Abstract
Rac GTPases, small G-proteins widely implicated in tumorigenesis and metastasis, transduce signals from tyrosine-kinase, G-protein-coupled receptors (GPCRs), and integrins, and control a number of essential cellular functions including motility, adhesion, and proliferation. Deregulation of Rac signaling in cancer is generally a consequence of enhanced upstream inputs from tyrosine-kinase receptors, PI3K or Guanine nucleotide Exchange Factors (GEFs), or reduced Rac inactivation by GTPase Activating Proteins (GAPs). In breast cancer cells Rac1 is a downstream effector of ErbB receptors and mediates migratory responses by ErbB1/EGFR ligands such as EGF or TGFα and ErbB3 ligands such as heregulins. Recent advances in the field led to the identification of the Rac-GEF P-Rex1 as an essential mediator of Rac1 responses in breast cancer cells. P-Rex1 is activated by the PI3K product PIP3 and Gβγ subunits, and integrates signals from ErbB receptors and GPCRs. Most notably, P-Rex1 is highly overexpressed in human luminal breast tumors, particularly those expressing ErbB2 and estrogen receptor (ER). The P-Rex1/Rac signaling pathway may represent an attractive target for breast cancer therapy.
1. Rac GTPases: isoforms and genes
The Rho/Rac GTPases are a family of small G-proteins widely implicated in normal physiology and disease. They play an important role in cytoskeleton rearrangements and are key regulators of cellular adhesion, migration, proliferation, survival, differentiation and malignant transformation [1]. Members of this family in humans are divided into 6 classes: Rho (RhoA, RhoB and RhoC), Rac (Rac1, Rac2, Rac3 and RhoG), Cdc42 (Cdc42, Tc10, TCL, Chp/Wrch-2 and Wrch-1), RhoBTB, Rnd and RhoT [2]. The most studied members are RhoA, Rac1 and Cdc42. Like most GTPases, Rho, Rac and Cdc42 function as molecular switches that cycle between an inactive state that binds GDP and an active state that binds GTP. GTP is hydrolyzed to GDP through their intrinsic GTPase activity to render the G-protein inactive [3]. The switch between GDP and GTP is primarily regulated by two types of proteins: GEFs (Guanine Nucleotide Exchange Factors) that facilitate GTP loading and thereby activate the small G-protein, and GAPs (GTPase Activating Proteins) that stimulate the hydrolysis of GTP by enhancing intrinsic GTPase activity, thus leading to G-protein inactivation. A third class of proteins known as Rho GDIs (GDP Dissociation Inhibitors) sequesters the inactive GTPases in the cytosol, preventing their translocation and subsequent activation. Dissociation from Rho GDI becomes essential for proper activation of the G proteins [4–6].
Rac isoforms have a very high degree of homology. The greatest divergence is in the C-terminal end, which is also the hypervariable region in Ras [7]. This domain is important for driving subcellular localization and binding to specific cellular regulators [8]. Rac2 shares significant nucleotide sequence identity (~88%) with the other Rac isoforms. At the nucleotide level Rac3 has 77% identity with Rac1, 83% identity with Rac2 and 69% identity with RhoG. At the amino acid level, Rac3 has 92% identity with Rac1 and 89% identity with Rac2.
The Rac1 gene localizes to chromosome 7 (7p22) and comprises 7 exons over a length of 29 kB [9]. Rac1, but not Rac2 or Rac3 genes, contains an additional exon 3b that is included by alternative splicing in the variant Rac1b, a constitutively active mutant that is expressed mainly in colon and breast cancer [10, 11]. Rac1 is ubiquitously expressed, and it is involved in signal transduction pathways that control proliferation, adhesion, and migration. Its inactivation by gene targeting in mice leads to embryonic lethality caused by both gastrulation defects and apoptosis of mesodermal cells [12].
The Rac2 gene contains 7 exons in chromosome 22 (22q13.1) [13] and its expression is silenced in non-hematopoietic cells by DNA methylation [14]. Rac2-deficient mice show defects in neutrophil, macrophage, mast cell, lymphocyte B and lymphocyte T function [15, 16]. Rac2 plays an important role in integrin-mediated hematopoietic stem-cell adhesion [17]. Patients with impaired Rac2 function display major alterations in hematopoiesis and an immunodeficiency syndrome [18, 19].
The Rac3 gene encompasses 6 exons in chromosome 17 (17q25.3). Rac3 is primarily expressed in brain, although its expression has been reported in some human cell lines including GM04155 (lymphoblastic leukemia), K562 (chronic myelogenous leukemia), 5838 (Ewing sarcoma), HL60 (promyelocytic leukemia) and DU4475 (breast cancer) [8]. Rac3−/− mice display slight motor coordination problems and hyperactive behaviour [20].
Rac proteins associate with membranes in order to carry out their biological functions. However, unlike other Ras superfamily proteins, this anchoring step is not achieved during biosynthesis but rather requires a combination of intrinsic and cooperative signaling. The first and most crucial signal is the post-translational modification of the “CAAX box” by incorporation of a geranyl-geranyl group or less frequently a farnesyl group. In cooperation with the CAAX box a closely located proline-rich domain contributes to the association of Rac with specific proteins in focal adhesion complexes [21, 22].
2. Regulation of Rac activity by GEFs and GAPs
As mentioned above, Rac cycles between inactive and active states, two conformations that depend on the binding of GDP and GTP, respectively. Guanine nucleotides have picomolar affinities for Rac, and as a consequence their dissociation rate from the G-protein is slow. In order to lead to fast responses such as actin cytoskeleton reorganization, GEFs accelerate GDP/GTP exchange by several orders of magnitude [23]. GEFs catalyze the dissociation of the nucleotide from the G-protein by modifying the nucleotide-binding site. Rac has similar affinities for both nucleotides, and GEF binding does not favour binding of GTP over GDP. The resulting increase in GTP-bound over GDP-bound in Rac is rather due to the higher cellular concentrations of GTP relative to GDP. The mechanism by which GEFs weaken the binding of the nucleotide has been investigated in detail. According to the currently accepted model, the bound nucleotide in Rac is sandwiched between two loops called switches 1 and 2. These regions together with the phosphate-binding loop interact with the phosphates and a coordinating magnesium ion [23, 24]. As the catalytic domains of GEFs display in many cases significant structural differences, it is possible to design drugs capable of interfering with their binding to Rac in a GEF-specific manner. One example is NSC23766, a compound that binds to a surface cleft between the switch 1 and switch 2 regions of Rac and prevents the binding of GEFs Tiam1 and Trio [25]. GEFs can be promiscuous in terms of small G-protein activation (for example Vav2 activates Rho, Cdc42 and Rac) or display selectivity, such as Tiam1 or P-Rex1 for Rac [26, 27].
Rac can be activated by a variety of stimuli, including growth factors (such as EGF, PDGF, and HGF) and G-protein-coupled receptor ligands (such as SDF-1α, sphingosine-1-phosphate, and bombesin). Tyrosine-kinase receptors may convey signals to Rac-GEFs via intermediate molecules, as described for P-Rex1 (Figure 1). As a consequence Rac-GEFs may change their subcellular localization, undergo conformational changes that disrupt autoinhibitory mechanisms, and/or go through allosteric changes in the catalytic domain. In several cases the activation of GEFs is mediated by phosphatidylinositol (3,4,5)-trisphosphate (PIP3), the product of the class I phosphoinositide 3-kinases (PI3K), which binds to the PH domain present in the Rac-GEFs [28]. PI3K-independent activation of GEFs may involve their direct binding to the tyrosine-kinase receptor, as described for Vav2 with EGFR [29]. GEFs can be also tightly regulated by tyrosine phosphorylation [30].
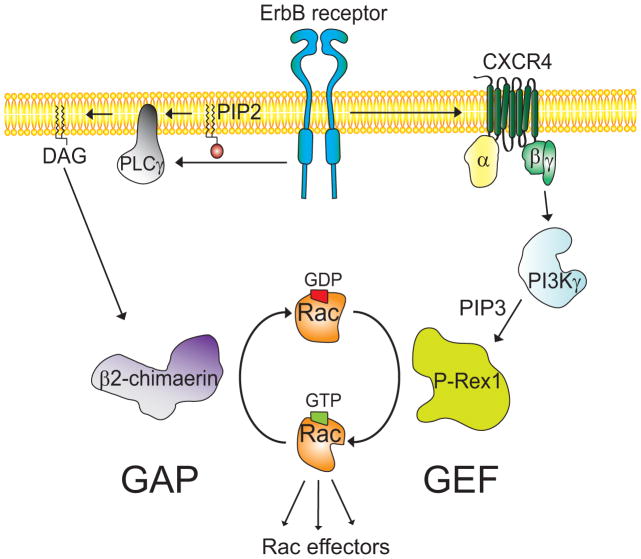
Figure 1. A model for the regulation of Rac activity by GEFs and GAPs in breast cancer.
ErbB receptors activate the PI3K-Gβγ-dependent P-Rex1 through transactivation of the GPCR CXCR4. Chimaerin Rac-GAPs are also regulated by ErbB receptors and act as a brake for the Rac activation.
While the mechanisms by which Rac-GEFs promote Rac activation have been extensively studied, much less is known about the basis for Rac inactivation. Rac-GAPs accelerate the intrinsic GTPase activity of small G-proteins by several orders of magnitude. Biochemical and structural analyses revealed that GAPs stabilize important residues of the intrinsically mobile catalytic machinery of the G-protein. Some Rac-GAPs are stringently regulated by receptor stimulation. For example, the chimaerin Rac-GAPs are activated by the lipid second messenger diacylglycerol (DAG) generated in response to growth factor receptor stimulation (Figure 1). DAG binds to the C1 domain present in chimaerins to promote their redistribution to the plasma membrane, where they bind to active GTP-bound Rac and accelerate the hydrolysis of the nucleotide. This receptor-regulated step represents a mechanism that limits Rac activation in response to stimuli [31–33]. β2-chimaerin is also regulated by protein kinase C (PKC) phosphorylation and through protein-protein interactions. Such mechanisms may be key for modulating subcellular localization and association to membranes [34–37].
