream primer paired with T-261G allele-specific forward primers (CCAGCTCTTAGCAACCCAGTT or CCAGCTCTTAGCAACCCAGTG) as described by Proudnikov et al. (2006) with genotype calls made by examining change in fluorescence as a function of cycle number for each sample. The resulting molecular 2-SNP allele calls were in agreement with haplotypes statistically determined using Phase 2.1. The 2-SNP haplotype – 261T/-161T was not observed. (Table I Here)
Table I.
HTR1B SNPs Examined in this University Student Cohort
| NCBI SNP (dbSNP build 126) |
Location relative to HTR1B1 |
PCR amplicon primers | TaqMan MGB probes (SNP position in bold) |
|---|---|---|---|
| rs115688177 | Promoter | AGCGGAGCAGGGCTCG | Vic- CCCAGGGCTAAGAC |
| T-261G | TTCCTCAATTATTCCTCCGCC | 6FAM- CCCAGGTCTAAGACC | |
| rs13058 | Promoter | GCGGCTTAGCTAGGCGCT | Vic- CACCCTTGACCTCTA |
| A-161T | CGCAGGTTTGTCCCCAGTT | 6FAM- CACCCATGACCTCT | |
| rs6296 | Exon 1 | CGTCCTCGGTCACCTCTATTAACT | Vic- CTCCTGTGTATGTGAAC |
| G861C | GTCGGAGACTCGCACTTTGACT | 6FAM- TCTCCTGTGTATGTCAAC | |
| rs6297 | Exon 1 | AAACAAGCATTCCATAAACTGATACG | Vic- ACTTGCCATTTGC |
| A1180G | GGTCCCCAAAGGTCGCTTAG | 6FAM- TGACTTGCCGTTTGCAGT | |
| rs13212041 | 3′UTR | AAAGTGACAGGTACATGAAATTAAGAGAA | Vic- TGCAGACTTTGGC |
| A1997G | CACAACTCTAACAAACAAACCATTATGTG | 6FAM- TGCAGACTTCGGC |
Nucleotide position relative to HTR1B ATG using the March 2006 version of the human genome chromosome 6 annotation (www.genome.ucsc.edu).
Data analysis
The survey data were screened prior to analyses to exclude participants who provided fewer than the minimum of 15 of 30 daily surveys for a given year following our standard protocol (Covault et al., 2007). Five of the 361 participants failed to meet this minimum standard in any of the years and were eliminated from all survey analyses. We also limited our analyses to years in which participants were undergraduate students to ensure that data were from a similar population. This screening did not compromise the sample, as the majority of participants were undergraduate students throughout the study. Thus, we retained data across an average of 3.1 years (+/− 0.9), reflecting an average of 79 reports per person (+/−27) (range 16 – 120 reports). The Hierarchical Linear Modeling statistical software program (HLM; 6.04 (Raudenbush, Bryk, & Congdon, 2007) was used to examine associations between the survey data and the genetic information. This approach took into account the unbalanced (e.g., varying number of daily and yearly reports) and nested nature of the data. We ran hierarchical linear models to test (i) whether average hostility scores differed across each genotype group; (ii) whether hostility scores differed across the common haplotype configurations; and (iii) whether hostility scores differed across diplotypes. Men and women were analyzed separately to provide separate effect size estimates. More detailed descriptions of the hierarchical linear models are found in Supplementary Materials.
Linkage disequilibrium (LD) plots, haplotype blocks, and haplotype frequencies for the sample of Caucasian students were generated using the software program Haploview v3.2.2 (Barrett et al., 2005). Best-estimate haplotype pairs for each individual subject were generated using Phase v2.1 software, which incorporates a Bayesian statistical method for reconstructing estimated haplotypes from population data (Stephens et al., 2001; Stephens & Donnelly, 2003).
Results
Genotype frequencies for the 361 Caucasian participants are presented in Table II. All frequencies were in Hardy-Weinberg Equilibrium. Allele frequencies for the 5 SNPs were consistent with prior reports in the literature.
Table II.
Marker Frequency Data for Caucasian University Student Participants
| Marker | Position relative to AUG |
Minor Allele |
Minor Allele Frequency |
Genotype Counts | Total N* |
HWE (χ2, p) |
||
|---|---|---|---|---|---|---|---|---|
| rs115688177 | −261 | G | .469 | TT (95) |
TG (191) |
GG (73) |
(359) | 1.66, 0.20 |
| rs130058 | −161 | T | .306 | AA (165) |
AT (168) |
TT (26) |
(359) | 3.66, 0.06 |
| rs6296 | 861 | C | .234 | GG (206) |
GC (138) |
CC (15) |
(359) | 1.88, 0.17 |
| rs6297 | 1180 | G | .142 | AA (261) |
AG (89) |
GG (6) |
(356) | 0.26, 0.61 |
| rs13212041 | 1997 | G | .199 | AA (232) |
AG (114) |
GG (15) |
(361) | 0.04, 0.83 |
Note. Frequencies and Hardy-Weinberg Equilibrium statistics (HWE) computed for the 361 Caucasian participants.
Varying N for each marker reflects differences in the % of individuals for whom genotypes were obtained in this Caucasian DNA sample.
Genotype Results
Genotype differences in hostility for men and women are shown in Table III. Only men showed significant differences in hostility as a function of genotypes. Of the five genotypes tested, the 3′UTR variant (rs13212041) showed the largest differences in hostility scores among the men, with the AA vs. G carrier difference accounting for 7.1% of the variance in hostility scores. Rs6297 also showed significant differences, with the AA vs. G carrier difference accounting for 4% of the variance. Analysis of rs130058 and rs6296 both yielded non-significant trends, and each accounted for approximately 2% of the variance in hostility ratings. Rs115688177 was unrelated to hostility. Genotypes were also related to sadness and anxiety for men only, although effect sizes were generally smaller for these measures than for hostility (see Supplementary Materials, Table S-1). For example, the 3′UTR variant (rs13212041) accounted for 4.0% and 4.9% of the variance in anxiety and sadness, compared to 7.1% of the variance in hostility. Genotypes were unrelated to happiness either for men or women.
Table III.
