Abstract
Besides its beneficial role in thermotolerance, the chaperone protein Hsp104 is involved in the inheritance of yeast Saccharomyces cerevisiae prions. Guanidine hydrochloride was previously shown to interfere with Hsp104 chaperone activity in vivo, thus impairing thermotolerance and resulting in prion curing. It was also reported that guanidine inhibits Hsp104 ATPase and disaggregation activity. We show that in vitro guanidine significantly inhibits the disaggregation activity of ClpB, the bacterial orthologue of Hsp104. However, guanidine exerts opposite effects on the ATPase activities of Hsp104 and ClpB. While the ATPase activity of Hsp104 is inhibited, the analogous ClpB activity is stimulated several-fold. Mutation of the universally conserved aspartic acid residue in position 184 to serine (D184S) in HSP104 and the analogous mutation in clpB (D178S) resulted in chaperones with lower disaggregating and ATPase activities. The activities of such changed chaperones are not influenced by guanidine, which suggests the role of this residue in the interaction with guanidine.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-011-0312-4) contains supplementary material, which is available to authorized users.
Keywords: Chaperone-dependent protein disaggregation, Protein aggregation, Prion curing agent—guanidine hydrochloride, Hsp70, Hsp100
Introduction
Severe heat shock conditions cause aggregation of many intracellular proteins. Hsp104 from Saccharomyces cerevisiae and ClpB from Escherichia coli, members of the Hsp100 family of molecular chaperones, are crucial factors required for thermotolerance (Squires et al. 1991; Kitagawa et al. 1991; Sanchez and Lindquist 1990), as well as the disaggregation of protein aggregates which occurs once the cell is returned to the physiological temperature (Parsell et al. 1994a; Laskowska et al. 1996; Mogk et al. 1999). To mediate protein disaggregation, chaperones of the Hsp100 family require the assistance of the Hsp70 chaperone system (reviewed in Liberek et al. (2008)). Recent studies postulate that the Hsp70 system remodels aggregates at the initial step of disaggregation, enabling subsequent action by the Hsp100 chaperones (Ziętkiewicz et al. 2004, 2006; Weibezahn et al. 2004).
Hsp104 and ClpB, both Hsp100 proteins, belong to the AAA+ (ATPase associated with a variety of cellular activities) family whose members possess characteristic ATPase domain(s). AAA+ proteins self-assemble into oligomeric structures and use the energy derived from ATP hydrolysis to remodel their protein substrates (Vale 2000; Bösl et al. 2006). Hsp104 and ClpB have two nucleotide binding domains (NBD1 and NBD2; AAA+ domains) which are essential for hexamerization and chaperone function (Krzewska et al. 2001a; Weibezahn et al. 2003; Schirmer et al. 2001; Watanabe et al. 2002; Mogk et al. 2003). The first AAA+ domain of both Hsp104 and ClpB contains an additional coiled-coil region called the “middle domain,” located C-terminally to the ATP binding site (Lee et al. 2003). This middle domain is characteristic of Hsp100 disaggregating chaperones.
In addition to its role in thermotolerance, Hsp104 plays a role in the propagation and inheritance of yeast prions [PSI+], [URE3], and [RNQ+] (Wickner 1994; Sondheimer and Lindquist 2000). It was shown that the [PSI+] prion is lost in a yeast strain overexpressing Hsp104, as well as in an hsp104 deletion strain (Chernoff et al. 1995). Remarkably, the addition of millimolar concentrations of guanidine hydrochloride (GuHCl), a chaotropic salt, to the yeast medium leads to prion curing (Tuite et al. 1981; Ferreira et al. 2001; Jung and Masison 2001; Jung et al. 2002). Due to this property, GuHCl became a tool which allows studying the propagation of yeast prions. Moreover, GuHCl-treated strains show a reduced heat stress tolerance and, similar to the hsp104 deletion strain, display impaired intracellular reactivation of heat-inactivated luciferase (Ferreira et al. 2001; Jung and Masison 2001). It was also shown that various substitutions of the aspartic acid residue in position 184 in Hsp104 affect both prion curing by guanidine and thermotolerance. However, the thermotolerance of cells expressing these mutant proteins is diversely influenced by guanidine (depending on the mutation), and the degree of thermotolerance does not correlate with prion stability (Jung et al. 2002).