3. Rac function and effectors
A main role for Rac is the regulation of cytoskeleton reorganization, as it promotes actin assembly required for the formation of lamellipodia and membrane ruffles [38]. Regulation of cytoskeleton dynamics is essential for the maintenance of cellular morphology, polarity, adhesion and migration [2]. The control of cytoskeleton reorganization via Rac involves at least two different mechanisms (Figure 2). One is through the activation of Arp2/3, which has a prominent role in actin polymerization through WAVE/Scar indirect activation. WAVE can be activated either by disassembly of the Rac-Nap1-PIR121 complex or through IRSp53 activation [39]. A second mechanism for Rac-mediated cytoskeleton reorganization is through Pak (p21-activated kinase), a family that comprises 6 isoforms (Pak1–6). Paks have an N-terminal GTPase binding domain (GBD) where Rac-GTP or Cdc42-GTP binds, and a C-terminal serine/threonine kinase domain. Binding of active Cdc42 or Rac to the GBD domain of group I Paks (Pak 1, 2 and 3) releases autoinhibition and enhances kinase activity. Some reports suggest that Cdc42 does not enhance the kinase activity of group II Paks (Pak 4, 5 and 6), suggesting significant differences in the regulation of these kinases [40, 41]. Overexpression and/or hyperactivation of Pak isoforms have been detected in several cancer types, such as in breast cancer [41]. Pak1 is implicated in the regulation of cytoskeleton dynamics and cell motility, transcription, survival, and cell-cycle progression [42]. Pak1 exerts its effects on cytoskeleton organization through phosphorylation of various proteins, including LIMK (LIM kinase), MLCK (myosin light chain kinase), cortactin, and the Arpc1b subunit of the Arp2/3 complex [43–45].
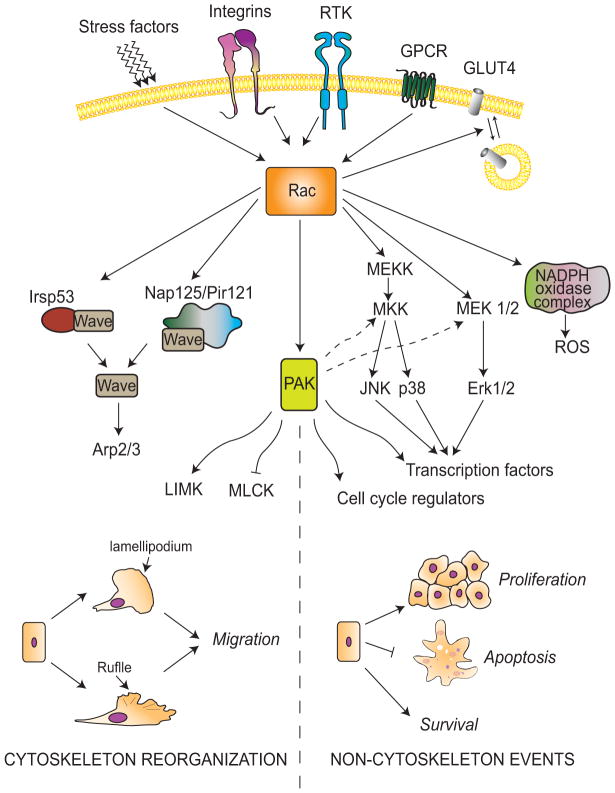
Figure 2. The Rac signaling pathway.
Rac is activated by tyrosine-kinase receptors (RTKs), GPCRs, integrins and stress. Rac activation promotes changes related to cytoskeleton reorganization (left) and other responses (right) through multiple mediators. Rac-GTP induces the activation of WAVE through either Irsp53 or Nap125-PIR121 complexes. WAVE activates Arp2/3, leading to changes in actin cytoskeleton necessary for ruffle formation and cell migration. Rac also activates PAK, which exerts its effects through Arp2/3 and LIMK activation or MLCK inhibition. In addition, Rac forms part of the NADPH oxidase complex that generates reactive oxygen species. Rac/PAK activates MAPKs implicated in stress response, mitogenesis, and survival.
In addition to its effects on actin cytoskeleton reorganization, Rac controls a number of additional functions, including cell cycle regulation, endocytosis and phagocytosis [46, 47]. Rac is also involved in integrin-mediated adhesion [48]. In addition, Rac forms part of the NADPH oxidase complex that generates reactive oxygen species and is involved in glucose uptake and insulin-dependent GLUT4 translocation [49–51]. Numerous studies established functional links between Rac and the regulation of MAPKs. Rac1 regulates transcription via JNK/c-Jun [52, 53], is involved in p38 activation by stress stimuli such as ionomycin, UV, hyperosmolarity, and LPS [54–57], activates the MKK3/6-p38MAPK pathway, and mediates invasion by H-Ras in mammary epithelial cells [58]. Rac1 cooperates with Raf-1 to activate the Erk pathway [59]. Although expression of active Rac does not activate Erk2, co-expression of Raf-1 and Rac1 synergizes for the activation of MEK1 and Erk [60]. Rac downstream effectors can also modulate the magnitude of the Rac response through feedback mechanisms [61–63].
4. Rac and cancer
Malignant cancer cells display abnormal migratory properties and have the ability to invade and metastasize. Since Rac modulates actin cytoskeleton reorganization and cell motility, impairment of Rac function by expressing a dominant-negative Rac mutants or Rac-GAPs markedly affects migration and invasiveness of cancer cells [32]. Of great interest, a recent study by Marshall and co-workers in melanoma models found that the Rac-specific GEF DOCK3 and the Rac effector WAVE2 are essential for promoting actin nucleation and an “elongated” cellular phenotype characteristic of mesenchymal behavior [64]. It is expected that the nature of the stimuli (i.e., integrins, oncogenes) in distinct cancer cell types leads to a differential utilization of Rac-GEFs to promote the motile and invasive phenotype.
Although functionally relevant Rac1 mutations have not been identified in tumors, a very recent study reported a “Rac1 risk allele” in patients with ulcerative colitis, who have a higher risk of developing colon cancer [65, 66]. Single nucleotide polymorphism in the Rac1 gene possibly augments Rac1 protein stability, which may enhance neutrophil migration and recruitment to the colon. Patients carrying the Rac1 risk allele display higher susceptibility to the development of colonic inflammatory bowel disease than those who do not carry that allele. Interestingly, neutrophil/macrophage-null Rac1 mice are less susceptible to dextrane sulphate sodium-induced colitis and have reduced neutrophil colonic infiltration and production of pro-inflammatory cytokines. One speculation is that enhanced Rac signaling in cancer cells or the surrounding stroma may facilitate neutrophil recruitment and inflammation. This study underscored a novel paradigm that may link inflammation and cancer development via Rac dysfunction.
Rac1 overexpression in tumors may play a role in cancer progression. A recent example has been described in pancreatic cancer [66, 67], in which a crucial role for Rac1 for preneoplastic lesion development has been established. Rac1 was found to be essential for early neoplasia-associated actin rearrangements, and ablation of the Rac1 gene in a K-ras murine model impairs the formation of pancreatic intraperitoneal neoplasia. Rac2 has been also linked to cancer, and both overexpression and decreased expression have been found in human brain tumors, head and neck squamous cell cancer, and leukemias. Somatic mutations in the Rac2 gene have been reported in human brain tumors [68]. Rac3 deregulation has been also linked to human cancer, including ovarian, breast, gastric, and brain cancer [69–73].
Despite reported changes in the expression of Rac isoforms in cancer, it is clear that the most common cause of Rac hyperactivation is the deregulation of upstream mechanisms, including excessive input from receptors or intermediate molecules such as PI3K. Ultimately, such signals will impact on the activation status of Rac-GEFs. Indeed, several Rac-GEFs, such as Tiam1, Ect2 and P-Rex1, play important roles in tumorigenesis and metastasis and are up-regulated or hyperactive in cancer [74–78], whereas on the other hand, there is less evidence for deregulation in the expression and/or function of Rac-GAPs. One example of a Rac-GEF involved in cancer is Tiam1. Tiam1 knock-out mice are resistant to the development of skin tumors in a model of DMBA-phorbol ester chemical carcinogenesis. Paradoxically, although the number of tumors in Tiam1−/− mice is smaller, they tend to progress to malignancy. Tiam1 deficiency is associated with an elevated apoptotic response during initiation and reduced proliferation during the promotion stage. In addition, Tiam1 mediates Ras-induced transformation in fibroblasts. Indeed, Tiam1 associates with activated Ras, and these two oncogenes cooperate to activate Rac. There is also substantial evidence that Tiam1 is implicated in invasiveness and metastasis [79, 80].
Another interesting Rac-GEF involved in cancer progression is Ect2, an oncogene that is aberrantly expressed in numerous cancer types, often as a consequence of gene amplification [75, 81–84]. Fields and coworkers established that Ect2 is important for cell proliferation, migration, invasion and tumorigenicity of human lung cancer cells, and that Rac1 is a critical effector of Ect2. The ability of Ect2 to support transformation in non-small cell lung cancer (NSCLC) is linked to the activation of a Rac1-Pak1-Erk signaling pathway that is regulated through an oncogenic PKCι-Par6-Ect2 complex. PKCι phosphorylation of Ect2 regulates the oncogenic activity of Ect2 in NSCLC cells [85, 86].
