Abstract
Background
Little is known about the structure and regulation of fungal α-L-rhamnosidase genes despite increasing interest in the biotechnological potential of the enzymes that they encode. Whilst the paradigmatic filamentous fungus Aspergillus nidulans growing on L-rhamnose produces an α-L-rhamnosidase suitable for oenological applications, at least eight genes encoding putative α-L-rhamnosidases have been found in its genome. In the current work we have identified the gene (rhaE) encoding the former activity, and characterization of its expression has revealed a novel regulatory mechanism. A shared pattern of expression has also been observed for a second α-L-rhamnosidase gene, (AN10277/rhaA).
Results
Amino acid sequence data for the oenological α-L-rhamnosidase were determined using MALDI-TOF mass spectrometry and correspond to the amino acid sequence deduced from AN7151 (rhaE). The cDNA of rhaE was expressed in Saccharomyces cerevisiae and yielded pNP-rhamnohydrolase activity. Phylogenetic analysis has revealed this eukaryotic α-L-rhamnosidase to be the first such enzyme found to be more closely related to bacterial rhamnosidases than other α-L-rhamnosidases of fungal origin. Northern analyses of diverse A. nidulans strains cultivated under different growth conditions indicate that rhaA and rhaE are induced by L-rhamnose and repressed by D-glucose as well as other carbon sources, some of which are considered to be non-repressive growth substrates. Interestingly, the transcriptional repression is independent of the wide domain carbon catabolite repressor CreA. Gene induction and glucose repression of these rha genes correlate with the uptake, or lack of it, of the inducing carbon source L-rhamnose, suggesting a prominent role for inducer exclusion in repression.
Conclusions
The A. nidulans rhaE gene encodes an α-L-rhamnosidase phylogenetically distant to those described in filamentous fungi, and its expression is regulated by a novel CreA-independent mechanism. The identification of rhaE and the characterization of its regulation will facilitate the design of strategies to overproduce the encoded enzyme - or homologs from other fungi - for industrial applications. Moreover, A. nidulans α-L-rhamnosidase encoding genes could serve as prototypes for fungal genes coding for plant cell wall degrading enzymes regulated by a novel mechanism of CCR.
Keywords: Aspergillus nidulans, Carbon catabolite repression, CreA-independent, Inducer exclusion, α-L-rhamnosidase, Transcriptional regulation
Background
The degradation of plant cell wall polysaccharides (i.e. cellulose, hemicellulose and pectins), and the subsequent utilization of their components as carbon sources is a key and highly regulated event when filamentous fungi grow on these substrates or infect plants. A number of those plant-derived substrates contain the neutral sugar L-rhamnose (6-deoxy-L-mannose) which is a component of the plant cell wall pectic polysaccharides rhamnogalacturonan I and rhamnogalacturonan II [1] and diverse secondary metabolites, including anthocyanins, flavonoids and triterpenoids (see reviews [2,3], and references therein). In addition, L-rhamnose (hereafter rhamnose) is present in animal tissues and in viruses, and is also a component of bacterial polysaccharides where it plays an important role in pathogenicity. The utilization, transformation or detoxification of these rhamnose-containing compounds involves different α-L-rhamnosidases (EC 3.2.1.40), which catalyze the hydrolysis of terminal non-reducing rhamnose residues in polysaccharides and α-L-rhamnosides including the artificial substrate p-nitrophenyl-α-L-rhamnopyranoside (pNPR) that is widely used to evaluate their activity. α-L-Rhamnosidases have been found in some plant and animal tissues as well as in a plethora of microorganisms including filamentous fungi (see reviews [2,3] and references therein). Based on primary sequence similarities these enzymes are classified within the Carbohydrate-Active Enzymes (CAZy) database [4] (http://www.cazy.org/) into three glycosyl hydrolase (GH) families. Whereas bacterial α-L-rhamnosidases are classified into either GH78 or GH106, fungal α-L-rhamnosidases (with the possible exception of the Aspergillus niger rhamnogalacturonan α-L-rhamnopyranohydrolase B [RgxB; EC 3.2.1.40] which is assigned to family GH28) belong to GH78.
α-L-Rhamnosidases are of considerable interest given their suitability in various applications within the food (e.g. citrus juice debittering, liberation of aromas and bioactive compounds), pharmaceutical (e.g. biotransformation of antibiotics and steroids), agro-/forestry (e.g. detoxification of rhamnose-conjugated plant secondary metabolites) and chemical industries (e.g. to produce rhamnose) ([2] and references therein). The biotechnological potentiality of these enzymes has led to the characterization of fourteen microbial α-L-rhamnosidase (GH78 family) encoding genes within the last decade (Additional file 1: Table S1). Of these only four were isolated from filamentous fungi: the genes encoding RhaA and RhaB of Aspergillus aculeatus [5], that encoding AkRha78 of Aspergillus kawachii [6] and the gene encoding the Aspergillus nidulans AN10277 protein [7], hereafter named rhaA.
In the presence of rhamnose as the sole carbon source, A. nidulans secretes at least one α-L-rhamnosidase of molecular mass ~102 kDa. Its biochemical properties make it suitable for oenological applications [8,9] as well as for improvement of the bioavailability of plant rhamnose-containing bioactive compounds. While this enzyme is active against naringin and hesperidin (major rhamnosylated flavonoids in citrus juices in which the rhamnose molecule is bound to glucose by α-(1,2) or α-(1,6) linkages respectively) and also the synthetic substrate pNPR (in which the aglycon is directly linked to the C1 position of rhamnose) [8,9], recombinant AN10277-His6/RhaA-His6 was not [7], strongly suggesting that these are two distinct enzymes. At least eight loci putatively encoding α-L-rhamnosidases have been annotated in the A. nidulans genome the products of which range from 62 to 169 kDa [10,11]. The locus encoding the α-L-rhamnosidase of oenological interest is however unknown.
The regulation of α-L-rhamnosidase genes is less well documented than that of many other genes encoding glycosyl hydrolases. It has been reported [12] that the Lactobacillus plantarum ram1 and ram2 genes are induced by rhamnose and repressed by D-glucose (hereafter glucose). However, the regulatory circuits controlling fungal α-L-rhamnosidase gene expression are unknown. In filamentous fungi it has been shown that the synthesis of a number of plant cell wall-degrading enzymes is regulated at the level of transcription i.e. induced when the plant polysaccharide or its degradation products are present and repressed by glucose (see reviews [13,14]). Glucose repression is a well-studied regulatory circuit in A. nidulans (reviewed in [15,16]), and the role of the wide-domain transcriptional repressor CreA in the expression of a number of genes encoding plant cell wall degrading enzymes has been widely characterized. Our previous studies [8] have shown that α-L-rhamnosidase activity in A. nidulans is induced by rhamnose and repressed by glucose. However, unlike other glycosyl hydrolase systems, relief of repression by glucose either by cultivation in the presence of the inducer and a so-called non-repressing carbon source (e.g. D-lactose or L-arabinose) or by the use of the creAd30 derepressed strain [17] was not sufficient for α-L-rhamnosidase production to be totally derepressed. It was therefore suggested that carbon catabolite repression (CCR) of A. nidulans α-L-rhamnosidase genes could involve a CreA-independent mechanism [8].
Here we show that rhamnose and glucose regulate α-L-rhamnosidase production at the transcriptional level in the model filamentous fungus A. nidulans. We have identified the A. nidulans gene encoding the previously characterized α-L-rhamnosidase [8,9] and confirmed that glucose repression of its expression, as well as that of the gene coding for the α-L-rhamnosidase AN10277/RhaA (rhaA), occurs by means of a CreA-independent mechanism. The data suggest that glucose prevents the entry of the inducing carbon source rhamnose into the cell and that this phenomenon does not involve CreA function. We thus hypothesize the existence of an inducer exclusion mechanism by which fungal α-L-rhamnosidase genes are controlled. In this regard, the lack of protection against the toxicity of the non-metabolizable glucose analogue 2-deoxy-D-glucose by a range of rhamnose concentrations suggesting that inducer exclusion is not due to competition for uptake of the inducer. Moreover, α-L-rhamnosidase gene expression was found to be repressed by sugars previously considered non-repressive as well as by some non-sugar carbon sources (e.g. ethanol). In contrast to glucose, ethanol did not prevent the uptake of rhamnose. Hence, these findings provide evidence for the existence of novel molecular mechanisms for exerting CCR on genes encoding plant cell wall degrading enzymes in filamentous fungi which could be of utility in modulating α-L-rhamnosidase production.