Recently, it has been shown that GuHCl reduces the ATPase activity of Hsp104 by approximately 50% (Grimminger et al. 2004). Guanidine selectively binds to the nucleotide-bound Hsp104 hexamer and increases the affinity of the chaperone for adenine nucleotides, thus promoting (or stabilizing) the oligomerization of the chaperone (Grimminger et al. 2004). However, it is not fully understood how the change in Hsp104 ATPase activity correlates with its ability to disaggregate protein aggregates. Wegrzyn et al. (2001) reported that prion curing in the presence of GuHCl was significantly slower than curing by removal or inactivation of Hsp104. Therefore, it has been suggested that GuHCl only partially inhibits various Hsp104 activities, as is the case for ATP hydrolysis (Grimminger et al. 2004).
The influence of GuHCl on the activity of ClpB, the E. coli orthologue of Hsp104, has not been investigated until now. We note that the Hsp104 NBD1 domain encompassing amino acid residue 184, critical for GuHCl-dependent prion curing and thermotolerance, is highly conserved among members of the Hsp100 family (Lee et al. 2003). Here, we show that GuHCl completely abolishes the disaggregation activity of Hsp104 and significantly inhibits the analogous activity of ClpB. However, GuHCl exerts opposite effects with respect to the ATPase activities of Hsp104 and ClpB. While the ATPase activity of Hsp104 is inhibited by GuHCl, the analogous ClpB activity is stimulated several-fold. Mutation in HSP104 changing the aspartic acid residue in position 184 to serine (D184S) as well as the analogous mutation in clpB (D178S) results in the formation of chaperones with lower disaggregating activities. However, the activities of such mutated chaperones are no longer influenced by GuHCl, indicating that this aspartic acid residue is important for interaction with guanidine.
Materials and methods
Protein purification
Published protocols were used for the purification of E. coli DnaK, DnaJ, GrpE (Zylicz et al. 1989), ClpB, and ClpB (D178S) (Woo et al. 1992). Yeast S. cerevisiae proteins Hsp104, Hsp104 (D184S), and Ydj1 were expressed in E. coli and purified as described in references Parsell et al. (1994b) and Cyr and Douglas (1994), respectively. Ssa1 protein was overexpressed in S. cerevisiae and purified according to the published protocol (Ziegelhoffer et al. 1995). GFP was expressed in E. coli and purified as described (Ziętkiewicz et al. 2004). Firefly luciferase (E 1701) was purchased from Promega. Protein concentrations were determined using the Bio-Rad Protein Assay system with BSA as a standard. Molar concentrations given are based on the monomeric structure for all chaperones.
Site-directed mutagenesis
Site-directed mutagenesis of pET15b-HSP104 and pET22b+clpB was performed using Pfu DNA polymerase and the oligonucleotide primers 5′-CGTCAAGGTAAACTTTCCCCTGTCATCGGCCGTG-3′ and 5′-GCCGAACAGGGCAAACTCTCTCCGGTGATTGGTCG-3′, respectively, as described in the QuickChange manual (Stratagene). All constructs were confirmed by sequencing.
Thermotolerance assay
Yeast cells were grown at 30°C in SC medium prepared as described (Sherman 1991) for W3031B, and in SC-His medium for other strains (Sherman 1991). Overnight cultures were diluted to an OD600 of 0.2. Heat shock was performed on exponentially growing cells adjusted to an OD600 of 0.4. Cells were pre-incubated at 37°C for 30 min, and then incubated with slight agitation at 50°C for 45 min. Aliquots were withdrawn at the indicated time points, cells were mixed by vortexing, serially diluted, and subsequently spotted on SC or SC-His solid media with or without 5 mM GuHCl.
ATPase assay
The ATPase activity of Hsp104, Hsp104 (D184S), ClpB, and ClpB (D178S) was measured using a coupled enzymatic assay (Norby 1988). This assay enables ATP regeneration and thus eliminates ADP, a potent competitive inhibitor of the reaction. Assays were carried out as described previously with minor modifications (Grimminger et al. 2004). Assays were performed in thermostated 200-μl cuvettes at 25°C using a Beckman DU650 spectrophotometer. Reactions were assembled in buffer A (50 mM Hepes/KOH, pH 7.5; 150 mM KCl; 20 mM MgAc) supplemented with 5 mM ATP. Reactions were started by addition of the protein of interest (1 μM Hsp104 or 4 μM ClpB in GuHCl titration experiment). ATP turnover was estimated from the slope of dA340/dt curve as described previously (Grimminger et al. 2004).