5. Rac and breast cancer
Rac1 was reported to be overexpressed or hyperactive in breast cancer tissues. Moreover, the active variant Rac1b is also expressed in human breast tumors [11]. Accumulating evidence indicates that the Rac effector Pak1 is implicated in breast cancer progression. Indeed, more than 50% of human breast tumors show overexpression and/or hyperactivation of Pak1 [87]. Early studies by Kumar and coworkers showed that expression of a kinase-dead Pak1 mutant reduced invasiveness in MDA-MB-231 breast cancer cells, and conversely, a constitutively active Pak1 mutant promotes MCF-7 cell migration, invasiveness and anchorage-independent growth [88]. Moreover, transgenic expression of an active Pak1 allele in the mouse mammary epithelium leads to the development of mammary tumors [89]. More recently, Chernoff and coworkers found a correlation between ErbB2 expression and Pak activation in estrogen receptor (ER)-positive human breast tumor samples. Activation of the Rac-Pak1 pathway by ErbB2 leads to a proliferative response and Erk activation [90], consistent with previous studies showing a link between Rac1/Pak1 and cyclin D1 induction [87, 91]. Overexpression of ErbB2 in MCF-10A human mammary epithelial cells and T-47D breast cancer cells elevates Rac1 activity [77, 92]. Actin and actinin are recruited to ErbB2 in response to TGF-β, which then co-localizes with the GEF Vav2 to activate Rac1 and Pak1 at cell protrusions [93]. As ErbB2 is overexpressed in a significant fraction of human breast cancers, it is conceivable that interfering with the Rac1/Pak1 pathway should have significantly impact on breast cancer progression. Proof-of-principle has been obtained by means of a specific Pak inhibitor, which delays tumor formation and impairs Erk activation in ErbB2 positive breast cancer cells [90]. In addition, Pak1 phosphorylates Ser305 in ERα, and the high Pak1 expression levels and its nuclear localization correlate with tamoxifen resistance in ERα-positive breast cancer [94–96], arguing for a potential benefit of Pak1 inhibitors in reversing antiestrogen resistance.
Studies in the last years revealed an important role for Rac1 in growth factor signaling in breast cancer. Our laboratory reported that the growth factor heregulinβ1 (HRG) causes a strong and sustained activation of Rac1 in breast cancer cells. While EGF also activates Rac1, this effect is rapid and short lived. Rac1 activation by HRG is mediated by ErbB3 and ErbB2 but is independent of ErbB4, and it involves the transactivation of EGFR. Ruffle formation and motility induced by HRG are sensitive to EGFR and PI3K inhibition [97]. Cyclin D1 induction by HRG in breast cancer cells is also dependent on Rac1 and NF-κB [98].
Emerging evidence supports a role for Rac-GEFs in the development and progression of breast cancer (Table 1). Early studies established a correlation between Tiam1 expression and high tumor grade in human breast carcinomas [99]. Moreover, highly migratory and metastatic breast cancer cell lines such as MDA-MB-231 cells express high Tiam1 levels, and the opposite is true for less invasive models such as MCF-7 or SK-BR-3 cells [100]. Tiam1 binds to the cytoskeletal protein ankyrin to stimulate breast tumor cell migration and invasion [101]. Recent studies showing an inverse correlation between Tiam1 and the progression of breast carcinomas [102] and a lack of correlation between Tiam1 expression and Rac activation status in breast cancer cells [103] strongly argue for the need to reassess the involvement of Tiam1 in breast cancer. Moreover, stromal Tiam1 may have a role in modulating malignant cell invasion and metastasis, as suppression of Tiam1 expression in stromal fibroblasts significantly increases invasiveness and metastatic potential of breast cancer cells [104].
Table 1.
Rac1 modulators in breast cancer. Rac-GEFs and Rac-GAPs have been implicated in the human breast cancer.
| GEF | Involvement in breast cancer | References |
|---|---|---|
| Dock4 | Mediates MDA-MB-231 motility and invasion | [150] |
|
| ||
| P-Rex1 | Overexpressed in luminal breast cancer and metastasis | [77, 78] |
| Mediates motility and tumorigenesis by ErbB receptors | [77, 78] | |
|
| ||
| P-Rex2a | Correlates with PTEN expression in tumors with PI3K pathway alterations | [110] |
| Associates with activating mutations in PI3K | [110] | |
| Physically interacts with PTEN and antagonize its tumor suppressor function | [110] | |
|
| ||
| Tiam1 | Correlates with high grade tumors | [101] |
| Mediates migration and invasion | [99] | |
| Mediates c-neu induced mammary tumor formation in mice | [104] | |
| Stromal Tiam1 modulates malignant cell invasion and metastasis | [151] | |
|
| ||
| Trio | Overexpressed in human breast tumors | [107] |
| Marker of poor prognosis | [107] | |
|
| ||
| Vav3 | Overexpressed in human breast tumors | [105] |
| Involved in ERα activation | [106] | |
|
| ||
| GAP | Involvement in breast cancer | References |
|
| ||
| ARHGAP10 | SNP associated with breast cancer survival | [152] |
|
| ||
| β2-chimerin | Down-regulated in breast cancer | [91] |
| Inhibits human breast cancer cell proliferation and motility | [91] | |
| Reduces tumor formation and metastasis in mice | [109] | |
|
| ||
| Other modulators | Involvement in breast cancer | References |
|
| ||
| BCAR3/AND-34 | Promotes antiestrogen resistance | [108] |
| Involved in cyclin D1 induction | [108] | |
| Affects cancer cell morphology | [108] | |
The proto-oncogene Vav3, a Rac/Rho GEF, is overexpressed in 81% of human breast tumors. In breast cancer cells, Vav3 transcriptionally activates ERα partially through PI3K/Akt [105, 106]. Trio, a Rac-GEF implicated in cell motility, is also overexpressed in human breast cancer, predominantly in tumors from patients with poor prognosis [107]. Overexpression of the Rac-activating protein AND-34/BCAR3 in breast cancer cells promotes antiestrogen resistance via cyclin D1 induction [108]. Enhanced Rac signaling may relate not only to Rac-GEF hyperactivation but also to deregulation of Rac-GAPs. For example, β2-chimaerin mRNA levels are significantly down-regulated in human breast cancer cell lines and tumors. Ectopic expression of β2-chimaerin in MCF-7 breast cancer cells reduces cyclin D1 levels, impairs G1/S cell cycle progression and migration, and reduces the tumorigenic potential of breast cancer cells [91, 109] (Table 1).
6. P-Rex Rac-GEFs: novel players in breast cancer
Recent studies identified the Rac-GEFs P-Rex1 and P-Rex2 as important players in cancer, particularly in breast and prostate cancer [76–78]. P-Rex1 mediates Rac activation and cell motility in breast cancer cells in response to stimulation of tyrosine-kinase receptors and GPCRs. Moreover, P-Rex1 is highly expressed in human breast tumors relative to normal mammary tissue, arguing for a potential role for this Rac-GEF in breast cancer progression [77]. In addition, P-Rex2 is a crucial regulator of the PTEN phosphatase and possibly exerts its effects in mammary cancer cells at least in part through a Rac-independent mechanism [110]. Before expanding into these novel paradigms in breast cancer, we will describe general aspects of P-Rex regulation and function.
6.a. P-Rex is dually regulated by PI3K and Gβγ subunits
A distinctive characteristic of P-Rex Rac-GEFs is that they are synergistically activated by the PI3K product PIP3 and βγ subunits of heterotrimeric G-proteins [27, 111–114]. In vitro assays have shown that PI3Kγ (a Gβγ regulated isoform of PI3K) specifically activates P-Rex1 [27, 111, 112]. This class IB PI3K seems to serve as a point of integration for upstream signals that converge on P-Rex1 [115]. Similar mechanisms appear to be necessary for the activation of P-Rex2 [112].
P-Rex1 is selectively regulated by a specific combination of Gβ and Gγ subunits but cannot be directly activated by Gα subunits. Studies using purified recombinant proteins reconstituted into synthetic lipid vesicles demonstrated that P-Rex1 is preferentially activated by β1–4 in complex with γ2. No activation can be observed by Gβ5γ2. Dimers containing the β1 subunit in complex with a panel of different Gγ subunits have varied ability to activate P-Rex1 [113]. Thus, the composition of the Gβγ dimer released upon receptor activation may be a determinant for the activation of Rac via P-Rex1.
P-Rex1 contains 28 putative PKA phosphorylation sites. It was reported that PKA inhibits the activation of P-Rex1 by PIP3 and Gβγ subunits, and treatment with λ-phosphatase enhances P-Rex1 Rac-GEF activity. Phosphorylation of P-Rex1 by stimulation of β-adrenergic receptors, which leads to activation of PKA via cAMP, diminishes its ability to activate Rac1 in cells [116]. The precise site(s) on P-Rex1 that become phosphorylated in response to stimuli, as well as putative PKA phosphorylation sites on P-Rex2 isoforms, remain to be identified.
P-Rex1 translocates to membranes in response to stimulation of GPCRs, a step that is required for Rac activation. Translocation is inhibited by wortmannin (PI3K inhibitor), M119 (inhibitor of Gβγ binding to effectors), tyrosine-kinase inhibitors, and PKA activators [117]. Gβγ subunits and PI3K activity are necessary and sufficient to promote P-Rex1 translocation and activation [114]. There has been speculation that phosphorylated P-Rex1 is inactive and resides mainly in the cytosol, whereas dephosphorylated P-Rex1 can redistribute to membranes in response to stimuli [114].
6.b. P-Rex domains and gene regulation
The human P-Rex1 gene (PREX1) is located in chromosome 20 (20q13.13). Montero et al. [78] suggested the presence of two additional P-Rex1 isoforms; however, despite bioinformatics predictions, the expression of these variants needs to be confirmed. The P-Rex1 protein is 1659 amino acids long (Mw=185 kDa) and contains a tandem DH (Dbl-homology)/PH (pleckstrin-homology) domain characteristic of Rho GEFs, two DEP (Disheveled, EGL-10, Pleckstrin) domains, two PDZ domains, and a C-terminal region with significant homology to inositol polyphosphate 4-phosphatase (IP4P) [27]. Through a panel of P-Rex-1 point mutations, deletions and truncations, the catalytic DH domain was initially found to mediate the Gβγ-dependent stimulation of GEF activity. Additionally, activation by PIP3 occurs via the PH domain of P-Rex1. A second putative binding site for PIP3 in P-Rex1 has been postulated, but its position has yet to be determined [118].