Results and discussion
The A. nidulans locus AN7151 encodes an α-L-rhamnosidase of potential oenological relevance
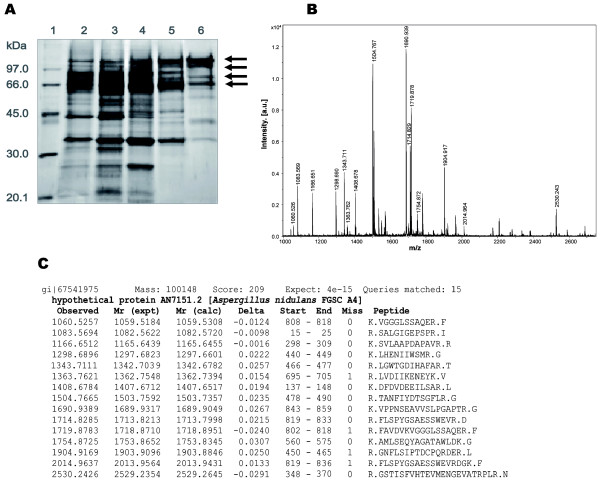
With a view to identifying the A. nidulans gene encoding the α-L-rhamnosidase previously characterized [8,9], we determined the sequence of this protein. α-L-Rhamnosidase was purified from a culture filtrate of the A. nidulans wild-type strain (biA1) grown on 1% w/v rhamnose as the sole carbon source. The purification scheme [9] mainly consisted of two anion and one cation exchange chromatographic steps. Protein samples from each stage of the purification were resolved using SDS-PAGE (Figure 1A). Four major bands of ~66 to > 90 kDa were finally observed and processed for mass fingerprint analysis using MALDI-TOF mass spectrometry. Figure 1B shows the MALDI-TOF mass spectrum of a tryptic digest of one selected band. Representative peptide mass fingerprints for fifteen of the peptides are shown (Figure 1C). Comparison of the mass spectrometry data with the ORFs predicted in the A. nidulans genome (http://www.broad.mit.edu) identified locus AN7151, designated rhaE, as the encoding gene. The majority of the peptide sequences obtained from the four SDS-PAGE bands analyzed were identical, suggesting that these bands arose from proteolysis and/or deglycosylation of the purified enzyme.
Figure 1.
Identification of the A. nidulans α-L-rhamnosidase gene by MALDI-TOF mass spectrometry. (A) Silver stained SDS-PAGE of the α-L-rhamnosidase enriched fractions selected after each purification step. Lane 1, molecular mass markers; lane 2, crude extract (12 μg); lane 3 DEAE Sephadex A-50 (fraction 2, 43 μg); lanes 4 and 5, Q-Sepharose Fast Flow (fractions 20-22 and fractions 20-23 from two consecutive chromatographies, 17.5 and 3.5 μg, respectively); lane 6, Resource S (fractions 14-18; 2.5 μg). Molecular masses (kDa) are shown in the left margin. (B) Representative MALDI-TOF spectrum of one of the indicated protein bands (arrowed) from panel A. (C) Identification details for the AN7151 encoded protein from MASCOT searches.
Gene structure of A. nidulans rhaE
Using RT-PCR a full-length cDNA corresponding to the ORF of the A. nidulans rhaE gene was generated and sequenced. Comparison of the cDNA sequence with the genomic sequence (Additional file 2: Figure S1) revealed the presence of two introns of 59 and 51 nucleotides which followed the canonical GT-AG splicing rule (E357/V358 and N504/Y505 within the bacterial alpha-L-rhamnosidase domain - Pfam PF05592 - of the A. nidulans predicted protein). The second intron is in-frame which explains its absence in automatic annotation of the gene. Glycosyl hydrolase genes are frequently found to be subjected to CCR and have CreA consensus motifs upstream of the ORF. Sequence analysis of the 1 kb sequence upstream of the proposed translational start site revealed the presence of 8 CreA (5'-SYGGRG-3') consensus target sites [18-20], 6 of them being located within the first 360 bp, suggesting that the transcription of rhaE could be directly repressed by CreA. However, it has previously been shown that the production of this α-L-rhamnosidase is repressed predominantly via a CreA-independent mechanism [8] which would indicate that the putative CreA targets in rhaE are not functional. Such a circumstance would not be without precedent for genes encoding plant cell wall degrading enzymes [21].
The polypeptide encoded by rhaE is 861 amino acids (aa) long and has a calculated molecular mass of 95205 Da. This value is in excellent agreement with the estimated molecular mass of the deglycosylated form of the purified α-L-rhamnosidase. The glycosylated form had a molecular mass of 102 kDa of which 7% was contributed by carbohydrates [9]. Intriguingly, while the A. nidulans α-L-rhamnosidase activity was detected and purified from the culture medium, the predicted RhaE protein appears to lack an N-terminal signal peptide (SignalP probability 0.081), suggesting that secretion of this enzyme may occur through a non-classical pathway [22]. In this regard, subcellular localization prediction using the SecretomeP program supports (NN/SecP score 0.608) this possibility.
The crystal structure of Bacillus sp. GL1 α-L-rhamnosidase RhaB has been resolved, and mutational studies have shown that four acidic residues (D567, E572, D579 and E841) are crucial for catalysis and/or substrate binding [23]. All four residues (D458, E464, D471 and E737) are conserved in the A. nidulans RhaE protein (Additional file 3: Figure S2) and are hence candidates for roles of functional importance.
Phylogenetic relationships between the A. nidulans RhaE and class A bacterial α-L-rhamnosidases
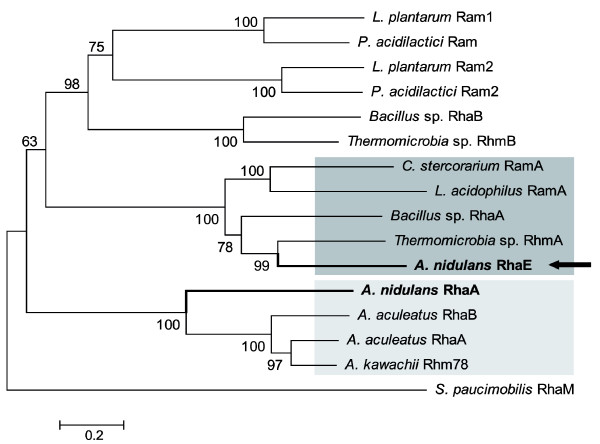
A phylogenetic tree based on aa sequence homologies was constructed for A. nidulans RhaE and those GH78 proteins encoded by experimentally verified genes (Figure 2). Alignment (ClustalW) was carried out with the program MEGA 3 [24] and the Neighbour-Joining algorithm with bootstrap values from 1000 trials was utilized. Interestingly, A. nidulans RhaE resides in a well-defined clade (boostrap value 100) containing the four class A bacterial α-L-rhamnosidases (i.e. Clostridium stercorarium RamA, Lactobacillus acidophilus RamA, Bacillus sp. GL1 RhaA and Thermomicrobia sp. PRI-1686 RhmA) whereas all other fungal α-L-rhamnosidases, including A. nidulans RhaA, segregated together in a highly divergent branch. This fungal cluster contains shorter enzymes (597-661 aa residues) whereas enzymes of the RhaE cluster are at least 200 residues longer (861-932 aa).
Figure 2.
Neighbour-Joining tree showing the phylogenetic relationships of the A. nidulans rhamnosidases RhaA and RhaE. MEGA3 software [24] was used to carry out the phylogenetic analysis. Amino acid sequences were aligned and the dendogram was determined using ClustalW and the Neighbour-Joining algorithm. Bootstrap values adjacent to each internal node represent percentages of 1000 replicates. The scale bar represents aa replacements per site. Sequences from the following UniProtKB database accessions were included: A. aculeatus Q9C1M9 (RhaA), A. aculeatus Q9HFW5 (RhaB), A. kawachii A9ZT55 (Rhm78), A. nidulans C8VMJ6 (AN10277/RhaA), Bacillus sp. GL1 Q93RE8 (RhaA), Bacillus sp. GL1 Q93RE7 (RhaB), C. stercorarium Q9S3L0 (RamA), L. acidophilus Q5FJ31 (RamA), L. plantarum Q88SF8 (Ram1), L. plantarum Q88SF6 (Ram2), Pediococcus acidilactici EONEV1 (Ram), P. acidilactici EONEK0 (Ram2), Thermomicrobia sp. PRI-1686 Q93RE8 (RhmA) and Thermomicrobia sp. PRI-1686 Q93RE7 (RhmB). Sphingomonas paucimobilis RhaM (Q76LC4), a GH106 rhamnosidase, was used to root the tree.
Conservation of RhaE among fungi
As RhaE is the first eukaryotic enzyme with structural similarity to bacterial α-L-rhamnosidases, we screened the fungal genome databases to see whether RhaE homologs could be found in other fungal species. BLASTP [25] searches using the RhaE sequence as a query confirmed its conservation in both the Ascomycota and Basidiomycota (Table 1). The most similar ortholog (69% identity over 869) was found in Aspergillus terreus and could correspond to the oenological α-L-rhamnosidase previously purified [26]. Orthologs with at least 50% identity were identified in Aspergillus flavus, Aspergillus oryzae, Magnaporthe grisea, Nectria haematococca, Fusarium graminearum, Fusarium verticilloides and Fusarium oxysporum. Less similar proteins (40-49% identity) were found in Verticillium albo-atrum, Penicillium chrysogenum, Aspergillus niger, Neosartorya fischeri, Tuber melanosporum, Penicillium marneffei, Aspergillus fumigatus, Stagonospora nodorum, the hemiascomycete yeast Clavispora lusitaniae and the Basidiomycetes Cryptococcus neoformans and Cryptococcus gatii. The A. nidulans genome specified two additional loci (AN12368 and AN11954) that could encode paralogs of RhaE (48% and 45% identity over 869 and 863 aa respectively). Additional studies are required to determine the catalytic activities of their products.
Table 1.