Refolding of urea-denatured luciferase
Firefly luciferase (4 μM) was denatured for 3 h at 30°C in buffer B (50 mM Tris, pH 7.4; 150 mM KCl; 20 mM MgAc; 5 mM DTT) containing 6 M urea. Refolding reactions were performed by addition of denatured luciferase (to the final concentration of 50 nM) to buffer A containing indicated chaperone proteins (2 μM Hsp104, 3 μM Ssa1p, 1 μM Ydj1p for yeast chaperones; 0.5 μM ClpB, 1 μM DnaK, 0.2 μM DnaJ, 0.1 μM GrpE for bacterial chaperones), 5 mM ATP, the ATP regenerating system (10 mM phosphocreatine, 100 μg/ml phosphocreatine kinase), and 0.15 mg/ml bovine serum albumin. Reaction mixtures were incubated for the indicated time at 24°C, and the luciferase activity was measured in a Beckman LS 6000 TA scintillation counter using the Luciferase Assay System (Promega E1500).
Oligomerization assay
In order to analyze the influence of GuHCl on the oligomeric state of both Hsp104 and Hsp104 (D184S), 20 μg of each protein diluted in buffer D (40 mM Hepes/KOH, pH 7.8; 150 mM NaCl; 10 mM MgCl2) were loaded onto a 3.5-ml 15–45% glycerol gradient in buffer C (40 mM Hepes/KOH, pH 7.8; 150 mM NaCl; 10 mM MgCl2; 5 mM 2-mercaptoethanol) supplemented with 5 mM ATP and (where indicated) with 5 mM GuHCl. The gradients were centrifuged at 4°C for 16 h in a Beckman SW 60 rotor at 46,000 rpm. Following centrifugation, 14 fractions (250 μl each) were collected from the top and analyzed by SDS-PAGE (12.5%) together with the remaining protein eluted from the bottom of the tube.
Partial trypsin digestion
Four micrograms of either ClpB or Hsp104 protein were incubated in 10 μl of buffer A (with 2 mM ATP) at 30°C for 10 min in the presence or absence of GuHCl (5 mM). Partial proteolysis was started by addition of 0.07 μg of trypsin (Sigma). At the indicated times, reactions were stopped by addition of SDS sample buffer. Proteolysis products were analyzed using SDS-PAGE (10%) and Coomassie Blue staining.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) was performed using a MicroCal AUTO-ITC (MicroCal-LLC, Northampton) as described previously (Grimminger et al. 2004). All experiments were performed at 25°C using assay buffer (50 mM Hepes/KOH, pH 7.5, 150 mM KCl, and 10 mM MgCl2). For the binding of GuHCl to Hsp104- or Hsp104 (D184S)-nucleotide complexes, experiments were carried out using 13.82 mM GuHCl in the injection syringe and an injection volume of 5 μl. The protein concentration in the cell was 20 μM, and the ADP concentration was 2 mM. Data analysis was performed using the Origin software package (Origin Lab, Northampton, MA, USA).
GFP disaggregation experiments
Experiments were performed as described previously (Ziętkiewicz et al. 2004). Briefly, GFP at 5 mg/ml concentration was heat-aggregated by incubation at 85°C for 10 min. The reactivation was started by addition of heat-aggregated GFP (9 μg) to the reaction mixture containing chaperone proteins (2 μM Hsp104, 3 μM Ssa1p, 1 μM Ydj1p for yeast chaperones; 0.5 μM ClpB, 1 μM DnaK, 0.2 μM DnaJ, 0.1 μM GrpE for bacterial chaperones) in buffer A supplemented with 5 mM ATP and the ATP regenerating system (10 mM phosphocreatine, 100 μg/ml phosphocreatine kinase). Reactions were carried out in thermostated 120-μl cuvettes at 25°C, and GFP reactivation (observed as an increase in GFP fluorescence) was monitored in real time using a Perkin Elmer LS 50B spectrofluorometer with excitation at 395 nm and emission at 510 nm. The excitation slit was set to 4 nm and the emission slit to 12 nm; 1% attenuator was used.