The DEP and PDZ domains are protein-protein interaction modules present in many signaling proteins. These domains seem to be required for Gβγ stimulation of P-Rex1, and deletion of these domains reduces the maximal activation by Gβγ subunits without affecting activation by PIP3 [118]. P-Rex1 interacts with mammalian target of rapamycin (mTOR) through the DEP domain, a mechanism that possibly integrates nutrient and growth factor signals. P-Rex1 can be activated through the mTOR complex 2 (mTORC2) to promote Rac activation and cell migration [119].
Inositol polyphosphate 4-phosphatase is an enzyme that catalyzes the 4′-dephosphorylation in phosphatidylinositols [120]. Even though key residues required for phosphatase activity of the IP4P domain are present both in P-Rex1 and P-Rex2a [27], the phosphatase activity has not been formally demonstrated in these proteins. This domain does not appear to be necessary for GEF activation [118], although the intramolecular interaction of the second DEP/first PDZ with the IP4P domain could be required for Gβγ-induced activation of P-Rex1. Moreover, PKA phosphorylation would prevent this domain-domain interaction, hence impeding Gβγ-dependent activation [121].
Recent studies by Tu and coworkers [122] established that expression of the P-Rex1 gene is regulated by a Sp1-HDAC complex. To determine the potential involvement of epigenetic mechanisms in the regulation of P-Rex1 expression, 22Rv1 non-metastatic prostate cancer cells were treated with either a DNA methyltransferase inhibitor (5-Aza-dC) or a histone deacetylase inhibitor (TSA). It was observed that histone deacetylation, but not DNA methylation, is involved in the suppression of P-Rex1 expression. HDAC inhibitors significantly increased levels of acetylated histone H4 associated with the P-Rex1 promoter in 22Rv1 cells and markedly increased P-Rex1 gene expression, without effects on the total amount of acetylated H4 compared to PC-3 metastatic cells. These results suggest that up-regulation of P-Rex1 in metastatic cells could be due to region-specific changes in histone acetylation within the P-Rex1 promoter rather than a global histone acetylation increase. The promoter region that regulates both basal and TSA-induced P-Rex1 expression has been identified. It contains a Sp1 binding site and may act as a regulatory cis element of P-Rex1 gene transcription. Indeed, ectopic overexpression of Sp1 significantly increased P-Rex1 promoter activity [122]. Sp1 is proposed to act as a docking site for HDACs, enabling them to suppress gene transcription via modulation of the chromatin structure. Dissociation of HDACs from Sp1 on the P-Rex1 promoter may contribute to the aberrant up-regulation of P-Rex1 in some cancer types.
The human PREX2 gene lies on chromosome 8 (8q13.2) and encodes two isoforms (P-Rex2a and PRex2b). P-Rex2a (Mw=183kDa) has strong sequence identity with P-Rex1 (59% identical) and a similar domain architecture. On the other hand, P-Rex2b (Mw=112 kDa) contains a much shorter C-terminus and lacks the IP4P domain. P-Rex2a is expressed in several tissues including heart, skeletal muscle, small intestine and placenta, whereas P-Rex2b expression was originally detected in the heart [111]. More recently, P-Rex2 was found in brain and lung, and at low levels in liver, thymus and spleen [123]. In vascular endothelial cells, P-Rex2b mediates direct Rac1 activation and cell migration in response to sphingosine-1-phosphate [124]. Unlike P-Rex1, P-Rex2 isoforms are not expressed in peripheral blood leukocytes [111, 112].
6.c. Functions of P-Rex1 and P-Rex2
P-Rex1, the first identified member of the family, was originally purified from neutrophils and found to have a fundamental role in the formation of reactive oxygen species (ROS) by NADPH oxidase, an enzymatic complex that requires Rac for activation [27]. Neutrophils from P-Rex1−/− mice display impaired GPCR-dependent activation of Rac2, ROS formation and motility, and have a defect in N-Formyl-Methionyl-Leucyl-Phenylalanine (fMLP)-induced F-actin formation and superoxide production [125, 126]. Remarkably, whereas P-Rex1 deficiency does not significantly affect Rac1 activation in mouse neutrophils, it impairs Rac2 activation in response to fMLP, possibly due to a higher affinity of P-Rex1 for Rac2 than for Rac1. This conclusion has been further supported by the fact that P-Rex1- and Rac2-deficient neutrophils have similar phenotypes [17, 127–129]. P-Rex1 is also a major regulator of Rac1 activation and chemotaxis in macrophages [130]. Additionally, P-Rex1 has been implicated in neuronal migration, neurite differentiation, and cerebellar long-term potentiation [131–133]. A role for P-Rex1 in angiogenesis has been recently established [134].
P-Rex2 deficient mice are viable, fertile and apparently healthy with some reduction in body weight. However, these mice have abnormal Purkinje cell dendrite morphology and develop a mild motor coordination disorder [123]. The stimuli and receptors that link P-Rex2 to Rac activation in Purkinje cells remain to be determined.
6.d. P-Rex1 is overexpressed in breast cancer
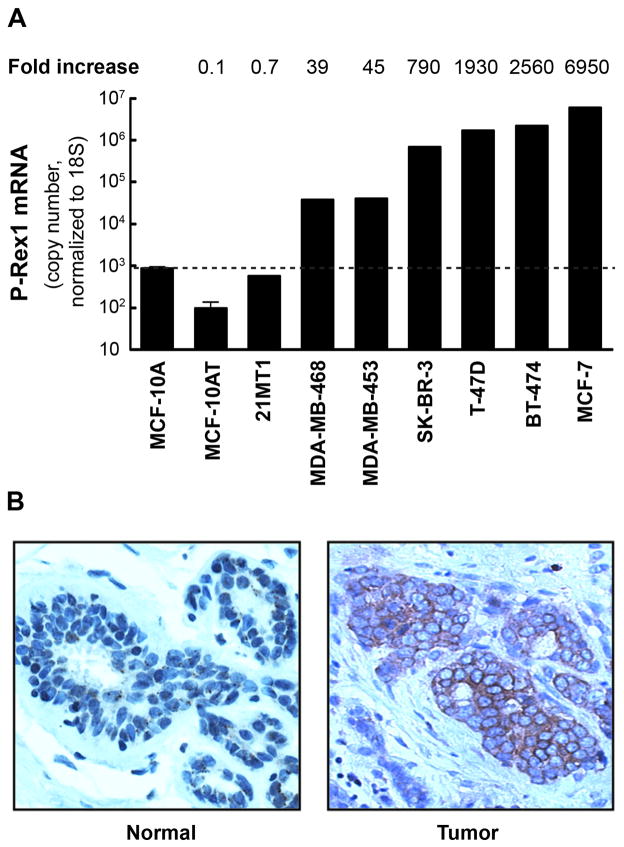
The hyperactivation of ErbB receptor signaling is a hallmark of breast cancer. Overexpression of ErbB2 and/or ErbB ligands such as TGF-α or HRG is a common feature of breast cancer and leads to uncontrolled growth and transformation [135–137]. Gain-of-function mutations of ErbB effectors such as PI3K or PTEN deletions have been also associated with breast cancer progression [138, 139]. The established role of Rac1 as a mediator of ErbB signaling prompted several laboratories to search for relevant Rac-GEFs implicated in this pathway. To address this issue, our laboratory designed a “Rac-GEF array” to determine the relative expression of Rac-GEFs in breast cancer cells, and found that P-Rex1 is highly expressed in MCF-7 and T-47D cells, which derive from ER-positive luminal breast tumors. In contrast, non-transformed MCF-10A cells have negligible P-Rex1 expression (Figure 3A). Basal breast cancer-derived cell lines such as MDA-MB-231 cells do not express P-Rex1. An extensive analysis of human breast cancer specimens revealed that >50% of tumors stain positive for P-Rex1 (Figure 3B), whereas no staining is observed in normal mammary tissue. P-Rex1 is expressed only in the tumor cells but not in the stroma. P-Rex1 expression is higher in primary tumors from patients that underwent metastasis relative to those that did not, and 67% of lymph nodes from breast cancer patients are P-Rex1 positive, suggesting a potential involvement of this Rac-GEF in metastatic dissemination of breast cancer cells. Strikingly, there is a marked difference in expression among breast cancer subtypes, since P-Rex1 is primarily elevated in luminal tumors but essentially undetected in basal-type tumors [77].
Figure 3. Overexpression of P-Rex1 in breast cancer.
Panel A. Expression of P-Rex1 mRNA by Q-PCR in human mammary cell lines. Values presented as “fold-increase” are relative to levels in MCF-10A cells. Panel B. Immunohistochemistry comparing P-Rex1 staining in a human breast tumor and normal mammary tissue.
The PREX1 locus is located in a region commonly amplified in breast tumors and cell lines [140–143]. The PREX1 gene is amplified in some breast cancer cell lines, such as MCF-7 BT-474, and HCC1419 cells, but not in T-47D cells [25], suggesting that mechanisms other than gene amplification may contribute, at least in part, to the overexpression of P-Rex1 in human breast tumors. An interesting observation is that P-Rex1 expression positively correlates with ER and ErbB2 expression. However, there is no correlation with PI3K mutations in the tumors. Notably, P-Rex1 depletion abolishes the tumorigenic activity of ErbB2 positive cells in nude mice [77].
6.e. P-Rex1 integrates ErbB receptor and CXCR4 signals in breast cancer cells
As mentioned above, P-Rex1 is a PI3K- and Gγβ-dependent Rac-GEF (see Figure 1). Activation of Rac1 by HRG in breast cancer cells is sensitive both to PI3Kγ inhibition and pertussis toxin. This surprising result strongly argued for the requirement of a Gi-coupled receptor in the activation of Rac1 by HRG. Moreover, ErbB receptor-mediated activation of Rac1 and migration was markedly reduced by inhibition/depletion of the Gβγ subunit-regulated PI3Kγ [77].