Selected fungal homologs of the A. nidulans AN7151/RhaE rhamnosidase
| Organism |
Accession no./Reference (NCBI/Broad Institute*) |
E value |
% Identity/% Similarity/ Aligned amino acids |
| ASCOMYCOTA | |||
| Eurotiomycetes | |||
| Aspergillus terreus | XP 001212100 | 0.0 | 69/81/869 |
| Aspergillus oryzae | XP 001827536 | 0.0 | 57/70/867 |
| Aspergillus flavus | XP 002384747 | 0.0 | 56/69/869 |
| Penicillium chrysogenum | XP 002560426 | 0.0 | 46/62/889 |
| Aspergillus nidulans | XP 658315 | 0.0 | 48/63/869 |
| Aspergillus niger | XP 001402266 | 0.0 | 45/62/893 |
| Aspergillus nidulans | XP 662910 | 0.0 | 45/62/863 |
| Neosartorya fischeri | XP 001258340 | 0.0 | 46/61/873 |
| Penicillium marneffei | XP 002152078 | 0.0 | 47/64/807 |
| Aspergillus fumigatus | XP 751335 | 0.0 | 43/59/886 |
| Dothideomycetes | |||
| Stagonospora nodorum | SNOG_14400* | 0.0 | 40/58/885 |
| Sordariomycetes | |||
| Magnaporthe grisea | XP 365490 | 0.0 | 54/70/876 |
| Nectria haematococca | XP 003041639 | 0.0 | 55/71/863 |
| Fusarium verticillioides | FVEG_12337* | 0.0 | 54/70/866 |
| Fusarium oxysporum | FOXG_13192* | 0.0 | 53/70/866 |
| Fusarium graminearum | FGSG_07974* | 0.0 | 53/69/869 |
| Verticilium albo-atrum | XP 003009324 | 0.0 | 49/65/867 |
| Pezizomycetes | |||
| Tuber melanosporum | XP 002839085 | 0.0 | 45/63/853 |
| Saccharomycetes | |||
| Clavispora lusitaniae | XP 002617481 | 4e-165 | 40/57/863 |
| BASIDIOMYCOTA | |||
| Tremellomycetes | |||
| Cryptoccocus neoformans | XP 571766 | 0.0 | 42/58/876 |
| Cryptoccocus gattii | CNBG_5170 | 0.0 | 43/61/769 |
Functional expression of A. nidulans RhaE α-L-rhamnosidase in S. cerevisiae
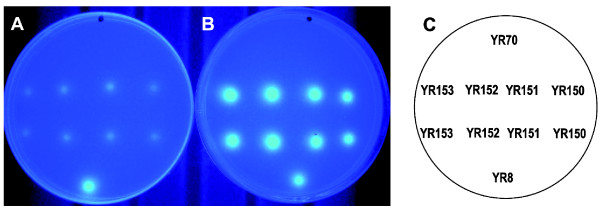
To further confirm that the product of rhaE has α-L-rhamnosidase activity, its cDNA was subcloned in a high copy number yeast vector (YEplac195) between the TDH3 promoter and the PGK1 terminator. The construct was used to transform the S. cerevisiae wine yeast strain T73-4. Uracil prototrophic transformants (T73-4, YEp195_RhaE) were obtained and grown on selective plates containing the artificial substrate 4-methylumbelliferyl α-L-rhamnopyranoside (MUR). Expression of rhaE in S. cerevisiae resulted in hydrolysis of MUR (Figure 3), confirming that it effectively encodes an α-L-rhamnosidase. As expected, no activity against MUR was found in a transformant (YR70) obtained with the original shuttle vector (T73-4, YEp195). Transformants YR150-YR153 containing the expression cassette for α-L-rhamnosidase were thus isolated. Evidence that in S. cerevisiae RhaE was not efficiently secreted to the media includes the observation that most of the activity in intact cells was observed after permeabilization of the cell walls with chloroform vapour (Figure 3B).
Figure 3.
in vivo detection of recombinant α-L-rhamnosidase activity. S. cerevisiae cells were grown at 30°C for about 5 h on SD plates containing the artificial substrate MUR. Differences in α-L-rhamnosidase activity between non permeabilized (A) and permeabilized (B) cells are shown. The names of the strains appear in panel C. Representative transformants (YR150-YR153) expressing the A. nidulans rhaE gene were tested. Strains harbouring an empty vector (YR70) or a plasmid expressing the A. aculeatus rhaA gene (YR8; [27]) were included as controls.
As RhaE could be secreted in A. nidulans via a non-classical mechanism whereas most of the activity in S. cerevisiae seems to be entrapped, we examined the cellular location of the recombinant enzyme in S. cerevisiae. In general, fungal glycosyl hydrolases bearing a secretion signal peptide in their aa sequences, including the α-L-rhamnosidase RhaA of A. aculeatus, are efficiently secreted when expressed in S. cerevisiae [27]. Transformant YR150 was cultured for 72 h and aliquots were taken every 24 h. Table 2 shows that at most 17% of the total activity was extracellular or cell-wall bound at any stage of the cultivation whereas intracellular activity was high throughout. In a similar way, the location of RhaE was predominantly intracellular in transformant YR151 (88.1 ± 1.4% intracellular, 8.0 ± 1.3% cell wall bound and 3.8% ± 0.6 extracellular at 72 h). These data suggest that cis determinants leading to secretion of RhaE in A. nidulans are not functional in S. cerevisiae.
Table 2.
Location of RhaE rhamnosidase activity in recombinant S. cerevisiae
| Time (h) | OD600 | Total activity (U ml-1) | % Intracellular | % Cell wall | % Extracellular |
|---|---|---|---|---|---|
| 24 | 15.1 ± 0.4 | 0.6 ± 0.02 | 90.9 ± 0.5 | 6.0 ± 0.3 | 3.1 ± 0.2 |
| 48 | 22.9 ± 0.1 | 1.2 ± 0.1 | 82.9 ± 1.9 | 11.2 ± 1.2 | 5.9 ± 0.7 |
| 72 | 23.0 ± 0.4 | 1.4 ± 0.3 | 82.9 ± 2.3 | 10.7 ± 1.5 | 6.4 ± 0.9 |
Rhamnose induction of A. nidulans α-L-rhamnosidase gene expression is subject to carbon catabolite repression independent of CreA
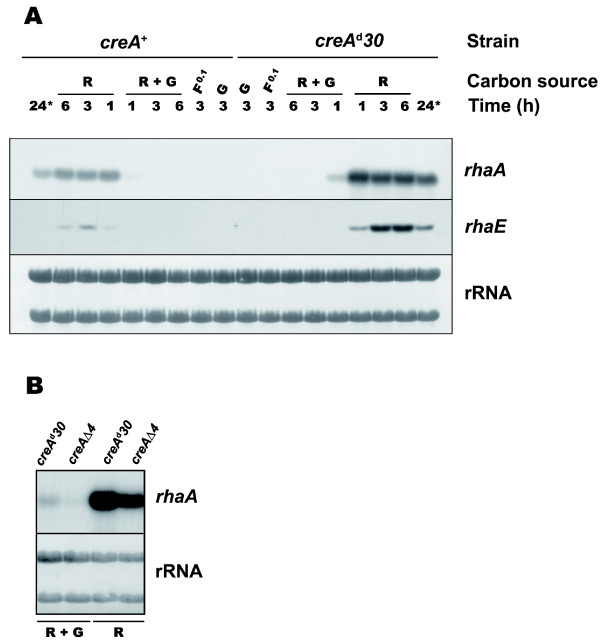
We have previously reported that α-L-rhamnosidase activity in A. nidulans is induced by rhamnose and repressed by glucose, and that the glucose repression was not relieved by the creAd30 mutation [8]. Thus we sought to assess whether the results obtained at the enzyme activity level correlated with those of rhaE mRNA levels. In addition, we also assessed whether the other A. nidulans α-L-rhamnosidase gene rhaA [7] co-regulated with rhaE. It should be mentioned that the artificial substrate pNPR was employed to evaluate α-L-rhamnosidase production in A. nidulans, therefore the activity of RhaA was not detected under our original experimental conditions [8,9]. To investigate the role of CreA in the expression of rhaE and rhaA, northern analyses were performed using both A. nidulans wild-type (creA+, biA1) and a strongly derepressed mutant (creAd30, biA1) [17]. Total RNAs were isolated from D-fructose grown (0.1% w/v) mycelia transferred in parallel to rhamnose (1% w/v) media containing or lacking glucose (1% w/v), as well as to media containing either D-fructose (0.1% w/v) or glucose (1% w/v) alone. RNAs were also obtained from mycelia grown for 24 h in rhamnose (1% w/v) without transfer. Figure 4A shows that rhaE and rhaA mRNAs accumulated upon transfer of the wild-type strain to 1% w/v rhamnose but not in mycelia transferred to 0.1% w/v D-fructose (the pre-growth condition) suggesting that rhamnose is required for induction. rhaE transcript was detected within 1 h after transfer and accumulated up to 3 h and then decreased; rhaE transcript was only observed after 24 h growth in rhamnose upon prolonged exposure of the blot (not shown). Likewise, induction of rhaA occurs within 1 h but the transcript level remained constant up to 6 h after transfer; less rhaA mRNA was detected after 24 h of growth on rhamnose. Induction of the α-L-rhamnosidase genes thus occurs rapidly after transfer, when the external rhamnose concentration is high (~1%), implying that this does not result from carbon source limitation. Rhamnose-induced expression of both genes in the creA+ strain was totally repressed in media containing both rhamnose and glucose or glucose alone (Figure 4A). These results demonstrate that both induction and repression of these genes occurs at the transcriptional level and suggest that they are co-regulated, though RhaA appeared less susceptible to modulation after the initial response to rhamnose.