HTR1B Genotype Differences in Hostility for Caucasian Young Men and Women
| Genotypes | Significance Tests | Effect Size | ||||||
|---|---|---|---|---|---|---|---|---|
| Men | Coding | b | SE | p | % var | |||
| rs115688177 | TT 1.31 (.31) |
TG 1.35 (.46) |
GG 1.39 (.36) |
0,1,2 | .037 | .039 | .335 | 0.5% |
| rs130058 | AA 1.28 (.28) |
AT 1.41 (.49) |
TT 1.37 (.35) |
0,1,1 | .113 | .059 | .057 | 2.2% |
| rs6296 | GG 1.30 (.36) |
GC 1.40 (.46) |
CC 1.49 (.49) |
0,1,1 | .114 | .066 | .084 | 2.5% |
| rs6297 | AA 1.39 (.43) |
AG 1.25 (.29) |
GG 1.05 (.04) |
0,1,1 | −.158 | .057 | .007 | 4.0% |
| rs13212041 | AA 1.42 (.46) |
AG 1.23 (.25) |
GG 1.22 (.31) |
0,1,1 | −.198 | .054 | .001 | 7.1% |
|
|
||||||||
| Women | Coding | b | SE | p | % var | |||
| rs115688177 | TT 1.32 (0.34) |
TG 1.24 (.23) |
GG 1.26 (0.36) |
0,1,2 | −.034 | .036 | .357 | 1.0% |
| rs130058 | AA 1.28 (0.27) |
AT 1.26 (0.32) |
TT 1.18 (0.18) |
0,1,1 | −.025 | .042 | .553 | 0.2% |
| rs6296 | GG 1.24 (0.28) |
GC 1.29 (0.30) |
CC 1.35 (0.28) |
0,1,1 | .058 | .043 | .182 | 1.3% |
| rs6297 | AA 1.26 (0.29) |
AG 1.28 (0.31) |
GG 1.24 (0.12) |
0,1,1 | .017 | .049 | .732 | 0.1% |
| rs13212041 | AA 1.25 (0.27) |
AG 1.31 (0.33) |
GG 1.22 (0.21) |
0,1,1 | .049 | .047 | .292 | 1.0% |
Note. Hostility scores reflect the averaged self-reports of anger and hostility rated on a 1 – 5 scale with higher numbers indicating greater intensity of emotion. Means (standard deviations) computed on raw data by averaging the hostility scores over 120 (max) days of reporting and then averaging across participants with identical genotypes. Significance tests and effect sizes used Hierarchical Linear Modeling. b = unstandardized coefficient testing for genotype differences using the corresponding coding scheme (linear = 0,1,2; dominant = 0,1,1); SE = standard error of thecoefficient; p = chance probability value (two-tailed). Effect Size, % var = the percentage of variance in hostility accounted for by genotype.
Haplotype Results
The 5 markers examined were in a single haplotype block with D”values of 0.87 to 1.0 (see Figure 1). Six haplotypes comprised >97% of chromosomes (See Table IV). The frequency of haplotypes as estimated for the entire sample by Haploview was in close agreement with the frequency based on individual subject best-estimate haplotype pairs generated using the Phase software application. The observed haplotype frequencies were comparable with frequencies reported using a subset of these markers (Duan et al., 2003; Proudnikov et al., 2006).
Figure 1.
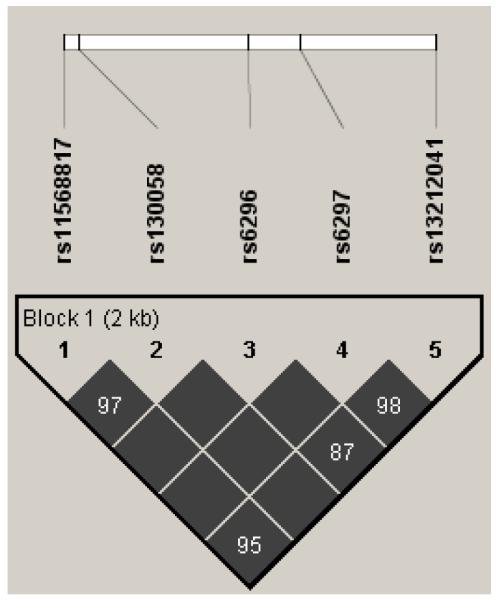
/I> Haploview generated LD plot illustration for Caucasian participants. Values shown represent 100 × ∣D”∣, empty boxes represent a value of 100 (i.e. D”=1). The location of 5 SNPs across the 2,258 bp HTR1B gene are schematically noted at the top beginning with the T–261G promoter region SNP rs115688177 at the far left and the 3′UTR microRNA site SNP rs13212041 at mRNA position 1,997 at the far right.
Table IV.
HTR1B Haplotype Frequencies for Caucasian Samples and Anticipated HTR1B Expression Levels for the Six Most Frequent Haplotypes
| Haplotype1 |
HTR1B mRNA Activity2 |
Haploview v3.2.2 Frequency |
Phase v2.1 Best- Estimate Frequency |
Haplotype Counts |
Frequency Observed by Duan et al.3 |
Frequency Observed by Proudnikov et al.4 |
|---|---|---|---|---|---|---|
| GTGAA | lower | 0.295 | 0.302 | 218 | 0.225 (GTGxx) | 0.345 (GTGAx) |
| TACAA | lower | 0.215 | 0.231 | 167 | 0.240 (TACxx) | 0.282 (TACAx) |
| GAGAA | higher | 0.160 | 0.162 | 117 | 0.214 (GAGxx) | 0.106 (GAGAx) |
| TAGGG | higher | 0.137 | 0.139 | 100 | 0.225 (TAGxx) | 0.141 (TAGGx) |
| TAGAA | lower | 0.112 | 0.098 | 71 | - | 0.127 (TAGAx) |
| TAGAG | higher | 0.054 | 0.055 | 40 | - | - |
|
|
||||||
| Total | 0.973 | 0.987 | 713 | - | - | |
Note5 SNP haplotype using markers rs11568817, rs130058, rs6296, rs6297, rs13212041 at positions −261, −161, 861, 1180, and 1999, respectively. Haplotypes shown are those with frequencies >0.02. SNPs associated with higher gene expression activity in vitro are underlined.
Predicted higher mRNA activity haplotypes included either the promoter −261G/−161A, or 3′UTR microRNA site 1997G.
Duan et al. did not examine markers rs6297 or rs13212041.
Proudnikov et al. did not examine marker rs13212041.
For the 6 common haplotypes, we classified three as having lower HTR1B gene expression (GTGAA, TACAA, and TAGAA) and three haplotypes as having higher HTR1B gene expression (GAGAA, TAGGG, and TAGAG). We based this classification on in vitro functional assays by Duan et al. (2003) who showed that the GA pairing at T-261G and A-161T had a 2.3-fold higher transcriptional activity and Jensen et al. (2008) who showed that the rs13212041 G-allele attenuated microRNA-mediated down regulation of gene expression relative to the A-allele. This classification combined the putative transcriptional and microRNA-mediated mRNA translation/stability effects contributed by the two separate functional polymorphisms.