CXCR4 is a Gi-coupled receptor for the chemokine SDF-1α/CXCL12. Both SDF-1α and its receptor have been widely implicated in the progression of breast cancer, particularly in metastatic dissemination. Moreover, there is a positive correlation between CXCR4 and ErbB2 expression in human breast tumors [144–146]. Our laboratory reported a striking functional association between CXCR4 and ErbB receptors for P-Rex1 activation [77]. CXCR4 becomes activated in response to ErbB ligands independently of SDF-1α and is required for P-Rex1/Rac1 activation and migration induced by HRG. EGFR is also required for the activation of CXCR4 by HRG, arguing for a model of transactivation of CXCR4 that involves multiple ErbB receptors. If PI3Kγ or Gβγ function is impaired, then activation of Rac1 by SDF-1α is fully abolished, whereas Rac1 activation by HRG is only partially reduced. Most probably, ErbB receptors signal to P-Rex1 through two cooperative mechanisms: one via transactivation of a CXCR4/Gβγ/PI3Kγ-dependent pathway, and the other by direct activation of type IA PI3K by ErbB receptors [77]. It would be of great interest to determine if other tyrosine-kinase receptors relevant in breast cancer, such as insulin-like growth factor receptor-1R (IGF-1R), Met or Ret signal via P-Rex1 using similar or distinct mechanisms.
Similarly to the previously described work, a subsequent study reported the involvement of P-Rex1 in HRG-induced Rac1 activation in breast cancer cells [78]. In addition to the effect of P-Rex1 on migration and invasiveness of breast cancer cells and its requirement in vivo for breast cancer cell tumorigenicity, this study demonstrated that treating breast cancer cells with HRG alters P-Rex1 phosphorylation status. Indeed, HRG promotes the dephosphorylation of Ser313 and Ser319 and phosphorylation of Ser605 and Ser1169 in P-Rex1. Phosphorylation in Ser313 and Ser319 may therefore restrain Rac1 activity, whereas phosphorylation in Ser605 and Ser1169 may facilitate Rac1 activation [78].
6.f. Role of P-Rex1 in breast cancer cell motility
An extensive functional analysis using P-Rex1 overexpressing breast cancer cell lines established the requirement of this GEF for Rac1 activation and cell migration induced by EGF, TGFα or HRG. Interestingly, silencing other GEFs, including Vav2, Vav3 or Tiam1, does not significantly affect Rac1 activation by HRG in T-47D cells. Most notably, P-Rex1 depletion essentially abolishes Rac1 activation and motility in ErbB2 positive cell lines such as BT-474, HCC1419 and MDA-MB-361 cells [77].
Subcellular localization analysis revealed that P-Rex1 translocates to the plasma membrane in response to HRG in a PI3Kγ-dependent manner [77]. The DH-PH domain is necessary and sufficient for translocation. There is a noticeable intracellular redistribution of P-Rex1 to membrane ruffles by HRG treatment. Moreover, P-Rex1-deficient T-47D cells fail to form membrane ruffles in response to HRG, strongly arguing for a role of the P-Rex1/Rac1 axis in actin cytoskeleton reorganization in response to ErbB3 receptor stimulation.
6.g. P-Rex2 and breast cancer
The PREX2 gene is located in a chromosomal region that has been linked to aggressive cancer and metastatic progression and is frequently amplified in breast and prostate cancers [147, 148]. Interestingly, analysis of a breast tumor dataset annotated for PI3K pathway alterations revealed a positive association between P-Rex2a and PTEN expression. Moreover, PTEN-expressing breast tumors display a significant positive association between increased P-Rex2a levels and activating mutations in PI3K [110].
The loss of PTEN has been linked to elevated Akt activity, cell survival, cell cycle progression and growth. Parsons and coworkers found that P-Rex2a interacts with PTEN and inhibits its lipid phosphatase activity, thereby antagonizing the tumor suppressor function of PTEN in breast cancer cells [110]. These effects depend on the P-Rex2a DH-PH domains and are independent of the GEF activity. Overexpression of P-Rex2a or its DH-PH domains in the MCF-10A cells, which expresses endogenous PTEN, increases Akt phosphorylation in Ser473 and proliferation. The single-acinar cell clusters that MCF-10A cells form in 3D Matrigel cultures are transformed to multiacinar epithelial structures upon expression of P-Rex2a. Moreover, co-expression of P-Rex2a and a constitutively active PI3K mutant leads to the formation of large branched, highly dysmorphic invasive structures. Expression of P-Rex2a in MCF-10A cells increases by 3-fold the number of colonies in soft agar formed by constitutively active PI3K. Furthermore, depletion of P-Rex2a from MCF-7 breast cancer cells that express wild-type PTEN and a constitutively active PI3K reduces phospho-Akt levels and impairs proliferation. On the other hand, these effects are not observed in BT-549, a PTEN-deficient cell line. Thus, the impact of P-Rex2a on the PI3K pathway and proliferation in breast cancer cells depend on the presence of a functional PTEN [110]. It became clear that P-Rex1 and P-Rex2 isoforms have differential effects on the PI3K/Akt pathway in breast cancer cells, since P-Rex1 RNAi depletion does not affect Akt1 activation [77], and therefore each P-Rex isoform may contribute in different ways to breast cancer progression. A recent report described Akt1-dependent effects of P-Rex1 in ovarian cancer cells [149], suggesting cell type-specific differences in P-Rex1 regulation. Even though PTEN regulation by P-Rex2a appears to be unrelated to Rac activation [110], additional studies are required to unambiguously establish the independence from Rac as well as to determine potential positive feedback loops that could be regulating P-Rex2a effects on the activity of PTEN.
7. Concluding remarks: targeting the Rac pathway as an approach for breast cancer treatment?
Rac GTPases function as tightly regulated signaling nodes that mediate inputs from receptors and oncogenes. The aberrant expression and/or activity of Rac regulators, particularly Rac-GEFs, offer a number of possibilities for targeting the Rac pathway. Based on current structural-function information on the interaction of Rac with GEFs it has been possible to identify small molecules that fit in the surface groove of Rac1 which determines GEF specification. One good example is the compound NSC23766 described above, a highly soluble and permeable inhibitor that prevents GTP loading onto Rac1 by Tiam1 and Trio [25]. This approach can be optimized to identify specific inhibitors for GEFs overexpressed in different cancer types, such as P-Rex1 in breast cancer. The rational design of compounds that target those GEFs with narrow tissue distribution should lead to drugs with limited off-target effects compared to traditional chemotherapeutic agents. Conceivably, it should be possible to design drugs that activate Rac-GAPs and counterbalance Rac hyperactivation in cancer, but such approach still requires a deeper understanding of the contributions of GAPs to cancer progression.
The discovery of P-Rex1 as a key factor in the progression of breast cancer has enormous implications from both prognostic and therapeutic standpoints. It is possible that P-Rex1 overexpression represents a biomarker for luminal breast cancer that predicts aggressiveness and metastasis. P-Rex1 is an effector of the ErbB network and therefore drugs that inhibit P-Rex1 may lead to the inactivation of ErbB receptor responses. This may have important implications for patients with ErbB2 positive breast tumors. As Rac1 and its downstream effectors have been implicated in antiestrogen resistance, it is possible that targeting the P-Rex1/Rac1/Pak1 pathway may also overcome the resistance of breast cancer cells to antiestrogens. Ultimately, deciphering the mechanistic insights that lead to P-Rex1 up-regulation and the signals that drive P-Rex1/Rac1 activation in breast cancer cells will allow for a better understanding of the mechanisms of mammary cell transformation and resistance to therapeutic agents.
Acknowledgments
This work was supported by grants CA74197, CA129133, and CA139120 from NIH, and KG090522 from Susan G. Komen for the Cure (M.G.K.).