Figure 4.
Glucose repression of A. nidulans rhamnosidase genes occurs via a CreA-independent mechanism. (A) Expression levels of rhaA and rhaE in wild-type (creA+) and creAd30 strains during cultivation on 1% w/v rhamnose (R), 0.1% w/v D-fructose (F0.1), 1% w/v glucose (G) and 1% w/v rhamnose + 1% w/v glucose (R + G) for the periods indicated. The asterisk (*) indicates growth in 1% w/v rhamnose without transfer. (B) Expression levels of rhaA in creAd30 and creAΔ4 strains during cultivation on 1% w/v rhamnose (R) and 1% w/v rhamnose + 1% w/v glucose (R + G) for 1 h after transfer. rRNA is shown as a loading control.
In the derepressed creAd30 strain (Figure 4A) induction of rhaE and rhaA likewise occurred within 1 h of transfer but transcript accumulation was considerably greater than in the wild-type. Contrary to the wild-type, the level of rhaE remained stable for at least 6 h after transfer. Interestingly, under inducing-repressing conditions (i.e. 1% w/v rhamnose + 1% w/v glucose), expression of rhaE was totally repressed, whilst a low transient expression of rhaA was observed 1 h after transfer. That glucose repressed the expression of rhaE and rhaA in both creA+ and creAd30 genetic backgrounds indicate the lack of a role for CreA in glucose repression of these genes. Moreover, the creAd30 mutation did not override the need for rhamnose for induction since transcripts were not observed in mycelia grown in 0.1% w/v D-fructose. Interestingly, in the creA derepressed mutant the transcript levels of rhaE and rhaA under inducing (1% w/v rhamnose) conditions were considerably greater than those seen in the wild-type indicating that some derepression occurs when functional CreA is absent and hence suggesting a role for CreA in rhaE and rhaA expression under inducing conditions. The presence of putative CreA binding sites in the rhaA and rhaE promoters suggesting direct repression by CreA under certain growth conditions.
To confirm the results with the creAd30 mutant, we also studied the expression of rhaA in a creA null mutant (strain creAΔ4). Figure 4B shows that glucose repression of rhaA expression was not relieved by the creAΔ4 mutation, strongly indicating the absence of second-site mutations that could have contributed to the repressed phenotype of the creAd30 strain, and thus leads weight to the hypothesis of glucose repression of the rha genes by a CreA-independent mechanism.
In filamentous fungi numerous genes encoding plant cell wall degrading enzymes have been shown to be subjected to CCR regulated by CreA/Cre1 ([14] and references therein) whereas there are few indications for the existence of CreA-independent mechanisms. Comparison of the levels of expression of reporter constructs in both creA+ and creAd30 backgrounds suggested the existence of both CreA-dependent and CreA-independent mechanisms of CCR regulating A. nidulans xylanolytic genes [21,28]. The occurrence in A. nidulans of a CCR mechanism independent of CreA has also been suggested for the pectinolytic gene pelA [29]. The regulation of the rhaA and rhaE genes shows similarities to that seen for pelA in a derepressed strain (creA204) in that the transcripts accumulated to higher levels than in the wild-type when grown under inducing conditions (1% w/v polygalacturonic acid) while glucose repression was not relieved by the loss-of-function creA204 mutation. It is possible that the same CreA-independent mechanism regulates these different systems.
Effect of other carbon sources on α-L-rhamnosidase gene induction
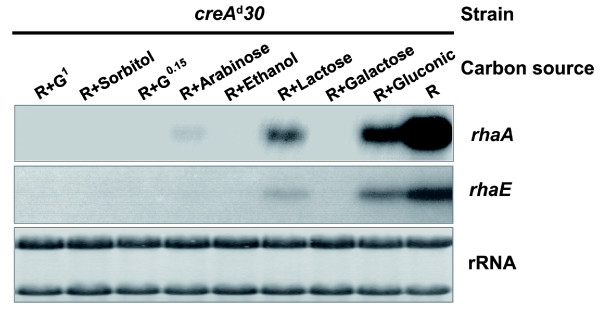
In A. nidulans, L-arabinose, ethanol, glycerol and D-lactose are considered to be non-repressing carbon sources whereas glucose and D-xylose are strongly repressing [30]. Concentration is also relevant since at low concentrations the repression by glucose decreases, suggesting that CCR is related to growth rate [31]. We have shown that simultaneous addition of rhamnose with glucose, glycerol, ethanol, L-arabinose or D-lactose resulted in a marked decrease in α-L-rhamnosidase activity [8]. We therefore studied how different carbon sources affect α-L-rhamnosidase gene expression in the creAd30 mutant. Northern blot analysis was used to study the expression of rhaE and rhaA in 1% w/v rhamnose-containing media supplemented with equimolar amounts of alternative carbon sources, i.e. D-gluconic acid, D-galactose, D-lactose, ethanol, L-arabinose and D-sorbitol as well as two different concentrations of glucose (0.15 and 1% w/v). The results are shown in Figure 5. The expression profiles of both genes were similar and whilst D-gluconic acid and D-lactose permitted induction of rhaE and rhaA when mycelia were transferred from 0.1% w/v D-fructose to the inducing media (1% w/v rhamnose) containing these additional carbon sources, they still exerted some degree of repression. A very low level of rhaA transcript was also observed in L-arabinose supplemented media. By contrast, D-galactose, ethanol, D-sorbitol and glucose (at high and low concentrations) totally prevented rhamnose induction of both genes. Similarly, only D-lactose and D-gluconic acid allowed rhamnose induction of both genes in the wild-type strain (data not shown). Thus, the sugar classification for CreA-mediated repression does not hold for the CreA-independent repression of α-L-rhamnosidase genes. L-Arabinose, ethanol, D-sorbitol and 0.15% w/v glucose, previously considered less repressive or even 'neutral' carbon sources, all greatly repress rhaA and rhaE transcription.
Figure 5.
Repression of rhamnosidase genes in the presence of rhamnose supplemented with different carbon sources. Northern blot analysis of the expression of the rhaA and rhaE genes in the creAd30 strain growing in the presence of rhamnose (R) and different carbon sources. Pregrown mycelia were transferred to inducing medium (1% w/v rhamnose; R) either with or without an equimolar amount of another carbon source and harvested after 3 h of incubation. G1 glucose 1% w/v, G0.15 glucose 0.15% w/v. rRNA is shown as a loading control.
Our previous results studying α-L-rhamnosidase production [8] coincide with the current data from northern analyses (Figures 4 and 5) and clearly show that CreA is not solely responsible for CCR of rhaE and rhaA gene expression, implying that α-L-rhamnosidase genes could be regulated via another repressor(s). It is possible for such a regulator to act directly on α-L-rhamnosidase gene promoters and/or indirectly by repression of the gene(s) encoding rhamnose permeases, rhamnose catabolic enzymes and/or rhamnose pathway specific activator(s). In S. cerevisiae it has been shown that glucose repression is mediated in part by the CreA related protein Mig1 and by two homologs (Mig2 and Mig3) which share almost identical zinc fingers and bind similar sites with different affinities [32,33]. In A. nidulans and other filamentous fungi only one carbon catabolite repressor CreA has been identified. BLASTP searches using the CreA protein sequence or the sequence of its two zinc fingers failed to find any clear alternative candidate within the zinc-finger family.
Glucose repression of rhaA and rhaE correlates with the lack of uptake of rhamnose
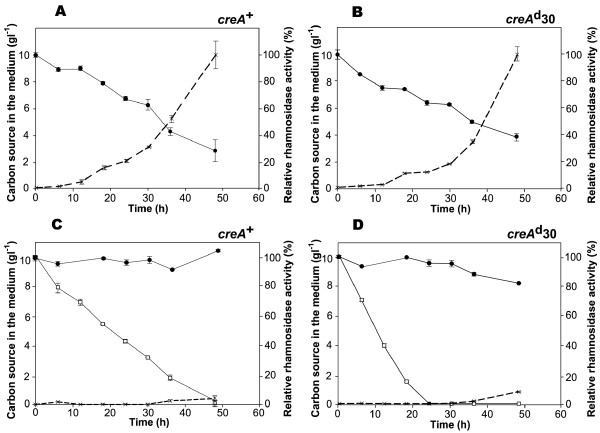
One possible mechanism by which glucose could prevent the induction of A. nidulans α-L-rhamnosidase genes would be by reducing the uptake of the inducing sugar rhamnose. To investigate this possibility A. nidulans wild-type (creA+) and mutant (creAd30) strains were grown for 18 h in the presence of 0.1% w/v D-fructose and mycelia were subsequently transferred to media containing either rhamnose (1% w/v) alone or a mixture of rhamnose (1% w/v) and glucose (1% w/v). Time-course measurements were made of the amounts of the carbon sources remaining in the culture filtrates (determined by high-performance liquid chromatography) and extracellular α-L-rhamnosidase activity (Figure 6). In the same way that the expression profiles of rhaA and rhaE were similar in the creA+ and creAd30 strains (Figures 4 and 5), the sugar utilization patterns of both strains were quite similar too. When grown in rhamnose as sole carbon source, consumption of this sugar was detectable after 8 h and its concentration decreased more or less linearly with time in both creA+ and creAd30 strains (Figure 6A and 6B respectively). After 48 h of growth about 30% (creA+) or 40% (creAd30) of the initial rhamnose was still available, demonstrating the paucity of this sugar as a carbon source compared to glucose which was consumed at a much greater rate and was exhausted after 48 h (Figure 6C and 6D). α-L-Rhamnosidase activity increased in parallel with rhamnose consumption, its initial detection being around 12 h after transfer.