With these haplotypes (see Table IV), it is interesting to note that the 861C allele, which had previously been reported to be associated with reduced HTR1B binding sites in post-mortem brain (Huang et al., 1999), was observed only in the lower expression TACAA haplotype containing the rs13212041 A-allele. Similarly, the −161T allele, which had previously been associated with increased hostility (Zouk et al., 2007), was observed only in the lower expression GTGAA haplotype containing the rs13212041 A-allele. Lastly, the rs6297 1180G allele, which had previously been shown to have a protective effect for heroin dependence (Proudnikov et al. 2006), was observed only in the higher expression TAGGG haplotype containing the rs13212041 G-allele.
We then examined the relation between the six common haplotypes and the hostility ratings from the daily survey data (see Table V). As predicted, but for men only, haplotypes expected to have lower HTR1B mRNA expression (TACAA, GTGAA, TAGAA) were associated with higher levels of self-reported hostility, whereas haplotypes expected to yield high HTR1B levels (TAGAG, TAGGG) were associated with lower levels of hostility. The GAGAA haplotype containing the high-activity GA promoter with the low-activity 3′UTR A-allele was associated with intermediate levels of hostility. It is notable that we did not observe the haplotype GAxxG containing the more inducible promoter (GA at T–261G/A–161T) paired with the microRNA binding site disrupted by the G-allele. Instead, we observed only the haplotype GAGAA, containing the more inducible promoter paired with the active microRNA binding site (A-allele). This pattern of inheritance coupled with the intermediate score for the hostile mood phenotype measure suggests that the functional increase in activity of the GA promoter may be ameliorated by microRNA-mediated down regulation of gene expression by the rs13212041 A-allele. As such, the haplotype GAGAA was classified as having intermediate HTR1B expression for consideration of diplotype associations with phenotype.
Table V.
The Six Frequent Haplotypes and Hostility Ratings for Men and Women
| Haplotypes | Hostility Mean(SD) |
Hostility Ranking |
Predicted mRNA activity1 |
N Haplotypes |
N Data2 |
Post hoc Reference - Highest Ranking Hostility Haplotype |
|---|---|---|---|---|---|---|
| Men | ||||||
| TACAA | 1.41(0.51) | 1 | lower | 76 | 228 | - |
| GTGAA | 1.38(0.50) | 2 | lower | 105 | 317 | ns |
| TAGAA | 1.33(0.40) | 3 | lower | 34 | 108 | ns |
| GAGAA | 1.31(0.40) | 4 | higher | 53 | 151 | p < .10 |
| TAGAG | 1.23(0.23) | 5 | higher | 20 | 61 | p < .001 |
| TAGGG | 1.20(0.29) | 6 | higher | 45 | 125 | p < .05 |
|
|
||||||
| Total | 333 | 990 | ||||
|
| ||||||
| Women | ||||||
| TAGAG | 1.41(0.51) | 1 | higher | 20 | 71 | - |
| TACAA | 1.31(0.35) | 2 | lower | 91 | 278 | ns |
| TAGGG | 1.26(0.30) | 3 | higher | 55 | 169 | ns |
| GAGAA | 1.25(0.32) | 4 | higher | 64 | 202 | ns |
| GTGAA | 1.23(0.29) | 5 | lower | 113 | 364 | ns |
| TAGAA | 1.22(0.24) | 6 | lower | 37 | 112 | ns |
|
|
||||||
| Total | 380 | 1196 | ||||
Note. Predicted higher activity mRNA haplotypes included either the promoter −261G/−161A, or 3′UTR microRNA site 1997G (underlined).
N Data = the number of data points analyzed for each haplotype (each participant contributed up to four hostility means- one per year of data collection- for each haplotype). Means (Standard Deviations) computed on raw data. Statistical comparisons used Hierarchical Linear Modeling.
Diplotype Results
We computed a diplotype score for each participant based on the predicted HTR1B activity value of both haplotypes. Each haplotype was assigned a value: “0” for lower-activity haplotypes (TACAA, GTGAA and TAGAA), “0.5′for the intermediate-activity haplotype (GAGAA), and “1′for higher-activity haplotypes (TAGAG and TAGGG). The two haplotype values were added, yielding five HTR1B diplotype activity levels (0, 0.5, 1, 1.5 or 2). Frequencies were as follows: 0 = 141 (40%); 0.5 = 81 (23%), 1 = 99 (28%); 1.5 = 21 (6%); 2 = 13 (3%). Six participants could not be assigned a diplotype due to missing haplotype information for one chromosome. The majority (92 of 99) of those with a diplotype score of 1 had one high and one low-activity haplotype; the remainder (7 of 99) had two intermediate activity GAGAA haplotypes containing both the −261G/A-161 promoter alleles and 3′UTR miR-96 binding site A-allele. Because of the low frequency of participants with two intermediate activity haplotypes (2 men; 5 women), we grouped them with the other 92 participants with a diplotype score of 1 in subsequent statistical analyses. Diplotype frequencies did not vary by gender, χ2 = .70, p = .951 [men: 0 = 67 (40%); 0.5 = 37 (22%), 1 = 46 (28%), 1.5 = 11 (7%), 2 = 5 (3%), plus two unassigned; women 0 = 74 (39%); 0.5 = 44 (23%); 1 = 53 (28%), 1.5 = 10 (5%); 2 = 8 (4%), plus four unassigned].
Mean hostility and other emotion scores as a function of diplotypes are shown in Table VI. The pattern of means showed that men (top rows) with no high-activity haplotypes (0s) reported the highest levels of hostility, compared to men with one or more higher-activity haplotypes (1s, 1.5s, 2s). Men with one intermediate activity haplotype containing the GAxxA sequence (0.5) reported intermediate levels of hostility. The similar hostility scores among men with one or more high activity haplotypes (1s, 1.5s, 2s), suggested a dominant effect of higher-activity haplotypes containing the G-allele of rs13212041. 1 In fact, the presence of one or more higher-activity haplotypes (vs. one intermediate vs. none) accounted for 8.4% of the variance in hostility corresponding to a Cohen”s d of 0.62, a medium effect size (Cohen, 1988). This effect size is stronger, although not substantively so, than the effect size found when testing rs13212041 alone (7.1%; d = .57).
Table VI.