Sedimentation experiments
Heat-aggregated GFP (9 μg) was incubated in buffer A containing the indicated chaperone proteins (2 μM Hsp104, 7.5 μM Ssa1p, 2.9 μM Ydj1p for yeast chaperones; 0.5 μM ClpB, 4.4 μM DnaK, 0.34 μM DnaJ, 0.45 μM GrpE for bacterial chaperones), 5 mM ATP, the ATP regenerating system and, where stated, 5 mM GuHCl. Disaggregation reactions were performed at 24°C for 15 min (when bacterial chaperones were used) or 45 min (for yeast chaperones). Reactions were stopped by addition of EDTA to the final concentration of 40 mM, and reaction mixtures (100 μl) were loaded onto a 3.5-ml 15–45% glycerol gradient in buffer C (40 mM Hepes/KOH, pH 7.8; 150 mM NaCl; 10 mM MgCl2; 5 mM 2-mercaptoethanol) supplemented with 1 mM ATP. The gradients were centrifuged for 2 h at 4°C in a Beckman SW 60 rotor at 40,000 rpm. Following centrifugation, 20 fractions (180 μl each) were collected from the top and, together with the remaining protein eluted from the bottom of the tube, analyzed by SDS-PAGE (12.5%) and subsequent western blotting using anti-GFP mouse monoclonal antibodies (Roche Applied Science).
Results
Residue D184 of Hsp104 is important for interaction with guanidine
It was shown that the presence of GuHCl in the medium abolishes thermotolerance and cures the prion phenotypes in yeast cells (Tuite et al. 1981; Ferreira et al. 2001; Jung and Masison 2001; Jung et al. 2002). Both effects are dependent on the presence of functional Hsp104 chaperone (Ferreira et al. 2001; Jung and Masison 2001). The aspartic acid residue in position 184 was identified as crucial for both prion curing and thermotolerance development. The specific amino acid substitution D184S results in a mutant protein that retains nearly normal thermotolerance but lacks prion curing ability in the presence of GuHCl (Jung et al. 2002).
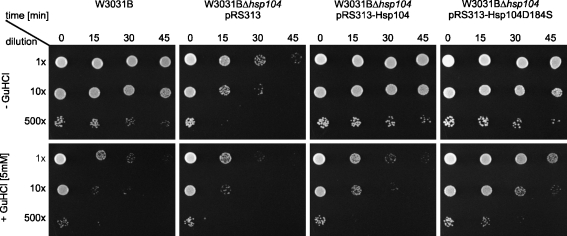
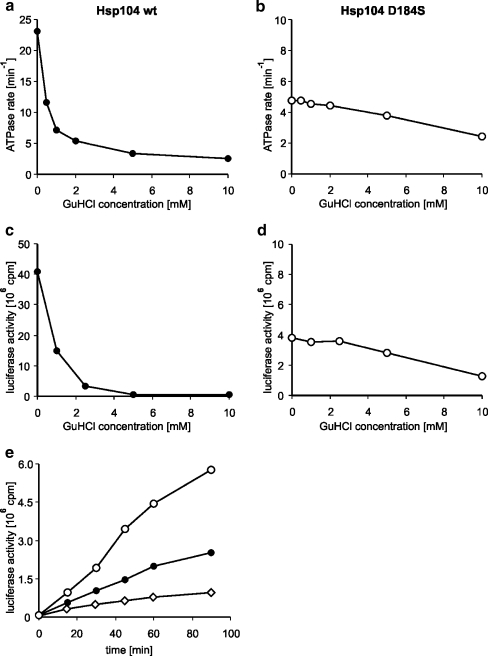
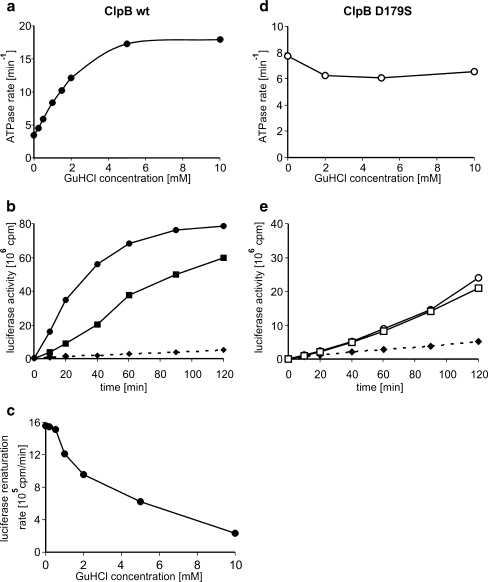
In our attempts to analyze the influence of GuHCl on Hsp104 activities, we first performed the thermotolerance assay for both wild type and the Hsp104 (D184S) mutant. As expected, addition of GuHCl completely abolished thermotolerance in the wild-type strain and the hsp104Δ yeast strain carrying the centromeric plasmid pRS313 with the HSP104 gene under its native promoter. In contrast, the hsp104Δ pRS313-Hsp104 (D184S) strain was only slightly affected by GuHCl (Fig. 1). We purified both Hsp104 and Hsp104 (D184S) to analyze the influence of GuHCl on their biochemical activities. First, we compared the ATPase activities of wild-type Hsp104 and the D184S mutant using a coupled enzymatic assay. The D184S mutant showed a 4.5-fold lower rate of ATP hydrolysis as compared to wild-type Hsp104. GuHCl present at 2 mM concentration only weakly influenced this rate (Fig. 2b) in contrast to wild-type Hsp104, which ATP hydrolysis rate was inhibited severely in these conditions (Fig. 2a), as reported previously (Grimminger et al. 2004). Only at high GuHCl concentrations, the rate of ATP hydrolysis observed for D184S mutant was inhibited twofold (Fig. 2b).