Figure 6.
Inhibition of rhamnose uptake by glucose. Time-course measurements of rhamnosidase activity and the amount of sugar remaining in the cultures during cultivation of the A. nidulans wild-type (creA+; panels A and C) and the carbon catabolite derepressed creAd30 (panels B and D) strains in MM containing either 1% w/v rhamnose (panels A and B) or a mixture of 1% w/v rhamnose and 1% w/v glucose (panels C and D). Rhamnosidase activities are presented as percentages of that observed at 48 h under inducing conditions. The concentrations of rhamnose and glucose in the growth media over time are indicative of the ability to take up these monosaccharides. Symbols: (•) concentration of rhamnose, (□) concentration of glucose, and (X) rhamnosidase activity. Data points are represented as the mean and standard deviation of three independent experiments.
In the mixed sugar cultures glucose was consumed preferentially compared to rhamnose in the wild-type strain and no α-L-rhamnosidase activity was detected (Figure 6C). Hence, a key event in the repression of α-L-rhamnosidase synthesis appears to be the lack of uptake of rhamnose. In concordance with the northern results (see above), CreA does not play a major role in this mechanism of repression since the uptake of rhamnose in the creAd30 strain was blocked by the presence of glucose, rhamnose only being taken up after the exhaustion of glucose, only after which α-L-rhamnosidase activity could be detected (Figure 6D).
Taken together, these results provide strong indications for the existence of a CreA-independent mechanism of 'inducer exclusion' by which the expression of α-L-rhamnosidase genes, and possibly other genes induced by rhamnose, is regulated. Whether rhamnose permeases are repressed by glucose at the transcriptional level by a factor other than CreA, or controlled post-transcriptionally (e.g. catabolite inactivation of the proteins effecting rhamnose uptake) or whether the rhamnose transport system(s) are competed by glucose - and possibly other carbon sources - is not known at present.
By way of antecedents, glucose repression of the A. nidulans proline catabolic cluster genes prnD and prnC is known to result from inducer exclusion since only the transcription of the proline permease gene (prnB) is directly repressed by CreA [34]. CreA-independent mechanisms of inducer exclusion have also been documented, an example of which is the phenylacetic acid (PhAc) uptake system: in the presence of glucose and PhAc, the PhAc transport system is absent in the wild-type as well as in the creAd30 mutant [35]. Finally, inducer exclusion arising from competitive inhibition of D-xylose uptake by the structurally related sugar glucose has also been suggested to explain at least part of the glucose repression of the xylanolytic gene xlnA [28,36]. With regard to the later however, two pieces of data cast doubt on competitive inhibition of inducer uptake as the means of effecting inducer exclusion of rhamnose. Firstly, α-L-rhamnosidase expression was completely repressed by both D-sorbitol and ethanol (Figure 5), carbon sources that are structurally distinct and unrelated to rhamnose. Secondly, the transient rhaA response to a mixture of glucose and rhamnose (Figure 4A) suggests that some rhamnose is taken up by the creAd30 mycelia immediately after medium transfer despite the presence of equimolar amounts of glucose; in the alc system the response to induction has been shown to be much faster than the establishment of CCR [36].
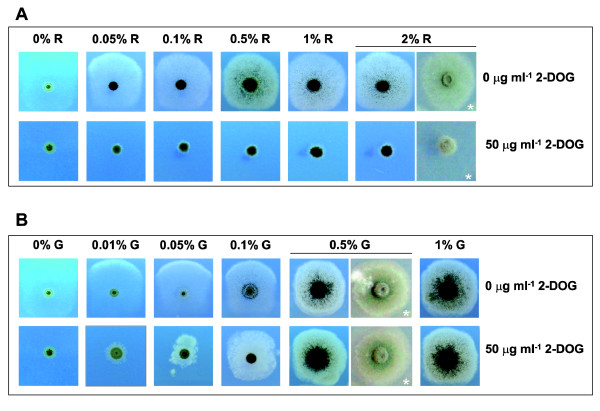
In A. nidulans, it is known that glucose and its non-metabolizable analogue 2-deoxy-D-glucose (2-DOG) are actively taken up by the same transporter(s) [37,38]. Therefore, one easy way to get an idea of whether rhamnose and glucose can compete at the uptake level is to see whether rhamnose can protect against the toxicity of the 2-DOG. Figure 7A shows that increasing amounts of rhamnose (from 0.05 to 2% w/v, equivalent to 2.75 mM to 110 mM respectively) did not reverse the toxicity of 50 μg ml-1 of 2-DOG (0.3 mM). As expected, glucose protection of 2-DOG toxicity was clearly observed under the same experimental conditions (Figure 7B). This result further supports the possibility of glucose effecting rhamnose uptake by repression and/or catabolite inactivation of a transporter(s) instead by competitive inhibition. Experiments are now in progress to characterize the molecular mechanism(s) by which glucose inhibits rhamnose induction in A. nidulans.
Figure 7.
Rhamnose does not protect from 2-DOG toxicity. Conidia-derived colonies 3 days after inoculation of the A. nidulans reference strain (biA1) on MM plates containing different amounts of rhamnose (R) (panel A) or glucose (G) (panel B) and supplemented (or not) with 50 μg ml-1 of 2-DOG. Photos - except those indicated with an asterisk - were taken against daylight to appreciate better colonies with low hyphal densities.
Repression by ethanol does not result from the lack of rhamnose transport
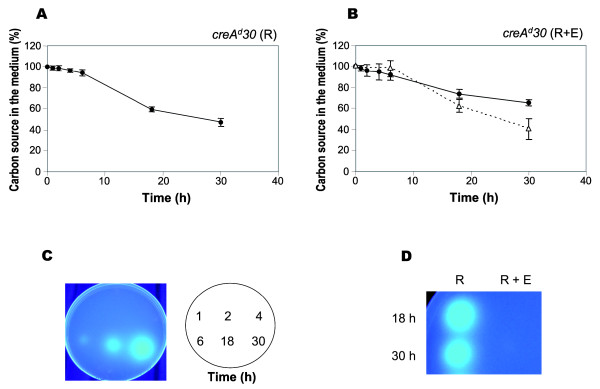
Bearing in mind the possibility that other carbon sources might negatively affect rhamnosidase gene induction through different mechanisms to that seen for glucose, the effect of ethanol in the uptake of rhamnose was assessed. The creAd30 strain was grown for 18 h in the presence of 0.1% w/v D-fructose and subsequently transferred to media containing either rhamnose (1% w/v, equivalent to 55 mM) alone or a mixture of rhamnose and ethanol (55 mM each), and the rates of disappearance of rhamnose from each transfer medium along the time were compared. In contrast to the sequential utilization of glucose and rhamnose (Figure 6), simultaneous disappearance of rhamnose and ethanol was observed (Figure 8B), albeit the rate of removal of rhamnose from the media is slightly faster in the absence of ethanol (Figure 8A). Extracellular rhamnosidase activities of these cultures were tested on MUR plates. Accordingly to the northern analyses (Figure 5), in the rhamnose-ethanol media rhamnosidase activity was undetected (Figure 8D) despite rhamnose was being taken up. In the absence of ethanol hydrolysis of MUR became detectable at 6 h (Figure 8C). These results indicate that the mechanism by which ethanol prevents rhamnosidase gene induction and consequently rhamnosidase activity is other than the transport step and thus, CCR of the rhamnosidase system can be achieved by different regulatory mechanisms.
Figure 8.
Co-utilization of rhamnose and ethanol. Time-course measurement of the amount of rhamnose or ethanol remaining in the cultures during cultivation of the A. nidulans creAd30 strain in MM containing either 1% w/v rhamnose (R) (Panel A) or a mixture of 1% w/v rhamnose and an equimolar amount of ethanol (E) (Panel B). Symbols: (•) concentration of rhamnose, (Δ) concentration of ethanol. Data points are represented as the mean and standard deviation of three independent experiments. α-L-rhamnosidase activity was tested by spotting 10 μl of the same cultures on agar plates containing MUR. (Panel C) α-L-rhamnosidase activity after transfer to the rhamnose media at the times indicated. (Panel D) A sector of a Petri dish showing the lack of α-L-rhamnosidase activity in the mixture cultures.
Conclusions
Using A. nidulans as a model system, we show for the first time that the production of α-L-rhamnosidases in eukaryotes is controlled at the transcriptional level by the carbon source (i.e. induction by L-rhamnose, and repression by glucose and other carbon sources). Interestingly, two loss-of-function mutations in the creA gene, which encodes the carbon catabolite repression (CCR) factor CreA, resulted in overproduction of rhaA and rhaE mRNAs by cells grown in medium containing L-rhamnose as sole carbon source, but had little or no effect on rhaA and rhaE mRNA levels in cells grown with glucose plus L-rhamnose as carbon source. We also show that the CreA-independent mechanism of glucose repression correlates with the absence of L-rhamnose uptake. A. nidulans α-L-rhamnosidase encoding genes could thus serve as a prototype for fungal genes regulated by a novel inducer exclusion, CreA-independent, mechanism of CCR.