Average Hostility, Anxiety, Sadness, and Happiness Scores for the HTR1B Receptor Diplotypes
| Number of Higher Activity HTR1B Haplotypes | Significance Tests | Effect Size | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Men | 0 | .5 | 1 | 1.5 | 2 | b | SE | p | % var |
| Hostility | 1.47 (0.51) | 1.31 (0.31) | 1.25 (0.28) | 1.19 (0.18) | 1.22 (0.31) | −.235 | .069 | .001 | 8.4% |
| Anxiety | 1.61 (0.52) | 1.38 (0.41) | 1.42 (0.36) | 1.25 (0.14) | 1.30 (0.24) | −.223 | .075 | .004 | 5.8% |
| Sadness | 1.47 (0.51) | 1.30 (0.33) | 1.27 (0.26) | 1.19 (0.12) | 1.29 (0.29) | −.206 | .068 | .003 | 6.3% |
| Happiness | 2.61 (0.64) | 2.76 (0.65) | 2.66 (0.79) | 2.60 (0.49) | 2.76 (1.11) | .049 | .124 | .697 | 0.1% |
| n = 67 | n = 35 | n = 45 | n = 11 | n = 5 | |||||
|
|
|
||||||||
| Women | 0 | .5 | 1 | 1.5 | 2 | b | SE | p | % var |
| Hostility | 1.23 (0.22) | 1.27 (0.33) | 1.32 (0.36) | 1.24 (0.22) | 1.22 (0.21) | .070 | .047 | .135 | 1.4% |
| Anxiety | 1.44 (0.33) | 1.47 (0.39) | 1.61 (0.55) | 1.40 (0.24) | 1.74 (0.44) | .148 | .071 | .038 | 2.9% |
| Sadness | 1.33 (0.27) | 1.37 (0.43) | 1.40 (0.39) | 1.25 (0.29) | 1.50 (0.40) | .069 | .056 | .225 | 1.0% |
| Happiness | 2.74 (0.72) | 2.81 (0.63) | 2.74 (0.61) | 3.08 (0.65) | 2.39 (0.80) | .007 | .113 | .949 | 0.0% |
| n = 73 | n = 43 | n = 53 | n = 10 | n = 8 | |||||
Note. Means (standard deviations) computed on raw data by averaging the emotion reports over 120 (max) days of reporting and then averaging across participants with identical diplotypes. Emotion reports were made on a 1 - 5 scale. Ns for diplotype groups were reduced by three men and two women due to insufficient survey data. Significance tests and effect sizes used Hierarchical Linear Modeling. b = unstandardized coefficient testing a three-group linear model (0s vs. 0.5s vs. 1 – 2s, recoded 0,0.5,1). SE = standard error of the genotype coefficient; p = chance probability value (two-tailed). Effect Size, % var = the percentage of variance in the outcome accounted for by genotype.
Men with no higher expression chromosomes also reported significantly more anxiety and sadness (see Table IV); however effect sizes were smaller than those observed for hostility (5.8% and 6.3% of the variance, ds = .50 and .52, respectively). Furthermore, when controlling for hostility2, the diplotype associations were substantially attenuated for anxiety and sadness (Anxiety % var = 4.0, d = .42; b(SE) = −.149(.060), p = .015; Sadness % var = 2.0, d = .32; b(SE) = −.072(.039), p = .067). By contrast, the diplotype association with hostility was minimally attenuated when controlling for anxiety (% var 7.9, d = .61; b(SE) = −.184 (.056), p = .002), sadness (% var 7.6, d = .60; b(SE) = −.121(.040), p = .003), or both sadness and anxiety together (% var 7.1%, d = .57; b(SE) = −.106(.037), p = .005). These patterns suggest that diplotype was associated most strongly with hostility, which partially accounted for the associations with anxiety and sadness, although not completely. Diplotype associations were not observed in reports of happiness for men.
Women showed no significant differences in daily self-reported hostility, sadness or happiness as a function of diplotype (see Table VI). Women with higher expression chromosomes did have higher anxiety than their peers. This finding was unexpected and warrants future replication.
Additional Moderators
We conducted exploratory analyses to examine whether diplotype effects varied with age (See Supplementary Analyses). Indeed, diplotype differences in hostility were strongest among 18-year-old men (b(SE) = −.314(.077), p < .001), followed by 19-year-old men (b(SE) = −.272(.069), p < .001), 20-year-old men (b (SE) = −.230 (.069), p = .001) and 21-year-old men (b(SE) = −.188(.075), p = .014). This pattern suggests that HTR1B genetic associations with hostility are stronger in younger men.
Discussion
These findings extend our understanding of the genetic basis of anger and hostility and suggest that common functional haplotypes in the serotonin 1B receptor gene are risk factors in the experience of hostile emotional states. Male university students with two low-activity HTR1B chromosomes reported feeling significantly more anger and hostility in their daily lives than their peers with at least one high-activity HTR1B chromosome. Male students with one low and one intermediate HTR1B activity chromosome reported intermediate levels of hostility. This three-group diplotype configuration accounted for 8.4% of the variance in men”s reports of hostility (Cohen”s d = .62)—a medium effect size that is substantially higher than many other single-gene association studies of a behavioral phenotype, which more typically account for 2– 4% of variance in phenotype (e.g., Covault et al., 2007; Lesch et al., 1996).
Much of the variance accounted for by the diplotype was attributable to the microRNA binding site polymorphism (rs13212041), which explained 7.1% of the variance in men”s reports of hostility (d = .57). Indeed, the G-allele of rs13212041 appeared to drive the dominant protective effect of high-activity diplotypes on men”s hostility. This pattern suggests that the microRNA binding site polymorphism has greater behavioral effects than the promoter variant. Yet, this microRNA site polymorphism alone did not completely account for HTR1B associated phenotypic differences. The addition of haplotype information, namely the inclusion of the GA-promoter to distinguish intermediate diplotypes, explained an additional 1.3% of the variance. Consequently, we believe that the GA-promoter information is useful to include, particularly given the frequency of the intermediate haplotype (GAGAA, which represented 16% of haplotypes).
The results of our haplotype examination inform prior studies examining other HTR1B markers linked to the two functional alleles examined here. The rs6296 861C allele previously associated with 20% fewer HTR1B binding sites (Huang et al., 1999) was only observed in the haplotype containing a low activity promoter together with the microRNA repressed rs13212041 A-allele (TACAA). In a study of heroin addiction (Proudnikov et al., 2006), the protective HTR1B rs6297 allele(1180G) was observed only in association with the inactive microRNA binding site 1997G allele (TAGGG). Lastly, the –161T allele at rs130058 reported to be associated with higher scores on the Buss–Durkee Hostility Inventory and overrepresented among suicide completers (Zouk et al., 2007) was only carried on a haplotype containing the microRNA-sensitive rs13212041 A-allele (GTGAA). In each case, the association of other SNPs with increased psychopathology or decreased HTR1B protein expression could be explained by the presence of the microRNA active A-allele (leading to less HTR1B activity), while the protective effect of the 1180G allele could be explained by the presence of the microRNA inactive G-allele (leading to more HTR1B activity).