Fig. 1.
Thermotolerance of Hsp104 (D184S) mutant is not inhibited by GuHCl. Yeast strains W3031B and W3031BΔhsp104 carrying pRS313, pRS313-Hsp104, or pRS313-Hsp104 (D184S) were tested for thermotolerance in the absence (upper panels) or presence (lower panels) of 5 mM GuHCl. Cells were pre-incubated at 37°C for 30 min, and then incubated with agitation at 50°C for 45 min. Aliquots were withdrawn at the indicated time points. Cells were serially diluted and subsequently spotted on the SC or SC-His solid media with or without 5 mM GuHCl
Fig. 2.
The Hsp104 (D184S) mutant possesses lower ATPase and disaggregating activity than the wild-type Hsp104, yet these activities are not influenced by GuHCl. The ATPase activity of Hsp104 (a) and Hsp104 (D184S) (b) in the presence of increasing concentrations of GuHCl was measured spectrophotometrically in a coupled enzymatic assay. The concentration of the proteins was 1 μM each. c, d Urea-denatured firefly luciferase (50 nM) was incubated with Hsp104 (c) or Hsp104 (D184S) (d) and the Ssa1p/Ydj1p chaperone system (2, 3, and 1 μM, respectively) for 1 h at 24°C in the presence of increasing concentrations of GuHCl. Luciferase reactivation was assessed by measuring its activity in a scintillation counter. e Urea-denatured firefly luciferase (50 nM) was incubated with Hsp104 (filled circles) or Hsp104 (D184S) (open circles) and the Ssa1p/Ydj1p chaperone system (2, 3, and 1 μM, respectively) or with the Ssa1p/Ydj1p chaperone system alone (diamonds) at 24°C in the presence of 5 mM GuHCl. Aliquots were withdrawn at the indicated time points, and the enzymatic activity of refolded luciferase was measured in a scintillation counter. Experiments presented in the figure are representative of data obtained from separate protein preparations
The influence of increasing concentrations of GuHCl on the efficiency of Hsp104-, Ssa1p-, and Ydj1p-dependent reactivation of urea-denatured firefly luciferase was also monitored. GuHCl efficiently inhibited the disaggregation of luciferase; 2 mM GuHCl nearly abolished luciferase disaggregation (Fig. 2c). Similar reactivation experiments performed with Hsp104 (D184S) showed that the mutant protein possesses a substantially lower disaggregation activity compared to wild type. However, in contrast to wild-type Hsp104, the activity of the mutant protein was hardly inhibited by 2 mM GuHCl (Fig. 2d). Only at high GuHCl concentration (10 mM), the efficiency of luciferase disaggregation was inhibited twofold (Fig. 2b). As a consequence, in the presence of 5 mM GuHCl, the ability of Hsp104 (D184S) to disaggregate and refold luciferase was substantially higher than that of wild-type Hsp104, which was efficiently inhibited under these conditions (Fig. 2e). Our in vitro analysis of Hsp104 and Hsp104 (D184S) activities is in good agreement with the in vivo thermotolerance assay showing that in the presence of GuHCl, the (D184S) mutant, but not the wild-type Hsp104, retains thermotolerance.