We have also identified the gene (rhaE) coding for an α-L-rhamnosidase of oenological relevance that we characterized previously. Phylogenetic analysis shows that this enzyme is the first eukaryotic α-L-rhamnosidase found to be closely related to bacterial rhamnosidases of class A (i.e. Thermomicrobia sp. RhmA, Bacillus sp. RhaA, Clostridium stecolarium RamA and Lactobacillus acidophilus RamA) rather than α-L-rhamnosidases from other fungi. The identification of rhaE and characterization of its regulation make it possible to design strategies to overproduce the encoded enzyme for industrial applications, as well as identify homologs in other fungi.
Methods
Strains and growth conditions
Escherichia coli strain DH5α [endA1, hsdR17, gyrA96, thi-1, relA1, supE44, recA1, ΔlacU169 (Φ80 lacZΔM15)] [39] was used as the recipient strain for cloning experiments and plasmid amplification. The S. cerevisiae wine strain T73-4 (ura3::470/ura3::470) [40] was used for functional expression of plasmids bearing the TDH3p::rha::PGK1t expression cassettes. The A. nidulans wild-type (biA1; Glasgow GO51), creAd30 [17] and ΔcreA (creA-Δ4, pantoB100. N.B. offspring of an outcross of the original mutant described by Shroff et al. [41]; M. Flipphi, unpublished data) strains were used.
E. coli was grown in LB medium (1% w/v tryptone, 0.5% w/v yeast extract, 1% w/v NaCl) with or without 100 μg ml-1 ampicillin. S. cerevisiae strains were grown in YPD-rich medium (1% w/v yeast extract, 2% w/v bacteriological peptone, 2% w/v glucose) or SD-minimal medium (0.17% w/v yeast nitrogen base without amino acids [Difco Laboratories, Detroit, USA], 2% w/v glucose, 0.5% w/v ammonium sulphate, with or without 20 mg l-1 uracil). All media used for A. nidulans strains were based on appropriately supplemented medium [40,42] containing 1% w/v of carbon source, unless otherwise specified. For creA mutants phosphate was included in the sporulation plates [43]. For solid media 1.5% w/v agar was added.
For transfer experiments mycelial biomass was generated from spores inoculated in minimal medium (MM) supplemented with 5 mM urea as nitrogen source, to which 0.1% w/v D-fructose was added as the sole carbon source. After ~18 h grown at 37°C with orbital shaking at 200 rpm, mycelia were harvested, washed with MM without carbon source and transferred. Induction medium was prepared by substituting D-fructose with rhamnose at 1% w/v, and induction-repression medium by substituting D-fructose with rhamnose plus glucose (1% w/v each). Other carbon sources were added instead of glucose at equimolar amounts.
Sensitivity to 2-DOG was tested by spotting the same number of spores (105 in 3 μl) on plates containing 5 mM urea as nitrogen source, 50 μg ml-1 of the toxic glucose analogue 2-DOG and different concentrations of glucose or rhamnose. Growth was monitored after incubation for 3 days at 37°C.
Production of α-L-rhamnosidase in S. cerevisiae was determined under the following growth conditions: the selected transformants were precultured overnight in SD medium lacking uracil, and cells were then used to inoculate (106 cells ml-1) 50-ml of complete (YPD) medium in 250-ml flasks. Yeast cultures were grown with continuous shaking (200 rpm) at 30°C. Recombinant α-L-rhamnosidase cellular location was determined as described [44].
DNA manipulations and transformations
E. coli plasmid isolation and general DNA manipulations were performed following standard protocols [39]. Transformation of the T73 strains was done using lithium acetate to permeabilize the cells as previously described [40,45]. Transformants were selected and maintained on SD plates without uracil. Restriction enzymes, Expand High Fidelity system, and T4 DNA ligase were purchased from Roche Diagnostics and used as recommended by the manufacturer. DNA and cDNA sequencing was carried out using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) and the ABI PRISM 310 Genetic Analyzer (Applied Biosystems, USA) at the DNA Sequencing Service of the University of Valencia. Chromatograms were analysed using the program Chromas LITE 2.01. Oligonucleotides used in the present study are listed in Additional file 1: Table S2.
RNA isolation, northern blot analysis and RT-PCR
Total RNA was isolated from powdered mycelia using RNA Plus (Qbiogene, USA) following the manufacturer's instructions. Northern gels were loaded with 15 μg of glyoxylated total RNA per track essentially as described [36,39]. Membranes (Hybond-N, Amersham Biosciences) were stained with 0.03% w/v methylene blue as a loading control. Gene specific fragments corresponding to rhaE, rhaA and gpdA (350 bp, 385 bp and 610 bp, respectively) were generated by PCR (oligonucleotide pairs: Rha122Ndir_Rha122Nrev; Rha35Ndir_Rha35STOP; gpdAu_gpdAd) and used as templates to generate 32P-dCTP radiolabeled probes using the High Prime system (Roche).
Reverse transcription was performed using 1 μg of total RNA, an oligo (dT)18 anchor primer and the M-MLV reverse transcriptase (USB). Total RNA was previously treated with RNase-free DNase (Roche) for 30 min at 37°C and the DNase was heat-inactivated afterwards. Amplification of the target cDNA by PCR was performed using internal primers (see below).
Heterologous expression of the A. nidulans rhaE gene
To demonstrate the presence of a full-length α-L-rhamnosidase structural gene, the S. cerevisiae wine yeast T73-4 was used for its heterologous expression. The open reading frame (ORF) of rhaE (GenBank accession no. FR873475) was amplified by PCR from A. nidulans cDNA using the primers Rha122dir and Rha122rev and subcloned into pGEM-T-Easy generating pGEM-Rha122. The resulting fragment was ligated between the S. cerevisiae glyceraldehyde-3-phosphate dehydrogenase gene promoter (TDH3p) and the phosphoglycerate kinase terminator (PGK1t) in the high-copy-number shuttle vector YEplac195 [46] in two subcloning steps. First, a 434 bp PstI-SalI fragment from pGEM-Rha122 was ligated into the PstI-SalI restriction sites of YEp195T that contains the phosphoglycerate kinase terminator and polyadenylation signals (PGK1t) (isolated from pG-1; [47]); oligonucleotides PGKt-1 and PGKt-2 and the selectable URA3 gene, yielding YEp195T-R122. To complete the expression cassette, the TDH3p HindIII-AflIII fragment (isolated from pR31; [27]) and the rest of the rha122 ORF (as a 2154 bp BspHI-PstI fragment recovered from pGEM-Rha122) were cloned into HindIII-PstI restricted YEp195T-R122 to generate YEplac195-Rha122. Fragments obtained by PCR were checked by sequencing to ensure the absence of PCR-induced mutations. Transformants were selected at 30°C on uracil-deficient media. Three of the resultant colonies (YR150-YR153) were randomly picked and selected for further studies.
α-L-Rhamnosidase assays
α-L-Rhamnosidase activity was measured using an assay based on the hydrolysis of p-nitrophenyl α-L-rhamnopyranoside (pNPR). Release of p-nitrophenol was measured spectrophotometrically at 400 nm. The assay was performed for 15 min at 50°C in a final volume of 250 μl using 1.4 mM substrate in 250 mM McIlvaine buffer (citrate-phosphate buffer) pH 4.0, essentially as described [9]. One unit (U) of enzyme activity was defined as the amount of enzyme that releases 1 μmol of p-nitrophenol (from pNPR) per minute in the same conditions. Protein concentrations were determined by the Bradford assay [48] using BSA as standard. SD or agar plates supplemented with 40 μM MUR and 1 mM McIlvaine buffer pH 4.0 were used for plate assays of α-L-rhamnosidase activities. After incubation, hydrolysis of MUR was assessed under uv light. Chloroform permeabilization of yeast cells was carried out following the method of Herman and Halvorson [49].
Protein production, purification and sequence
Protein purification was carried out following a similar protocol to that previously described [9]. Polypeptides remaining in the α-L-rhamnosidase active pool after the final purification step were resolved by SDS-PAGE, manually excised and stored at 4°C in ultrapure water. Protein sequence analyses were performed at the Centro Nacional de Investigaciones Cardiovasculares (CNIC), CSIC, Madrid, Spain. Polypeptides were digested automatically using a Proteineer DP protein digestion station (Bruker-Daltonics, Bremen, Germany) following the protocol described by Schevchenko et al. [50] with minor variations. For peptide mass fingerprinting and LIFT-TOF/TOF spectra acquisition [51] an aliquot of α-cyano-4 hydroxycinnamic acid in 33% v/v aqueous acetonitrile and 0.1% v/v trifluoroacetic acid was mixed with an aliquot of the above digestion solution and the mixture was deposited onto an AnchorChip MALDI probe (Bruker-Daltonics). MALDI-mass spectra were generated using an Ultraflex MALDI TOF/TOF system (Bruker-Daltonics). MALDI-MS and MS/MS data were combined using the BioTools program (Bruker-Daltonics) to search the NCBInr (nonredundant) and SwissProt databases using Mascot software (Matrix Science).