We also found an association between HTR1B diplotype and both anxiety and sadness. Although not predicted, this association is consistent with several lines of research. First, both anxiety and depression have been consistently linked to hostility (Kennedy, Morris, Pedley, & Schwab, 2001; Swan, Carmelli & Roseman, 1989; Suls & Bunde, 2005). Second, the pharmacological treatment literature reveals that anxiolytics and serotonergic antidepressants are effective not only in the treatment of anxiety and depression, respectively, but also in the treatment of aggressive behaviors (reviewed by Lavine, 1997). The psychological treatment literature also demonstrates that interventions directed toward anger reduction also reduce anxiety and depression (Deffenbacher, Dahlen, Lynch, Morris & Gowensmith, 2000). That the associations to anxiety and depression emerged among men is consistent with the notion that men are vulnerable to masked or hidden depression in the form of aggressive behavior (Fischer & Good, 1997; see also Hankin & Abramson, 2001) and that gender norms lead some depressed men to exhibit externalizing symptoms (but see Addis, 2008 for a critique).
The present findings have several implications. They highlight the importance of a comprehensive molecular genetic approach linking functional variants in candidate genes to complex phenotypes. In this way, our findings build on previous research by Duan et al. (2003) and Jensen et al. (2008) suggesting that there are multiple functional HTR1B polymorphisms and several common haplotypes in the population. Our results also demonstrate an important role for microRNA directed repression of HTR1B in the modulation of hostile emotions. The A-allele at rs13212041 appears to predispose individuals to a more hostile emotional style via microRNA-directed repression of the HTR1B mRNA, consistent with our previous study (Jensen et al., 2008). By contrast, the G-allele appears to predispose young men to a less hostile emotional style by the blocking microRNA-directed repression. Whereas two A-alleles are necessary for increased hostility risk, one G-allele is enough to reduce such risk. We also found that there is linkage between the GA promoter of Duan et al. and the microRNA polymorphism of Jensen et al. Interestingly, the −261G/A-161 promoter variant does not appear to act together with the microRNA G-allele to create a “super high expressing” chromosome. Instead, the GA promoter occurred only with the low expression microRNA A-allele (GAGAA). These findings suggest that when inherited together, the high-activity GA promoter and low-activity A-allele temper one another, resulting in an intermediate HTR1B behavioral phenotype. Additional research is necessary to validate these patterns and demonstrate their presence in other populations. Findings may also implicate these functional haplotypes in other health and psychological conditions linked to hostility. Hostility has been independently related to increased cardiovascular reactivity (e.g., Räikkönen et al., 1999), impaired immune function (e.g., Kiecolt-Glaser et al., 2005), substance dependence (e.g., Costanzo et al., 2007; De Moja & Spielberger, 1997) and suicide (e.g., Baud, 2005; McGirr & Turecki, 2007). Genetic research in these areas may profit from investigating the 3”UTR microRNA binding site variant (rs13212041) and associated HTR1B haplotypes.
It is interesting that in our four-year longitudinal study, the strongest genetic differences for males occurred at age 18, and decreased somewhat thereafter. This pattern suggests the existence of developmental effects; however, given the small age range in this sample (18 – 21 years), we could examine these age effects only in an exploratory fashion. Future research needs to examine age effects in a sample with a larger age range (e.g., 18 – 50+). If age-related effects were found, it might suggest that phenotypic manifestation of these genetic variants depend on developmental factors, i.e. prefrontal cortical function, hormone levels, etc. Future research combining genotyping with fMRI, hormone assays, and/or additional psychosocial questionnaires would help to disambiguate key moderators. Moreover, examination of the effects of HTR1B haplotype differences on hostility in individuals outside college-age individuals—for example, in adolescent boys or older adult men—could help determine the broader implications of these genetic variants. If differences were observed among adolescent boys, the HTR1B haplotypes could be considered a risk factor for teen aggression and its correlates (fights, arrests, drug and alcohol use, etc.). If genetic differences persisted among adult men, HTR1B haplotypes could be confirmed as a potential cardiovascular risk factor (e.g., Type 1 personality: Miller et al., 1996).
No genotype differences were observed among women. This finding could be interpreted biologically, for example, that only men with low-expression haplotypes are especially prone to aggression-related emotional states due to hormonal or other Y chromosome-linked neural circuitry differences. However, this interpretation is tempered by the fact that these same women showed significant an association of the rs13212041 SNP with self-reported earlier lifetime behaviors (Jensen et al., 2008). A-allele homozygous women at rs13212041 reported a history of more aggression-like behaviors (starting fires, damaging property, and fighting) than G-allele carrier women. Therefore, a more parsimonious explanation is that women with low HTR1B expressing chromosomes may manifest the aggressive phenotype, but they do so earlier in their development. Research has shown that genetic influences on aggressive behaviors are stronger earlier in development among female than male children (e.g., for antisocial behaviors: Jacobson et al., 2002). Indeed our own data indicate that the genotype difference in hostility among males was highest at age 18. If so, then a retrospective history checklist covering the period including childhood to late adolescence could reveal further genetic associations of these HTR1B haplotypes with hostile emotional states among women. Yet, other work has shown that genetic influences on aggression tend to increase for both boys and girls from childhood to young adulthood (Jacobson et al., 2002; Miles & Carey, 1997). Additional research is needed to replicate and determine the mechanism for this gender-specific effect.
There are several methodological strengths in the present study. First, design considerations reduced the likelihood of Type 1 error (moderate sample size; robust measurement of emotional outcomes; demonstrated consistency in findings longitudinally). Second, our choice of HTR1B as a candidate gene was based on studies in animals and humans that implicate this gene in aggressive behavior. Third, three of the five SNPs we examined have previously been shown to modulate two key steps in gene expression: transcription and mRNA translation (Duan et al., 2003; Jensen et al., 2008). Nonetheless, because of the risk for Type 1 error in genetic association studies, we strongly advocate efforts to replicate these findings.
There are also limitations to this study that should be considered. As a genetic association study, our study was correlational in design. Although Duan et al. (2003) and Jensen et al. (2008) established functional effects of three of the SNPs studied, these polymorphisms could also be in LD with other yet to be identified genetic variants that are functional. Our use of haplotype analysis partially addressed this issue by examining linkage among previously identified and commonly studied SNPs. Also, we assumed a static effect of these functional variants on HTR1B gene expression, yet it is possible that these polymorphic regulatory elements have a dynamic effect on the phenotype. For instance, the emotional effects of the HTR1B 3′UTR A/G polymorphism should depend on the presence and activity level of the complementary microRNA (miR-96), yet microRNAs are not continuously active—their amount and activity are regulated by a variety factors still being discovered (Davis et al., 2008, Newman et al., 2008; Viswanathan et al., 2008; Zhou et al., 2008). As a result, it is possible that miR-96 does not exert its effect in all individuals with the rs132120411041 A/A genotype because of other factors that regulate the production and activation of miR-96. Future studies may profit from identifying factors that control the expression and activity of miR-96 and ascertain their potential interaction with genotype to predict hostility. Similarly, the functional effects of the GA promoter haplotype may be conditional on the presence of Ap1-mediated transcription inducers and its effect may be most evident under specific developmental, environmental or life experience conditions. Because our analysis was limited to non-Hispanic Caucasians, it is important to examine these associations in more diverse samples.