Abbreviations
- DH
Dbl-homology
- EGF
Epidermal growth factor
- ERα
estrogen receptor alpha
- Erk
Extracellular signal-regulated kinases
- fMLP
N-formyl-methionine-leucine-phenylalanine
- GAP
GTPase Activating Proteins
- GBD
GTPase binding domain
- GDI
GDP Dissociation Inhibitors
- GDP
Guanosine diphosphate
- GEF
Guanine nucleotide Exchange Factors
- GPCR
G-protein-coupled receptors
- GTP
Guanosine-5′-triphosphate
- HDAC
Histone Deacetylase
- HRG
heregulin
- IGF-1R
Insulin-like growth factor 1 receptor
- IP4P
inositol polyphosphate 4-phosphatase
- LPS
lipopolysaccharide
- MAPK
Mitogen-activated protein kinase
- mTOR
Mammalian target of rapamycin
- NSCLC
Non-small cell lung cancer
- Pak
p21-activated Protein Kinase
- PH
pleckstrin-homology
- PI3K
phosphoinositide 3-kinases
- PIP3
phosphatidylinositol (3,4,5)-trisphosphate
- P-Rex
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger
- PTEN
Phosphatase and tensin homolog
- ROS
reactive oxygen species
- TGFα
Transforming growth factor alpha
- TSA
Trichostatin A
References
- 1.Manser E. Dev Cell. 2002;3(3):323–328. doi: 10.1016/s1534-5807(02)00268-x. [DOI] [PubMed] [Google Scholar]
- 2.Bustelo XR, Sauzeau V, Berenjeno IM. Bioessays. 2007;29(4):356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wennerberg K, Der CJ. J Cell Sci. 2004;117(Pt 8):1301–1312. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- 4.Del Pozo MA, Kiosses WB, Alderson NB, Meller N, Hahn KM, Schwartz MA. Nat Cell Biol. 2002;4(3):232–239. doi: 10.1038/ncb759. [DOI] [PubMed] [Google Scholar]
- 5.Price LS, Langeslag M, ten Klooster JP, Hordijk PL, Jalink K, Collard JG. J Biol Chem. 2003;278(41):39413–39421. doi: 10.1074/jbc.M302083200. [DOI] [PubMed] [Google Scholar]
- 6.Etienne-Manneville S, Hall A. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 7.Didsbury J, Weber RF, Bokoch GM, Evans T, Snyderman R. J Biol Chem. 1989;264(28):16378–16382. [PubMed] [Google Scholar]
- 8.Haataja L, Groffen J, Heisterkamp N. J Biol Chem. 1997;272(33):20384–20388. doi: 10.1074/jbc.272.33.20384. [DOI] [PubMed] [Google Scholar]
- 9.Matos P, Skaug J, Marques B, Beck S, Verissimo F, Gespach C, Boavida MG, Scherer SW, Jordan P. Biochem Biophys Res Commun. 2000;277(3):741–751. doi: 10.1006/bbrc.2000.3743. [DOI] [PubMed] [Google Scholar]
- 10.Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E. Oncogene. 1999;18(48):6835–6839. doi: 10.1038/sj.onc.1203233. [DOI] [PubMed] [Google Scholar]
- 11.Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, Harbeck N, Schmitt M, Lengyel E. Oncogene. 2000;19(26):3013–3020. doi: 10.1038/sj.onc.1203621. [DOI] [PubMed] [Google Scholar]
- 12.Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H, Sakagami H, Kondo H, Nozawa S, Aiba A, Katsuki M. Oncogene. 1998;17(26):3427–3433. doi: 10.1038/sj.onc.1202595. [DOI] [PubMed] [Google Scholar]
- 13.Courjal F, Chuchana P, Theillet C, Fort P. Genomics. 1997;44(2):242–246. doi: 10.1006/geno.1997.4871. [DOI] [PubMed] [Google Scholar]
- 14.Ladd PD, Butler JS, Skalnik DG. Gene. 2004;341:323–333. doi: 10.1016/j.gene.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Leitenberg D, Li B, Flavell RA. J Exp Med. 2001;194(7):915–926. doi: 10.1084/jem.194.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdel-Latif D, Steward M, Lacy P. Can J Physiol Pharmacol. 2005;83(1):69–75. doi: 10.1139/y04-123. [DOI] [PubMed] [Google Scholar]
- 17.Roberts AW, Kim C, Zhen L, Lowe JB, Kapur R, Petryniak B, Spaetti A, Pollock JD, Borneo JB, Bradford GB, Atkinson SJ, Dinauer MC, Williams DA. Immunity. 1999;10(2):183–196. doi: 10.1016/s1074-7613(00)80019-9. [DOI] [PubMed] [Google Scholar]
- 18.Ambruso DR, Knall C, Abell AN, Panepinto J, Kurkchubasche A, Thurman G, Gonzalez-Aller C, Hiester A, deBoer M, Harbeck RJ, Oyer R, Johnson GL, Roos D. Proc Natl Acad Sci U S A. 2000;97(9):4654–4659. doi: 10.1073/pnas.080074897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams DA, Tao W, Yang F, Kim C, Gu Y, Mansfield P, Levine JE, Petryniak B, Derrow CW, Harris C, Jia B, Zheng Y, Ambruso DR, Lowe JB, Atkinson SJ, Dinauer MC, Boxer L. Blood. 2000;96(5):1646–1654. [PubMed] [Google Scholar]
- 20.Corbetta S, Gualdoni S, Albertinazzi C, Paris S, Croci L, Consalez GG, de Curtis I. Mol Cell Biol. 2005;25(13):5763–5776. doi: 10.1128/MCB.25.13.5763-5776.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinsella BT, Erdman RA, Maltese WA. Proc Natl Acad Sci U S A. 1991;88(20):8934–8938. doi: 10.1073/pnas.88.20.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Cell. 1999;98(1):69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 23.Vetter IR, Wittinghofer A. Science. 2001;294(5545):1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 24.Bos JL, Rehmann H, Wittinghofer A. Cell. 2007;129(5):865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Proc Natl Acad Sci U S A. 2004;101(20):7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michiels F, Habets GG, Stam JC, van der Kammen RA, Collard JG. Nature. 1995;375(6529):338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 27.Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, Tempst P, Hawkins PT, Stephens LR. Cell. 2002;108(6):809–821. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- 28.Welch HC, Coadwell WJ, Stephens LR, Hawkins PT. FEBS Lett. 2003;546(1):93–97. doi: 10.1016/s0014-5793(03)00454-x. [DOI] [PubMed] [Google Scholar]
- 29.Tamas P, Solti Z, Bauer P, Illes A, Sipeki S, Bauer A, Farago A, Downward J, Buday L. J Biol Chem. 2003;278(7):5163–5171. doi: 10.1074/jbc.M207555200. [DOI] [PubMed] [Google Scholar]
- 30.Bonnefoy-Berard N, Munshi A, Yron I, Wu S, Collins TL, Deckert M, Shalom-Barak T, Giampa L, Herbert E, Hernandez J, Meller N, Couture C, Altman A. Stem Cells. 1996;14(3):250–268. doi: 10.1002/stem.140250. [DOI] [PubMed] [Google Scholar]
- 31.Caloca MJ, Wang H, Kazanietz MG. Biochem J. 2003;375(Pt 2):313–321. doi: 10.1042/BJ20030727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Yang C, Leskow FC, Sun J, Canagarajah B, Hurley JH, Kazanietz MG. Embo J. 2006;25(10):2062–2074. doi: 10.1038/sj.emboj.7601098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kazanietz MG. Mol Carcinog. 2000;28(1):5–11. doi: 10.1002/(sici)1098-2744(200005)28:1<5::aid-mc2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 34.Griner EM, Caino MC, Soledad Sosa M, Colon-Gonzalez F, Chalmers MJ, Mischak H, Kazanietz MG. J Biol Chem. 2010;285(22):16931–16941. doi: 10.1074/jbc.M109.099036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siliceo M, Merida I. J Biol Chem. 2009;284(17):11354–11363. doi: 10.1074/jbc.M806098200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Kazanietz MG. Mol Biol Cell. 2010;21(8):1398–1408. doi: 10.1091/mbc.E09-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Kazanietz MG. J Biol Chem. 2002;277(6):4541–4550. doi: 10.1074/jbc.M107150200. [DOI] [PubMed] [Google Scholar]
- 38.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. Cell. 1992;70(3):401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 39.Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T. J Cell Biol. 2006;173(4):571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whale A, Hashim FN, Fram S, Jones GE, Wells CM. Front Biosci. 16:849–864. doi: 10.2741/3724. [DOI] [PubMed] [Google Scholar]
- 41.Dummler B, Ohshiro K, Kumar R, Field J. Cancer Metastasis Rev. 2009;28(1–2):51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rayala SK, Molli PR, Kumar R. Cancer Res. 2006;66(12):5985–5988. doi: 10.1158/0008-5472.CAN-06-0978. [DOI] [PubMed] [Google Scholar]
- 43.Bokoch GM. Immunol Res. 2000;21(2–3):139–148. doi: 10.1385/IR:21:2-3:139. [DOI] [PubMed] [Google Scholar]
- 44.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Nat Cell Biol. 1999;1(5):253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 45.Vadlamudi RK, Li F, Barnes CJ, Bagheri-Yarmand R, Kumar R. EMBO Rep. 2004;5(2):154–160. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiel DA, Reeder MK, Pfaff A, Coleman TR, Sells MA, Chernoff J. Curr Biol. 2002;12(14):1227–1232. doi: 10.1016/s0960-9822(02)00931-4. [DOI] [PubMed] [Google Scholar]
- 47.Wells CM, Walmsley M, Ooi S, Tybulewicz V, Ridley AJ. J Cell Sci. 2004;117(Pt 7):1259–1268. doi: 10.1242/jcs.00997. [DOI] [PubMed] [Google Scholar]
- 48.Kiosses WB, Shattil SJ, Pampori N, Schwartz MA. Nat Cell Biol. 2001;3(3):316–320. doi: 10.1038/35060120. [DOI] [PubMed] [Google Scholar]
- 49.Sundaresan M, Yu ZX, Ferrans VJ, Sulciner DJ, Gutkind JS, Irani K, Goldschmidt-Clermont PJ, Finkel T. Biochem J. 1996;318 (Pt 2):379–382. doi: 10.1042/bj3180379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khayat ZA, Tong P, Yaworsky K, Bloch RJ, Klip A. J Cell Sci. 2000;113(Pt 2):279–290. doi: 10.1242/jcs.113.2.279. [DOI] [PubMed] [Google Scholar]