Sugar analyses
Sugars present in the A. nidulans culture filtrates were quantified by high-performance anion-exchange chromatography (HPAEC) using a Dionex (DX500 or ICS 3000) system (Sunnyvale, CA, USA) equipped with a CarboPac PA-1 column and a pulse electrochemical detection unit in the pulsed amperometric detection mode. Separation was performed by isocratic elution with 16 mM NaOH at a flow rate of 1 ml min-1 and the data were analyzed using the Peaknet or the Chromeleon software packages (Dionex). Calibration curves were made using standard solutions of sugars of final concentrations of 0, 1, 2, 5, 10 and 25 mg l-1. Ethanol concentrations were measured by an enzymatic method using a kit (Roche).
Bioinformatic analyses
Protein sequences were obtained using BLAST programs [25] at NCBI (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). Retrieved sequences were subsequently used in additional searches against the A. nidulans database at the Broad Institute of MIT and Harvard (http://www.broad.mit.edu/annotation/fgi/). Other bioinformatic tools and software packages were provided by the ExPASy Proteomics Server (http://www.expasy.org/): DNA alignments were done using ClustalW [52]; protein localization was predicted using SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP/; [53]) or SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP/; [22]) using eukaryote or mammalian networks respectively.
Nucleotide sequence accession number
The A. nidulans AN7151 (rhaE) cDNA/mRNA sequence has been deposited in the EMBL database under the accession number FR873475.
Abbreviations
aa: amino acid; CCR: Carbon catabolite repression; 2-DOG: 2-deoxy-D-glucose; GH: Glycosyl hydrolase; MALDI-TOF: Matrix-assisted laser desorption/ionization time-of-flight; MM: Minimal medium; MUR: 4-methylumbelliferyl α-L-rhamnopyranoside; pNPR: p-nitrophenyl-α-L-rhamnopyranoside; ORF: Open reading frame; RT-PCR: Reverse transcription-polymerase chain reaction; SDS-PAGE: Sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JATR, MF and EP carried out the experimental work and were involved in data analyses and interpretation. PM participated in RhaE purification and sugar analyses. MO conceived and designed the study, coordinated the work, was involved in data analyses and interpretation of results and wrote the manuscript. All authors have read and approve the final manuscript.
Supplementary Material
Table S1. α-L-Rhamnosidases (GH78) for which encoding genes have been experimentally characterized [5-7,54-59]. Table S2. Primers used in the present study.
Figure S1. Nucleotide and amino acid sequences of the A. nidulans α-L-rhamnosidase AN7151/rhaE. The deduced amino acid sequence of the AN7151/rhaE gene product is indicated in green boldface. Introns are shown in red. Sequences used for primer design to amplify cDNA sequences are underlined. An asterisk denotes the stop codon. Underlined amino acids correspond to peptides identified by MALDI-TOF mass fingerprinting. Conserved putative catalytic residues Asp/E458, Glu/D464, Asp/E471 and Glu/E737-experimentally characterized in Bacillus sp. RhaB [23]-are highlighted in yellow. The cDNA sequence is deposited in Genbank under the accession number FR873475.
Figure S2. Amino acid alignment. Amino acid sequences of Bacillus sp. GL1 RhaB (accession no. Q93RE7) and A. nidulans AN7151/RhaE rhamnosidases were aligned using ClustalW and default conditions.* indicates identity, : indicates high similarity, and . indicates low similarity. Conserved catalytic residues Asp, Glu, Asp, and Glu-experimentally characterized in RhaB [23]-are highlighted in yellow boldface.
Contributor Information
Juan A Tamayo-Ramos, Email: juan.tamayoramos@wur.nl.
Michel Flipphi, Email: drir.michelflipphi@gmail.com.
Ester Pardo, Email: epardo@iata.csic.es.
Paloma Manzanares, Email: pmanz@iata.csic.es.
Margarita Orejas, Email: morejas@iata.csic.es.
Acknowledgements
This work was supported by the Spanish Ministerio de Ciencia e Innovación/FEDER [grant numbers AGL2002-01906, BIO2008-00228 and CSD2007-0063/Consolider-Ingenio]. J.A.T., M.F. and E.P. were the recipients of a FPI predoctoral fellowship from the Comisión Interministerial de Ciencia y Tecnología, a Ramón y Cajal contract [RYC2004-003005] and a Juan de la Cierva [JCI2007-123/733] contract respectively. The group also participates in the EC COST Action FA0907_BIOFLAVOUR.
We wish to thank Pilar Miró and Claudia Palotti for preliminary RT-PCR experiments, Emilio Camafeita for performing MALDI-TOF assays, Julia Marín for assistance with the Dionex ICS 3000 system, Andrew MacCabe for critical revision of the manuscript prior to submission and to an anonymous referee for helpful suggestions.
References
- Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Manzanares P, Vallés S, Ramón D, Orejas M. In: Industrial Enzymes. Polaina J, MacCabe AP, editor. Berlin-Heidelberg: Springer Verlag; 2007. α-L-Rhamnosidases: old and new insights; pp. 117–140. [Google Scholar]
- Yadav V, Yadav PK, Yadav S, Yadav KDS. α-L-Rhamnosidase: a review. Process Biochem. 2010;45:1226–1235. doi: 10.1016/j.procbio.2010.05.025. [DOI] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares P, van den Broeck HC, de Graaff LH, Visser J. Purification and characterization of two different α-L-rhamnosidases, RhaA and RhaB, from Aspergillus aculeatus. Appl Environ Microbiol. 2001;67:2230–2234. doi: 10.1128/AEM.67.5.2230-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseki T, Mese Y, Nishibori N, Masaki K, Fujii T, Handa T, Yamane Y, Shiono Y, Murayama T, Lefuji H. Characterization of an α-L-rhamnosidase from Aspergillus kawachii and its gene. Appl Microbiol Biotechnol. 2008;80:1007–1013. doi: 10.1007/s00253-008-1599-7. [DOI] [PubMed] [Google Scholar]
- Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci USA. 2006;103:11417–11422. doi: 10.1073/pnas.0604632103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orejas M, Ibáñez E, Ramón D. The filamentous fungus Aspergillus nidulans produces an α-L-rhamnosidase of potential oenological interest. Lett Appl Microbiol. 1999;28:383–388. [Google Scholar]
- Manzanares P, Orejas M, Ibáñez E, Vallés S, Ramón D. Purification and characterization of an α-L-rhamnosidase from Aspergillus nidulans. Lett Appl Microbiol. 2000;31:198–202. doi: 10.1046/j.1365-2672.2000.00788.x. [DOI] [PubMed] [Google Scholar]
- de Vries RP, van Grieken C, van Kuyk PA, Wösten HAB. The value of genome sequences in the rapid identification of novel genes encoding specific plant cell wall degrading enzymes. Curr Genomics. 2005;6:157–187. doi: 10.2174/1389202053971974. [DOI] [Google Scholar]
- Coutinho PM, Andersen MR, Kolenova K, van Kuyk PA, Benoit I, Gruben BS, Trejo-Aguilar B, Visser H, van Solingen P, Pakula T, Seiboth B, Battaglia E, Aguilar-Osorio G, de Jong JF, Ohm RA, Aguilar M, Henrissat B, Nielsen J, Stålbrand H, de Vries RP. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol. 2009;46(Suppl 1):S161–S190. doi: 10.1016/j.fgb.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Ávila M, Jaquet M, Moine D, Requena T, Peláez C, Arigoni F, Jankovic I. Physiological and biochemical characterization of the two α-L-rhamnosidases of Lactobacillus plantarum NCC245. Microbiology. 2009;155:2739–2749. doi: 10.1099/mic.0.027789-0. [DOI] [PubMed] [Google Scholar]
- de Vries RP. Regulation of Aspergillus genes encoding plant cell wall polysaccharide-degrading enzymes; relevance for industrial production. Appl Microbiol Biotechnol. 2003;61:10–20. doi: 10.1007/s00253-002-1171-9. [DOI] [PubMed] [Google Scholar]
- Aro N, Pakula T, Penttilä M. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol Rev. 2005;29:719–739. doi: 10.1016/j.femsre.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Flipphi M, Felenbok B. In: The Mycota III. Biochemistry and Molecular Biology. 2. Brambl R, Marzluf GA, editor. Berlin-Heidelberg: Springer Verlag; 2004. The onset of carbon catabolic repression and interplay between specific induction and carbon catabolite repression in Aspergillus nidulans; pp. 403–420. [Google Scholar]
- Kelly JM. In: The Mycota III. Biochemistry and Molecular Biology. Brambl R, Marzluf GA, editor. Berlin-Heidelberg: Springer Verlag; 2004. The regulation of carbon metabolism in filamentous fungi; pp. 386–401. [Google Scholar]
- Arst HN Jr, Tollervey D, Dowzer CEA, Kelly JM. An inversion truncating the cre gene of Aspergillus nidulans results in carbon catabolite derepression. Mol Microbiol. 1990;4:851–854. doi: 10.1111/j.1365-2958.1990.tb00656.x. [DOI] [PubMed] [Google Scholar]