Finally, future studies of the association of this SNP with psychological phenotypes might also include standard trait measures that target anger and hostility. However, we anticipate that the effect sizes may be attenuated with standard trait measures as compared with real-time data capture techniques. Although we have assessed emotional states repeatedly over four years, our aggregation of these repeated state assessments essentially created a trait indicator of anger/hostility. We believe that this aggregated indicator of emotional states may be more reliable and less vulnerable to recall error and bias than traditional, once-administered, questionnaire-based trait measures. At the same time, our two-item indicator lacked the breadth of standard trait measures.
Conclusion
Our study broadens the genetic bases of emotionality and suggests a link between common haplotypes in the serotonin 1B receptor gene and angry and hostile emotions in daily life among young men. Phenotypes were best explained by haplotypes that varied in the microRNA binding polymorphism (rs13212041) and, to a lesser extent, the promoter variants (rs11568817 and rs130058). These findings add to a growing literature showing that polymorphisms that influence gene expression related to the serotonin 1B receptor gene play an important role in phenotypic variation—particularly that which is part of a broad aggression-related cluster, which includes anger and hostile emotions. Additional research is needed on the correlates of these haplotypes and the mechanisms by which they exert effects on phenotype, particularly the brain-based processing differences and affective mechanisms that may underlie gene-based emotional differences, e.g., using neuroimaging (e.g., Canli et al., 2006; Hariri et al., 2002). Finally, the apparent gender-specific and potentially developmental manifestations of the association should be examined further and taken into account in subsequent studies of the SNPs examined in this study. Use of real-time data capture techniques may help to increase effect sizes in future genetic association studies.
Supplementary Material
Acknowledgements
This study was supported by NIH grants P50AA03510 (Alcohol Research Center), M01 RR06192 (University of Connecticut General Clinical Research Center), K24 AA13736 (to HRK), F31 DA023341 (to KPJ), and T32 AA07290 (NIAAA postdoctoral training grant for TSC). We gratefully acknowledge the assistance of Nicholas Maltby for web programming.
Footnotes
This effect was the same when excluding the two men with two intermediate activity haplotypes and retaining only the men with one high/one low activity diplotype (all with diplotype scores of 1). Interestingly, the two men with two intermediate activity haplotypes evidenced greater hostility (M(SD) = 1.52(.48) than the other men with one high/one low activity haplotype (M(SD) = 1.24(.27). This pattern is consistent with the observation that the Gallele in the 3′UTR binding site, present in those with one high/one low activity haplotype, exerts a dominant effect on ameliorating hostility.
Following guidelines by Raudenbush & Bryk (2002, pp. 33), hostility was entered as a grand-mean centered level-1 predictor. Similar to ANCOVA, this approach yields an adjusted intercept reflecting the expected value of anxiety (or sadness) for an individual at the grand mean of hostility.
References
- Addis ME. Gender and depression in men. Clin Psychol: Sci Pr. 2008;15:153–168. [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Armeli S, Conner TS, Covault J, Tennen H, Kranzler HR. The serotonin transporter gene polymorphism (5-HTTLPR), drinking motives and negative life events among college students. J Stud Alc Drugs. 2008;69(6):814–823. doi: 10.15288/jsad.2008.69.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Baud P. Personality traits as intermediary phenotypes in suicidal behavior: Genetic issues. Am J Med Genet C Semin Med Genet. 2005;133:34–42. doi: 10.1002/ajmg.c.30044. [DOI] [PubMed] [Google Scholar]
- Berkowitz L. Aggression: Its causes, consequences, and control. Temple University Press; Philadelphia PA; USA: 1983. [Google Scholar]
- Bouchard TJ., Jr Genetic influence on human psychological traits: A survey. Cur Dir Psychol Sci. 2004;13(4):148–151. [Google Scholar]
- Boyle SH, Jackson WG, Suarez EC. Hostility, anger, and depression predict increases in C3 over a 10-year period. Brain Behav Immun. 2007;21(6):816–23. doi: 10.1016/j.bbi.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss AH, Perry M. The aggression questionnaire. J Pers Soc Psychol. 1992;63:452–459. doi: 10.1037//0022-3514.63.3.452. [DOI] [PubMed] [Google Scholar]
- Canli T, Qiu M, Omura K, Congdon E, Haas BW, Amin Z, Herrmann MJ, Constable RT, Lesch KP. Neural correlates of epigenesis. Proc Natl Acad Sci. 2006;103:16033–16038. doi: 10.1073/pnas.0601674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cates DS, Houston BK, Vavak CR, Crawford MH. Heritability of hostility-related emotions, attitudes, and behaviors. J Behav Medicine. 1993;16(3):237–256. doi: 10.1007/BF00844758. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447(7146):823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Cigler T, LaForge KS, McHugh PF, Kapadia SU, Leal SM, Kreek MJ. Novel and previously reported single-nucleotide polymorphisms in the human 5-HT(1B) receptor gene: No association with cocaine or alcohol abuse or dependence. Am J Med Genet. 2001;105:489–497. doi: 10.1002/ajmg.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MS, Neumaier JF. The 5-HT1B receptor: Behavioral implications. Psychopharmacol Bull. 2001;4:170–185. [PubMed] [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. 2nd Edition Lawrence Erlbaum Associates; Hillsdale, N.J.: 1988. [Google Scholar]
- Costanzo PR, Malone PS, Belsky D, Kertesz S, Pletcher M, Sloan FA. Longitudinal differences in alcohol use in early adulthood. J Stud Alcohol Drugs. 2007;68(5):727–37. doi: 10.15288/jsad.2007.68.727. [DOI] [PubMed] [Google Scholar]