- 51.JeBailey L, Wanono O, Niu W, Roessler J, Rudich A, Klip A. Diabetes. 2007;56(2):394–403. doi: 10.2337/db06-0823. [DOI] [PubMed] [Google Scholar]
- 52.Hill CS, Wynne J, Treisman R. Cell. 1995;81(7):1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 53.Coso OA, Chiariello M, Yu JC, Teramoto H, Crespo P, Xu N, Miki T, Gutkind JS. Cell. 1995;81(7):1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 54.Eom YW, Yoo MH, Woo CH, Hwang KC, Song WK, Yoo YJ, Chun JS, Kim JH. Biochem Biophys Res Commun. 2001;285(3):825–829. doi: 10.1006/bbrc.2001.5233. [DOI] [PubMed] [Google Scholar]
- 55.Eom YW, Cho SH, Hwang JS, Yoon SB, Na DS, Kang IJ, Kang SS, Song WK, Kim JH. Biochem Biophys Res Commun. 2001;284(1):126–132. doi: 10.1006/bbrc.2001.4937. [DOI] [PubMed] [Google Scholar]
- 56.Woo CH, Kim JH. Mol Cells. 2002;13(3):470–475. [PubMed] [Google Scholar]
- 57.Uhlik MT, Abell AN, Johnson NL, Sun W, Cuevas BD, Lobel-Rice KE, Horne EA, Dell’Acqua ML, Johnson GL. Nat Cell Biol. 2003;5(12):1104–1110. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- 58.Shin I, Kim S, Song H, Kim HR, Moon A. J Biol Chem. 2005;280(15):14675–14683. doi: 10.1074/jbc.M411625200. [DOI] [PubMed] [Google Scholar]
- 59.Khosravi-Far R, Solski PA, Clark GJ, Kinch MS, Der CJ. Mol Cell Biol. 1995;15(11):6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frost JA, Steen H, Shapiro P, Lewis T, Ahn N, Shaw PE, Cobb MH. Embo J. 1997;16(21):6426–6438. doi: 10.1093/emboj/16.21.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DerMardirossian C, Schnelzer A, Bokoch GM. Mol Cell. 2004;15(1):117–127. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 62.Callow MG, Zozulya S, Gishizky ML, Jallal B, Smeal T. J Cell Sci. 2005;118(Pt 9):1861–1872. doi: 10.1242/jcs.02313. [DOI] [PubMed] [Google Scholar]
- 63.Zuluaga S, Gutierrez-Uzquiza A, Bragado P, Alvarez-Barrientos A, Benito M, Nebreda AR, Porras A. FEBS Lett. 2007;581(20):3819–3825. doi: 10.1016/j.febslet.2007.06.078. [DOI] [PubMed] [Google Scholar]
- 64.Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Cell. 2008;135(3):510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 65.Muise AM, Walters T, Xu W, Shen-Tu G, Guo CH, Fattouh R, Lam GY, Wolters VM, Bennitz J, van Limbergen J, Renbaum P, Kasirer Y, Ngan BY, Turner D, Denson LA, Sherman PM, Duerr RH, Cho J, Lees CW, Satsangi J, Wilson DC, Paterson AD, Griffiths AM, Glogauer M, Silverberg MS, Brumell JH. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.04.057. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wertheimer E, Kazanietz MG. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.06.027. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 67.Heid I, Lubeseder-Martellato C, Sipos B, Mazur PK, Lesina M, Schmid RM, Siveke JT. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.04.043. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 68.Hwang SL, Lieu AS, Chang JH, Cheng TS, Cheng CY, Lee KS, Lin CL, Howng SL, Hong YR. Acta Neurochir (Wien) 2005;147(5):551–554. doi: 10.1007/s00701-005-0515-5. discussion 554. [DOI] [PubMed] [Google Scholar]
- 69.Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Proc Natl Acad Sci U S A. 2000;97(1):185–189. doi: 10.1073/pnas.97.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baugher PJ, Krishnamoorthy L, Price JE, Dharmawardhane SF. Breast Cancer Res. 2005;7(6):R965–974. doi: 10.1186/bcr1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morris CM, Haataja L, McDonald M, Gough S, Markie D, Groffen J, Heisterkamp N. Cytogenet Cell Genet. 2000;89(1–2):18–23. doi: 10.1159/000015583. [DOI] [PubMed] [Google Scholar]
- 72.Pan Y, Bi F, Liu N, Xue Y, Yao X, Zheng Y, Fan D. Biochem Biophys Res Commun. 2004;315(3):686–691. doi: 10.1016/j.bbrc.2004.01.108. [DOI] [PubMed] [Google Scholar]
- 73.Hwang SL, Chang JH, Cheng TS, Sy WD, Lieu AS, Lin CL, Lee KS, Howng SL, Hong YR. J Clin Neurosci. 2005;12(5):571–574. doi: 10.1016/j.jocn.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 74.Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, Collard JG. Cell. 1994;77(4):537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 75.Miki T, Smith CL, Long JE, Eva A, Fleming TP. Nature. 1993;362(6419):462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- 76.Qin J, Xie Y, Wang B, Hoshino M, Wolff DW, Zhao J, Scofield MA, Dowd FJ, Lin MF, Tu Y. Oncogene. 2009;28(16):1853–1863. doi: 10.1038/onc.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM, Luo J, Benovic JL, Klein-Szanto A, Yagi H, Gutkind JS, Parsons RE, Kazanietz MG. Mol Cell. 2010;40(6):877–892. doi: 10.1016/j.molcel.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Montero JC, Seoane S, Ocana A, Pandiella A. Oncogene. 2011;30(9):1059–1071. doi: 10.1038/onc.2010.489. [DOI] [PubMed] [Google Scholar]
- 79.Mertens AE, Roovers RC, Collard JG. FEBS Lett. 2003;546(1):11–16. doi: 10.1016/s0014-5793(03)00435-6. [DOI] [PubMed] [Google Scholar]
- 80.Strumane K, Rygiel TP, Collard JG. Methods Enzymol. 2006;407:269–281. doi: 10.1016/S0076-6879(05)07023-0. [DOI] [PubMed] [Google Scholar]
- 81.Saito S, Tatsumoto T, Lorenzi MV, Chedid M, Kapoor V, Sakata H, Rubin J, Miki T. J Cell Biochem. 2003;90(4):819–836. doi: 10.1002/jcb.10688. [DOI] [PubMed] [Google Scholar]
- 82.Salhia B, Tran NL, Chan A, Wolf A, Nakada M, Rutka F, Ennis M, McDonough WS, Berens ME, Symons M, Rutka JT. Am J Pathol. 2008;173(6):1828–1838. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sano M, Genkai N, Yajima N, Tsuchiya N, Homma J, Tanaka R, Miki T, Yamanaka R. Oncol Rep. 2006;16(5):1093–1098. [PubMed] [Google Scholar]
- 84.Zhang ML, Lu S, Zhou L, Zheng SS. Hepatobiliary Pancreat Dis Int. 2008;7(5):533–538. [PubMed] [Google Scholar]
- 85.Justilien V, Fields AP. Oncogene. 2009;28(41):3597–3607. doi: 10.1038/onc.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Justilien V, Jameison L, Der CJ, Rossman KL, Fields AP. J Biol Chem. 2011;286(10):8149–8157. doi: 10.1074/jbc.M110.196113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balasenthil S, Sahin AA, Barnes CJ, Wang RA, Pestell RG, Vadlamudi RK, Kumar R. J Biol Chem. 2004;279(2):1422–1428. doi: 10.1074/jbc.M309937200. [DOI] [PubMed] [Google Scholar]
- 88.Vadlamudi RK, Adam L, Wang RA, Mandal M, Nguyen D, Sahin A, Chernoff J, Hung MC, Kumar R. J Biol Chem. 2000;275(46):36238–36244. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- 89.Wang RA, Zhang H, Balasenthil S, Medina D, Kumar R. Oncogene. 2006;25(20):2931–2936. doi: 10.1038/sj.onc.1209309. [DOI] [PubMed] [Google Scholar]
- 90.Arias-Romero LE, Villamar-Cruz O, Pacheco A, Kosoff R, Huang M, Muthuswamy SK, Chernoff J. Oncogene. 2010;29(43):5839–5849. doi: 10.1038/onc.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang C, Liu Y, Leskow FC, Weaver VM, Kazanietz MG. J Biol Chem. 2005;280(26):24363–24370. doi: 10.1074/jbc.M411629200. [DOI] [PubMed] [Google Scholar]
- 92.Ueda Y, Wang S, Dumont N, Yi JY, Koh Y, Arteaga CL. J Biol Chem. 2004;279(23):24505–24513. doi: 10.1074/jbc.M400081200. [DOI] [PubMed] [Google Scholar]
- 93.Wang SE, Shin I, Wu FY, Friedman DB, Arteaga CL. Cancer Res. 2006;66(19):9591–9600. doi: 10.1158/0008-5472.CAN-06-2071. [DOI] [PubMed] [Google Scholar]
- 94.Rayala SK, Talukder AH, Balasenthil S, Tharakan R, Barnes CJ, Wang RA, Aldaz CM, Khan S, Kumar R. Cancer Res. 2006;66(3):1694–1701. doi: 10.1158/0008-5472.CAN-05-2922. [DOI] [PubMed] [Google Scholar]
- 95.Bostner J, Ahnstrom Waltersson M, Fornander T, Skoog L, Nordenskjold B, Stal O. Oncogene. 2007;26(49):6997–7005. doi: 10.1038/sj.onc.1210506. [DOI] [PubMed] [Google Scholar]
- 96.Holm C, Rayala S, Jirstrom K, Stal O, Kumar R, Landberg G. J Natl Cancer Inst. 2006;98(10):671–680. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- 97.Yang C, Liu Y, Lemmon MA, Kazanietz MG. Mol Cell Biol. 2006;26(3):831–842. doi: 10.1128/MCB.26.3.831-842.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang C, Klein EA, Assoian RK, Kazanietz MG. Biochem J. 2008;410(1):167–175. doi: 10.1042/BJ20070781. [DOI] [PubMed] [Google Scholar]
- 99.Adam L, Vadlamudi RK, McCrea P, Kumar R. J Biol Chem. 2001;276(30):28443–28450. doi: 10.1074/jbc.M009769200. [DOI] [PubMed] [Google Scholar]
- 100.Minard ME, Kim LS, Price JE, Gallick GE. Breast Cancer Res Treat. 2004;84(1):21–32. doi: 10.1023/B:BREA.0000018421.31632.e6. [DOI] [PubMed] [Google Scholar]
- 101.Bourguignon LY, Zhu H, Shao L, Chen YW. J Cell Biol. 2000;150(1):177–191. doi: 10.1083/jcb.150.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stebel A, Brachetti C, Kunkel M, Schmidt M, Fritz G. Oncol Rep. 2009;21(1):217–222. [PubMed] [Google Scholar]