- Kulmburg P, Mathieu M, Dowzer C, Kelly J, Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993;7:847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- Cubero B, Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13:407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeso EA, Peñalva MA. In vitro binding of the two-finger repressor CreA to several consensus and non-consensus sites at the ipnA upstream region is context dependent. FEBS Lett. 1994;342:43–48. doi: 10.1016/0014-5793(94)80581-4. [DOI] [PubMed] [Google Scholar]
- Orejas M, MacCabe AP, Pérez-González JA, Kumar S, Ramón D. The wide-domain carbon catabolite repressor CreA indirectly controls expression of the Aspergillus nidulans xlnB gene, encoding the acidic endo-β-(1,4)-xylanase X24. J Bacteriol. 2001;183:1517–1523. doi: 10.1128/JB.183.5.1517-1523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- Cui Z, Maruyama Y, Mikami B, Hashimoto W, Murata K. Crystal structure of glycoside hydrolase family 78 α-L-rhamnosidase from Bacillus sp. GL1. J Mol Biol. 2007;374:384–398. doi: 10.1016/j.jmb.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego MV, Piñaga F, Ramón D, Vallés S. Purification and characterization of an α-L-rhamnosidase from Aspergillus terreus of interest in winemaking. J Food Sci. 2001;66:204–209. doi: 10.1111/j.1365-2621.2001.tb11317.x. [DOI] [Google Scholar]
- Manzanares P, Orejas M, Gil JV, de Graaff LH, Visser J, Ramón D. Construction of a genetically modified wine yeast strain expressing the Aspergillus aculeatus rhaA gene, encoding an α-L-rhamnosidase of enological interest. Appl Environ Microbiol. 2003;69:7558–7562. doi: 10.1128/AEM.69.12.7558-7562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orejas M, MacCabe AP, Pérez-González JA, Kumar S, Ramón D. Carbon catabolite repression of the Aspergillus nidulans xlnA gene. Mol Microbiol. 1999;31:177–184. doi: 10.1046/j.1365-2958.1999.01157.x. [DOI] [PubMed] [Google Scholar]
- Dean RA, Timberlake WE. Regulation of the Aspergillus nidulans pectate lyase gene pelA. Plant Cell. 1989;1:275–284. doi: 10.1105/tpc.1.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C, Arst HN Jr. Carbon catabolite repression in Aspergillus nidulans. Eur J Biochem. 1975;51:573–577. doi: 10.1111/j.1432-1033.1975.tb03958.x. [DOI] [PubMed] [Google Scholar]
- Ilyés H, Fekete E, Karaffa L, Fekete É, Sándor E, Szentirmai A, Kubicek CP. CreA-mediated carbon catabolite repression of β-galactosidase formation in Aspergillus nidulans is growth rate dependent. FEMS Microbiol Lett. 2004;235:147–151. doi: 10.1016/j.femsle.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Lutfiyya LL, Iyer VR, DeRisi J, DeVit MJ, Brown PO, Johnston M. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics. 1998;150:1377–1391. doi: 10.1093/genetics/150.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Nordberg N, Murén R, Ameur A, Komorowski J, Ronne H. Combinatorial control of gene expression by the three yeast repressors Mig1, Mig2 and Mig3. BMC Genomics. 2008;9:601. doi: 10.1186/1471-2164-9-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero B, Gómez D, Scazzocchio C. Metabolite repression and inducer exclusion in the proline utilization gene cluster of Aspergillus nidulans. J Bacteriol. 2000;182:233–235. doi: 10.1128/JB.182.1.233-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Cañón JM, Luengo JM. The phenylacetic acid uptake system of Aspergillus nidulans is under a cre-independent model of catabolic repression which seems to be mediated by acetyl-CoA. J Antibiot. 1997;50:45–52. doi: 10.7164/antibiotics.50.45. [DOI] [PubMed] [Google Scholar]
- Flipphi M, van de Vondervoort PJI, Ruijter GJG, Visser J, Arst HN Jr, Felenbok B. Onset of carbon catabolite repression in Aspergillus nidulans. Parallel involvement of hexokinase and glucokinase in sugar signaling. J Biol Chem. 2003;278:11849–11857. doi: 10.1074/jbc.M209443200. [DOI] [PubMed] [Google Scholar]
- Elorza MV, Arst HN Jr. Sorbose resistant mutants of Aspergillus nidulans. Mol Gen Genet. 1971;111:185–193. doi: 10.1007/BF00267792. [DOI] [PubMed] [Google Scholar]
- Mark CG, Romano AH. Properties of the hexose transport systems of Aspergillus nidulans. Biochim Biophys Acta. 1971;249:216–226. doi: 10.1016/0005-2736(71)90098-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor NJ: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Puig S, Ramón D, Pérez-Ortín JE. Optimized method to obtain stable food-safe recombinant wine yeast strains. J Agric Food Chem. 1998;46:1689–1693. doi: 10.1021/jf9706538. [DOI] [Google Scholar]
- Shroff RA, O'Connor SM, Hynes MJ, Lockington RA, Kelly JM. Null alleles of cre, the regulator of cabron catabolite repression in Aspergillus nidulans. Fungal Genet Biol. 1997;22:28–38. doi: 10.1006/fgbi.1997.0989. [DOI] [PubMed] [Google Scholar]
- Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- Tamayo EN, Villanueva A, Hasper AA, de Graaff LH, Ramón D, Orejas M. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet Biol. 2008;45:984–993. doi: 10.1016/j.fgb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- González-Candelas L, Cortell LA, Ramón D. Construction of a recombinant wine yeast strain expressing a fungal pectate lyase gene. FEMS Microbiol Lett. 1995;126:263–269. doi: 10.1111/j.1574-6968.1995.tb07428.x. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;14:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Schena M, Picard D, Yamamoto KR. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink GR, editor. Vol. 194. San Diego: Academic; 1991. Vectors for constitutive and inducible gene expression in yeast; pp. 389–398. Methods in Enzymology. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Herman A, Halvorson H. Identification of the structural gene for β-glucosidase in Saccharomyces lactis. J Bacteriol. 1963;85:895–900. doi: 10.1128/jb.85.4.895-900.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal Bioanal Chem. 2003;376:952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Zverlov VV, Hertel C, Bronnenmeier K, Hroch A, Kellermann J, Schwarz WH. The thermostable α-L-rhamnosidase RamA of Clostridium stercorarium: biochemical characterization and primary structure of a bacterial α-L-rhamnoside hydrolase, a new type of inverting glycoside hydrolase. Mol Microbiol. 2000;35:173–179. doi: 10.1046/j.1365-2958.2000.01691.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto W, Miyake O, Nankai H, Murata K. Molecular identification of an α-L-rhamnosidase from Bacillus sp. strain GL1 as an enzyme involved in complete metabolism of gellan. Arch Biochem Biophys. 2003;415:235–244. doi: 10.1016/S0003-9861(03)00231-5. [DOI] [PubMed] [Google Scholar]
- Birgisson H, Hreggvidsson GO, Fridjónsson OH, Mort A, Kristjánsson JK, Mattiasson B. Two new thermostable α-L-rhamnosidases from a novel thermophilic bacterium. Enzyme Microb Technol. 2004;34:561–571. doi: 10.1016/j.enzmictec.2003.12.012. [DOI] [Google Scholar]
- Beekwilder J, Marcozzi D, Vecchi S, de Vos R, Janssen P, Francke C, van Hylckama Vlieg J, Hall RD. Characterization of rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus. Appl Environ Microbiol. 2009;75:3447–3454. doi: 10.1128/AEM.02675-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ávila M, Jaquet M, Moine D, Requena T, Peláez C, Arigoni F, Jankovic I. Physiological and biochemical characterization of the two α-L-rhamnosidases of Lactobacillus plantarum NCC245. Microbiology. 2009;155:2739–2749. doi: 10.1099/mic.0.027789-0. [DOI] [PubMed] [Google Scholar]
- Michlmayr H, Brandes W, Eder R, Scümann C, del Hierro AM, Kulbe KD. Characterization of two distinct GH Family 78 α-L-rhamnosidases from Pediococcus acidilactici. Appl Environ Microbiol. 2011;77:6524–6530. doi: 10.1128/AEM.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. α-L-Rhamnosidases (GH78) for which encoding genes have been experimentally characterized [5-7,54-59]. Table S2. Primers used in the present study.
Figure S1. Nucleotide and amino acid sequences of the A. nidulans α-L-rhamnosidase AN7151/rhaE. The deduced amino acid sequence of the AN7151/rhaE gene product is indicated in green boldface. Introns are shown in red. Sequences used for primer design to amplify cDNA sequences are underlined. An asterisk denotes the stop codon. Underlined amino acids correspond to peptides identified by MALDI-TOF mass fingerprinting. Conserved putative catalytic residues Asp/E458, Glu/D464, Asp/E471 and Glu/E737-experimentally characterized in Bacillus sp. RhaB [23]-are highlighted in yellow. The cDNA sequence is deposited in Genbank under the accession number FR873475.
Figure S2. Amino acid alignment. Amino acid sequences of Bacillus sp. GL1 RhaB (accession no. Q93RE7) and A. nidulans AN7151/RhaE rhamnosidases were aligned using ClustalW and default conditions.* indicates identity, : indicates high similarity, and . indicates low similarity. Conserved catalytic residues Asp, Glu, Asp, and Glu-experimentally characterized in RhaB [23]-are highlighted in yellow boldface.