- Covault J, Tennen H, Armeli S, Conner TS, Herman A, Cillessen A, Kranzler H. Interactive effects of the serotonin transporter 5-HTTLPR polymorphism and stressful life events on college student drinking and drug use. Biol Psychiatry. 2007;61(5):609–616. doi: 10.1016/j.biopsych.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Davidge KM, Atkinson L, Douglas L, Lee V, Shapiro S, Kennedy JL, Beitchman JH. Association of the serotonin transporter and 5-HTlDβ receptor genes with extreme, persistent and pervasive aggressive behaviour in children. Psychiatr Genet. 2004;14:143–146. doi: 10.1097/00041444-200409000-00004. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moja CA, Spielberger CD. Anger and drug addiction. Psychol Rep. 1997;81(1):152–154. doi: 10.2466/pr0.1997.81.1.152. [DOI] [PubMed] [Google Scholar]
- Deffenbacher JL, Dahlen ER, Lynch RS, Morris CD, Gowensmith WN. An application of Beck”s cognitive therapy to general anger reduction. Cognitive Ther Res. 2000;24:689–697. [Google Scholar]
- Duan J, Sanders AR, Molen JE, Martinolich L, Mowry BJ, Levinson DF, Crowe RR, Silverman JM, Gejman PV. Polymorphisms in the 5′-untranslated region of the human serotonin receptor 1B (HTR1B) gene affect gene expression. Mol Psychiatry. 2003;8(11):901–910. doi: 10.1038/sj.mp.4001403. [DOI] [PubMed] [Google Scholar]
- Fischer AR, Good GE. Men and psychotherapy: An investigation of alexithymia, intimacy, and masculine gender roles. Psychother-Theor Res. 1997;34:160–170. [Google Scholar]
- Gacek P, Conner TS, Tennen H, Kranzler H, Covault J. Tryptophan hydroxylase 2 gene and alcohol use among college students. Addict Biol. 2008;13(3-4):440–448. doi: 10.1111/j.1369-1600.2008.00118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbacher EM, Matthews KA. Are psychological characteristics related to risk of the metabolic syndrome? A review of the literature. Ann Behav Med. 2007;34(3):240–252. doi: 10.1007/BF02874549. [DOI] [PubMed] [Google Scholar]
- Gouin JP, Kiecolt-Glaser JK, Malarkey WB, Glaser R. The influence of anger expression on wound healing. Brain Behav Immun. 2008;22(5):699–708. doi: 10.1016/j.bbi.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunthert K, Conner TS, Armeli S, Tennen H, Covault J, Kranzler H. The serotonin transporter gene polymorphism (5-HTTLPR) and anxiety reactivity in daily life: A daily process approach to gene-environment interaction. Psychosom Med. 2007;69(8):762–768. doi: 10.1097/PSY.0b013e318157ad42. [DOI] [PubMed] [Google Scholar]
- Hankin BL, Abramson LY. Development of gender differences in depression: An elaborated cognitive vulnerability-transactional stress theory. Psychol Bull. 2001;127(6):773–796. doi: 10.1037/0033-2909.127.6.773. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- Hawi Z, Dring M, Kirley A, Foley D, Kent L, Craddock N, Asherson P, Curran S, Gould A, Richards S, et al. Serotonergic system and attention deficit hyperactivity disorder (ADHD): A potential susceptibility locus at the 5-HT(1B) receptor gene in 273 nuclear families from a multi-centre sample. Mol Psychiatry. 2002;7(7):718–725. doi: 10.1038/sj.mp.4001048. [DOI] [PubMed] [Google Scholar]
- Huang YY, Grailhe R, Arango V, Hen R, Mann JJ. Relationship of psychopathology to the human serotonin 1B genotype and receptor binding kinetics in postmortem brain tissue. Neuropsychopharmacol. 1999;21:238–246. doi: 10.1016/S0893-133X(99)00030-5. [DOI] [PubMed] [Google Scholar]
- Huang YY, Oquendo MA, Friedman JM, Greenhill LL, Brodsky B, Malone KM, Khait V, Mann JJ. Substance abuse disorder and major depression are associated with the human 5-HT1B receptor gene (HTR1B) G861C polymorphism. Neuropsychopharmacol. 2003;28(1):163–169. doi: 10.1038/sj.npp.1300000. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Ickowicz A, Feng Y, Wigg K, Quist J, Pathare T, Roberts W, Malone M, Schachar R, Tannock R, Kennedy JL, Barr CL. The serotonin receptor HTR1B: gene polymorphisms in attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(1):121–125. doi: 10.1002/ajmg.b.30398. [DOI] [PubMed] [Google Scholar]
- Jacobson KC, Prescott CA, Kendler KS. Sex differences in the genetic and environmental influences on the development of antisocial behavior. Dev and Psychopathology. 2002;14(2):395–316. doi: 10.1017/s0954579402002110. [DOI] [PubMed] [Google Scholar]
- Jensen KP, Covault J, Conner TS, Tennen H, Kranzler HR, Furneaux HM. A common polymorphism in serotonin receptor 1B mRNA moderates regulation by miR-96 and associates with aggressive human behaviors. Mol Psychiatr. 2008 doi: 10.1038/mp.2008.15. E-pub ahead of print February 19, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BL, Morris RL, Pedley LL, Schwab JJ. The ability of the symptom checklist SCL-90 to differentiate various anxiety and depressive disorders. Psychiat Quart. 2001;72:277–288. doi: 10.1023/a:1010357216925. [DOI] [PubMed] [Google Scholar]
- Kiecolt-Glaser JK, Loving TJ, Stowell JR, Malarkey WB, Lemeshow S, Dickinson SL, Glaser R. Hostile marital interactions, proinflammatory cytokine production, and wound healing. Arch Gen Psychiatry. 2005;62(12):1377–1384. doi: 10.1001/archpsyc.62.12.1377. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129(6):1141–51. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Kranzler HR, Hernandez-Avila C, Gelernter J. Polymorphism of the 5-HT1B receptor gene (HTR1B) and antisocial substance dependence. Neuropsychopharmacol. 2002;26:115–122. doi: 10.1016/S0893-133X(01)00283-4. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Long JC, Eggert M, Ozaki N, Robin RW, Brown GL, Naukkarinen H, Virkkunen M, Linnoila M, Goldman D. Linkage of antisocial alcoholism to the serotonin 5-HT1B receptor gene in 2 populations. Arch Gen Psychiatry. 1998;55(11):989–994. doi: 10.1001/archpsyc.55.11.989. [DOI] [PubMed] [Google Scholar]
- Lavine R. Psychopharmacological treatment of aggression and violence in the substance using population. J Psychoactive Drugs. 1997;29:321–329. [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller-Reible CR, Hamer DH, Murphy DL. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- Manuck SB, Kaplan JR, Clarkson TB. Behaviorally induced heart rate reactivity and atherosclerosis in cynomolgus monkeys. Psychosom Med. 1983;45(2):95–108. doi: 10.1097/00006842-198305000-00002. [DOI] [PubMed] [Google Scholar]