- 103.Adams HC, 3rd, Chen R, Liu Z, Whitehead IP. Breast Cancer Res. 2010;12(5):R69. doi: 10.1186/bcr2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu K, Rajagopal S, Klebba I, Dong S, Ji Y, Liu J, Kuperwasser C, Garlick JA, Naber SP, Buchsbaum RJ. Oncogene. 2010;29(50):6533–6542. doi: 10.1038/onc.2010.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee K, Liu Y, Mo JQ, Zhang J, Dong Z, Lu S. BMC Cancer. 2008;8:158. doi: 10.1186/1471-2407-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rosenblatt AE, Garcia MI, Lyons L, Xie Y, Maiorino C, Desire L, Slingerland J, Burnstein KL. Endocr Relat Cancer. 2011;18(2):207–219. doi: 10.1677/ERC-10-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lane J, Martin TA, Watkins G, Mansel RE, Jiang WG. Int J Oncol. 2008;33(3):585–593. [PubMed] [Google Scholar]
- 108.Near RI, Zhang Y, Makkinje A, Vanden Borre P, Lerner A. J Cell Physiol. 2007;212(3):655–665. doi: 10.1002/jcp.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Menna PL, Skilton G, Leskow FC, Alonso DF, Gomez DE, Kazanietz MG. Cancer Res. 2003;63(9):2284–2291. [PubMed] [Google Scholar]
- 110.Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, Hopkins B, Keniry M, Sulis ML, Mense S, Hibshoosh H, Parsons R. Science. 2009;325(5945):1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Donald S, Hill K, Lecureuil C, Barnouin R, Krugmann S, John Coadwell W, Andrews SR, Walker SA, Hawkins PT, Stephens LR, Welch HC. FEBS Lett. 2004;572(1–3):172–176. doi: 10.1016/j.febslet.2004.06.096. [DOI] [PubMed] [Google Scholar]
- 112.Rosenfeldt H, Vazquez-Prado J, Gutkind JS. FEBS Lett. 2004;572(1–3):167–171. doi: 10.1016/j.febslet.2004.06.097. [DOI] [PubMed] [Google Scholar]
- 113.Mayeenuddin LH, McIntire WE, Garrison JC. J Biol Chem. 2006;281(4):1913–1920. doi: 10.1074/jbc.M506034200. [DOI] [PubMed] [Google Scholar]
- 114.Barber MA, Donald S, Thelen S, Anderson KE, Thelen M, Welch HC. J Biol Chem. 2007;282(41):29967–29976. doi: 10.1074/jbc.M701877200. [DOI] [PubMed] [Google Scholar]
- 115.Stephens LR, Eguinoa A, Erdjument-Bromage H, Lui M, Cooke F, Coadwell J, Smrcka AS, Thelen M, Cadwallader K, Tempst P, Hawkins PT. Cell. 1997;89(1):105–114. doi: 10.1016/s0092-8674(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 116.Mayeenuddin LH, Garrison JC. J Biol Chem. 2006;281(4):1921–1928. doi: 10.1074/jbc.M506035200. [DOI] [PubMed] [Google Scholar]
- 117.Zhao T, Nalbant P, Hoshino M, Dong X, Wu D, Bokoch GM. J Leukoc Biol. 2007;81(4):1127–1136. doi: 10.1189/jlb.0406251. [DOI] [PubMed] [Google Scholar]
- 118.Hill K, Krugmann S, Andrews SR, Coadwell WJ, Finan P, Welch HC, Hawkins PT, Stephens LR. J Biol Chem. 2005;280(6):4166–4173. doi: 10.1074/jbc.M411262200. [DOI] [PubMed] [Google Scholar]
- 119.Hernandez-Negrete I, Carretero-Ortega J, Rosenfeldt H, Hernandez-Garcia R, Calderon-Salinas JV, Reyes-Cruz G, Gutkind JS, Vazquez-Prado J. J Biol Chem. 2007;282(32):23708–23715. doi: 10.1074/jbc.M703771200. [DOI] [PubMed] [Google Scholar]
- 120.Norris FA, Ungewickell E, Majerus PW. J Biol Chem. 1995;270(1):214–217. doi: 10.1074/jbc.270.1.214. [DOI] [PubMed] [Google Scholar]
- 121.Urano D, Nakata A, Mizuno N, Tago K, Itoh H. Cell Signal. 2008;20(8):1545–1554. doi: 10.1016/j.cellsig.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 122.Wong CY, Wuriyanghan H, Xie Y, Lin MF, Abel PW, Tu Y. J Biol Chem. 2011;286(29):25813–25822. doi: 10.1074/jbc.M110.211292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Donald S, Humby T, Fyfe I, Segonds-Pichon A, Walker SA, Andrews SR, Coadwell WJ, Emson P, Wilkinson LS, Welch HC. Proc Natl Acad Sci U S A. 2008;105(11):4483–4488. doi: 10.1073/pnas.0712324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Z, Paik JH, Wang Z, Hla T, Wu D. Prostaglandins Other Lipid Mediat. 2005;76(1–4):95–104. doi: 10.1016/j.prostaglandins.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 125.Welch HC, Condliffe AM, Milne LJ, Ferguson GJ, Hill K, Webb LM, Okkenhaug K, Coadwell WJ, Andrews SR, Thelen M, Jones GE, Hawkins PT, Stephens LR. Curr Biol. 2005;15(20):1867–1873. doi: 10.1016/j.cub.2005.09.050. [DOI] [PubMed] [Google Scholar]
- 126.Dong X, Mo Z, Bokoch G, Guo C, Li Z, Wu D. Curr Biol. 2005;15(20):1874–1879. doi: 10.1016/j.cub.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 127.Gu Y, Filippi MD, Cancelas JA, Siefring JE, Williams EP, Jasti AC, Harris CE, Lee AW, Prabhakar R, Atkinson SJ, Kwiatkowski DJ, Williams DA. Science. 2003;302(5644):445–449. doi: 10.1126/science.1088485. [DOI] [PubMed] [Google Scholar]
- 128.Glogauer M, Marchal CC, Zhu F, Worku A, Clausen BE, Foerster I, Marks P, Downey GP, Dinauer M, Kwiatkowski DJ. J Immunol. 2003;170(11):5652–5657. doi: 10.4049/jimmunol.170.11.5652. [DOI] [PubMed] [Google Scholar]
- 129.Li S, Yamauchi A, Marchal CC, Molitoris JK, Quilliam LA, Dinauer MC. J Immunol. 2002;169(9):5043–5051. doi: 10.4049/jimmunol.169.9.5043. [DOI] [PubMed] [Google Scholar]
- 130.Wang Z, Dong X, Li Z, Smith JD, Wu D. Prostaglandins Other Lipid Mediat. 2008;87(1–4):9–13. doi: 10.1016/j.prostaglandins.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoshizawa M, Kawauchi T, Sone M, Nishimura YV, Terao M, Chihama K, Nabeshima Y, Hoshino M. J Neurosci. 2005;25(17):4406–4419. doi: 10.1523/JNEUROSCI.4955-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Waters JE, Astle MV, Ooms LM, Balamatsias D, Gurung R, Mitchell CA. J Cell Sci. 2008;121(Pt 17):2892–2903. doi: 10.1242/jcs.030353. [DOI] [PubMed] [Google Scholar]
- 133.Jackson C, Welch HC, Bellamy TC. PLoS One. 2010;5(8):e11962. doi: 10.1371/journal.pone.0011962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carretero-Ortega J, Walsh CT, Hernandez-Garcia R, Reyes-Cruz G, Brown JH, Vazquez-Prado J. Mol Pharmacol. 77(3):435–442. doi: 10.1124/mol.109.060400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hynes NE, Lane HA. Nat Rev Cancer. 2005;5(5):341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 136.Citri A, Skaria KB, Yarden Y. Exp Cell Res. 2003;284(1):54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 137.Yarden Y, Sliwkowski MX. Nat Rev Mol Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 138.Bose S, Crane A, Hibshoosh H, Mansukhani M, Sandweis L, Parsons R. Hum Pathol. 2002;33(4):405–409. doi: 10.1053/hupa.2002.124721. [DOI] [PubMed] [Google Scholar]
- 139.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH. Cancer Biol Ther. 2004;3(8):772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 140.Hodgson JG, Chin K, Collins C, Gray JW. Breast Cancer Res Treat. 2003;78(3):337–345. doi: 10.1023/a:1023085825042. [DOI] [PubMed] [Google Scholar]
- 141.Mastracci TL, Shadeo A, Colby SM, Tuck AB, O’Malley FP, Bull SB, Lam WL, Andrulis IL. Genes Chromosomes Cancer. 2006;45(11):1007–1017. doi: 10.1002/gcc.20368. [DOI] [PubMed] [Google Scholar]
- 142.Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM. Proc Natl Acad Sci U S A. 1994;91(6):2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jonsson G, Staaf J, Olsson E, Heidenblad M, Vallon-Christersson J, Osoegawa K, de Jong P, Oredsson S, Ringner M, Hoglund M, Borg A. Genes Chromosomes Cancer. 2007;46(6):543–558. doi: 10.1002/gcc.20438. [DOI] [PubMed] [Google Scholar]
- 144.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 145.Akekawatchai C, Holland JD, Kochetkova M, Wallace JC, McColl SR. J Biol Chem. 2005;280(48):39701–39708. doi: 10.1074/jbc.M509829200. [DOI] [PubMed] [Google Scholar]
- 146.Li YM, Pan Y, Wei Y, Cheng X, Zhou BP, Tan M, Zhou X, Xia W, Hortobagyi GN, Yu D, Hung MC. Cancer Cell. 2004;6(5):459–469. doi: 10.1016/j.ccr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 147.Fejzo MS, Godfrey T, Chen C, Waldman F, Gray JW. Genes Chromosomes Cancer. 1998;22(2):105–113. doi: 10.1002/(sici)1098-2264(199806)22:2<105::aid-gcc4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 148.Sun J, Liu W, Adams TS, Sun J, Li X, Turner AR, Chang B, Kim JW, Zheng SL, Isaacs WB, Xu J. Prostate. 2007;67(7):692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 149.Kim EK, Yun SJ, Ha JM, Kim YW, Jin IH, Yun J, Shin HK, Song SH, Kim JH, Lee JS, Kim CD, Bae SS. Oncogene. 2011;30(26):2954–2963. doi: 10.1038/onc.2011.22. [DOI] [PubMed] [Google Scholar]
- 150.Hiramoto-Yamaki N, Takeuchi S, Ueda S, Harada K, Fujimoto S, Negishi M, Katoh H. J Cell Biol. 190(3):461–477. doi: 10.1083/jcb.201005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Strumane K, Rygiel T, van der Valk M, Collard JG. J Cancer Res Clin Oncol. 2009;135(1):69–80. doi: 10.1007/s00432-008-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Azzato EM, Pharoah PD, Harrington P, Easton DF, Greenberg D, Caporaso NE, Chanock SJ, Hoover RN, Thomas G, Hunter DJ, Kraft P. Cancer Epidemiol Biomarkers Prev. 19(4):1140–1143. doi: 10.1158/1055-9965.EPI-10-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]