- Martin R, Watson D, Wan CK. A three-factor model of trait anger: Dimensions of affect, behavior, and cognition. J Pers. 2000;68:869–897. doi: 10.1111/1467-6494.00119. [DOI] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF, Sonenberg N. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317(5845):1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- McGirr A, Turecki G. The relationship of impulsive aggressiveness to suicidality and other depression-linked behaviors. Curr Psychiatry Rep. 2007;9(6):460–466. doi: 10.1007/s11920-007-0062-2. [DOI] [PubMed] [Google Scholar]
- McGrath RE, Meyer GJ. When effect sizes disagree. The case of r and d. Psychol Methods. 2006;11(4):386–301. doi: 10.1037/1082-989X.11.4.386. [DOI] [PubMed] [Google Scholar]
- Miczek KA, de Almeida RM. Oral drug self-administration in the home cage of mice: alcohol-heightened aggression and inhibition by the 5-HT1B agonist anpirtoline. Psychopharmacology (Berl) 2001;157(4):421–429. doi: 10.1007/s002130100831. [DOI] [PubMed] [Google Scholar]
- Miles DR, Carey G. Genetic and environmental architecture of human aggression. J Pers Soc Psychol. 1997;72(1):207–217. doi: 10.1037//0022-3514.72.1.207. [DOI] [PubMed] [Google Scholar]
- Miller TQ, Smith TW, Turner CW, Guijarro ML, Hallet AJ. A meta-analytic review of research on hostility and physical health. Psychol Bull. 1996;119:322–348. doi: 10.1037/0033-2909.119.2.322. [DOI] [PubMed] [Google Scholar]
- New AS, Gelernter J, Goodman M, Mitropoulou V, Koenigsberg H, Silverman J, Siever LJ. Suicide, impulsive aggression, and HTR1B genotype. Biol Psychiatry. 2001;50(1):62–65. doi: 10.1016/s0006-3223(01)01108-8. [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. RNA Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. 2008. June 19 E-pub ahead of print. [DOI] [PMC free article] [PubMed]
- Nishiguchi N, Shirakawa O, Ono H, Nishimura A, Nushida H, Ueno Y, Maeda K. No evidence of an association between 5HT1B receptor gene polymorphism and suicide victims in a Japanese population. Am J Med Genet. 2001;105(4):343–345. doi: 10.1002/ajmg.1347. [DOI] [PubMed] [Google Scholar]
- Norlander B, Eckhardt C. Anger, hostility, and male perpetrators of intimate partner violence: a meta-analytic review. Clin Psychol Rev. 2005;25(2):119–152. doi: 10.1016/j.cpr.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Oliver B, van Oorschot R. 5-HT1B receptors and aggression: A review. Eur J Pharmacol. 2005;526:207–217. doi: 10.1016/j.ejphar.2005.09.066. [DOI] [PubMed] [Google Scholar]
- Popova NK. From genes to aggressive behavior: The role of serotonergic system. BioEssays. 2006;28:495–503. doi: 10.1002/bies.20412. [DOI] [PubMed] [Google Scholar]
- Proudnikov D, LaForge KS, Hofflich H, Levenstien M, Gordon D, Barral S, Ott J, Kreek MJ. Association analysis of polymorphisms in serotonin 1B receptor (HTR1B) gene with heroin addiction: a comparison of molecular and statistically estimated haplotypes. Pharmacogenet Genomics. 2006;16(1):25–36. doi: 10.1097/01.fpc.0000182782.87932.d6. [DOI] [PubMed] [Google Scholar]
- Quist JF, Barr CL, Schachar R, Roberts W, Malone M, Tannock R, Basile VS, Beitchman J, Kennedy JL. The serotonin 5-HT1B receptor gene and attention deficit hyperactivity disorder. Mol Psychiatry. 2003;8(1):98–102. doi: 10.1038/sj.mp.4001244. [DOI] [PubMed] [Google Scholar]
- Räikkönen K, Matthews KA, Flory JD, Owens JF. Effects of hostility on ambulatory blood pressure and mood during daily living in healthy adults. Health Psychol. 1999;18(1):44–53. doi: 10.1037//0278-6133.18.1.44. [DOI] [PubMed] [Google Scholar]
- Raudenbush S, Bryk A, Congdon R. Hierarchical linear and nonlinear modeling software (HLM 6.04) SSI Scientific Software; Lincolnwood, IL: 2007. [Google Scholar]
- Raudenbush S, Bryk A. Hierarchical linear models: Applications and data analysis methods. 2nd ed Sage; Thousand Oaks: 2002. [Google Scholar]
- Rujescu D, Giegling I, Sato T, Möller HJ. Lack of association between serotonin 5-HT1B receptor gene polymorphism and suicidal behavior. Am J Med Genet B Neuropsychiatr Genet. 2003;116B(1):69–71. doi: 10.1002/ajmg.b.10732. [DOI] [PubMed] [Google Scholar]
- Sanders AR, Cao Q, Taylor J, Levin TE, Badner JA, Cravchik A, Comeron JM, Naruya S, Del Rosario A, Salvi DA, Walczyk KA, Mowry BJ, Levinson DF, Crowe RR, Silverman JM, Gejman PV. Genetic diversity of the human serotonin receptor 1B (HTR1B) gene. Genomics. 2001;72(1):1–14. doi: 10.1006/geno.2000.6411. [DOI] [PubMed] [Google Scholar]
- Saudou F, Amara DA, Dierich A, LeMeur M, Ramboz S, Segu L, Buhot MC, Hen R. Enhanced aggressive behavior in mice lacking 5-HT1B receptor. Science. 1994;265:1875–1878. doi: 10.1126/science.8091214. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Smoller JW, Biederman J, Arbeitman L, Doyle AE, Fagerness J, Perlis RH, Sklar P, Faraone SV. Association between the 5HT1B receptor gene (HTR1B) and the inattentive subtype of ADHD. Biol Psychiatry. 2006;59(5):460–467. doi: 10.1016/j.biopsych.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone A, Shiffman S, Atienza A, Nebeling L. The science of real-time data capture: Self-reports in health research. Oxford University Press; Oxford (UK): 2007. [Google Scholar]
- Suls J, Bunde J. Anger, anxiety, and depression as risk factors for cardiovascular disease: The problems and implications of overlapping affective dispositions. Psychol Bull. 2005;131(2):260–300. doi: 10.1037/0033-2909.131.2.260. [DOI] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Rosenman RH. Psychological correlates of two measures of coronary-prone hostility. Psychosomatics. 1989;30:270–278. doi: 10.1016/S0033-3182(89)72271-4. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21(15):1857–62. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, Damschroder-Williams P, Du J, Chen G, Manji HK. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacol. 2008 doi: 10.1038/npp.2008.131. ePub 13 August 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouk H, McGirr A, Lebel V, Benkelfat C, Rouleau G, Turecki G. The effect of genetic variation of the serotonin 1B receptor gene on impulsive aggressive behavior and suicide. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(8):996–1002. doi: 10.1002/ajmg.b.30521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