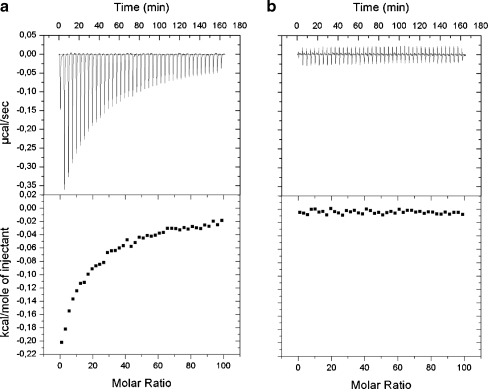
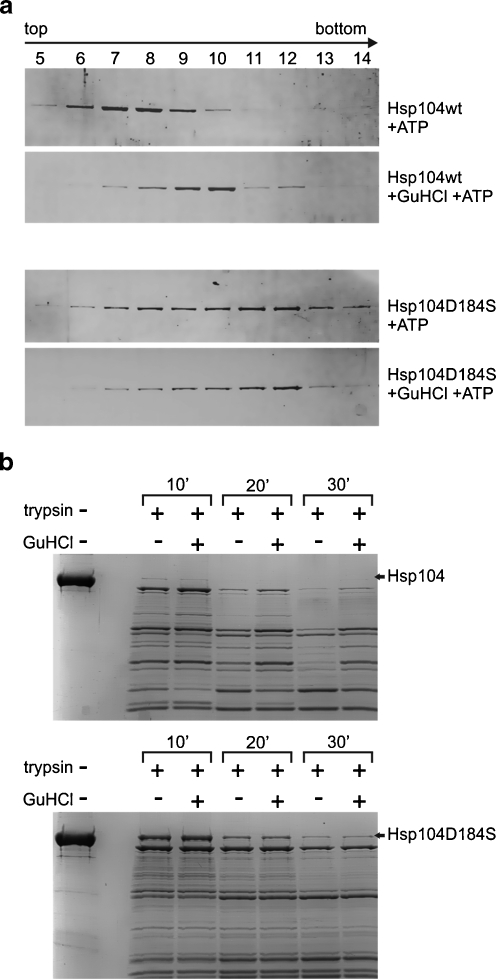
It was shown previously by isothermal titration calorimetry that GuHCl interacts only with nucleotide-bound Hsp104 (Grimminger et al. 2004). We performed a similar experiment with the Hsp104 (D184S) protein variant. No interaction of GuHCl with nucleotide-bound Hsp104 (D184S) was found, as compared to the wild-type Hsp104 used as a control (Fig. 3). This suggests that the aspartic acid residue in position 184 in Hsp104 plays a role in the interaction with guanidine. The work of Grimminger et al. (2004) also showed that GuHCl promotes the nucleotide-dependent oligomerization of Hsp104. We performed similar experiments analyzing the sedimentation profile of wild-type Hsp104 and its D184S mutant. In accordance with the report by Grimminger et al. (2004), Hsp104 sedimented faster in the presence of nucleotide and GuHCl, as compared to its sedimentation profile in the presence of nucleotide only (Fig. 4a). Under the same conditions, the Hsp104 (D184S) sedimentation profile was not changed by the addition of GuHCl, and the mutant protein sedimented faster than wild-type Hsp104 (Fig. 4a).
Fig. 3.
GuHCl does not interact with Hsp104 (D184S). Binding of GuHCl to Hsp104 (a) and Hsp104 (D184S) (b) in the presence of 2 mM ADP was measured using isothermal titration calorimetry. A stock solution of GuHCl (13.82 mM) was titrated to a solution of Hsp104 or Hsp104 (D184S) (20 μM) in assay buffer at 25°C, and the associated heat change was monitored (upper panels). The resulting binding curves (lower panels) were obtained after integration of the injection peaks
Fig. 4.
Mutation (D184S) in Hsp104 influences its oligomeric state. a Sedimentation analysis of the Hsp104 and Hsp104 (D184S) oligomeric state. Hsp104 (upper panels) and Hsp104 (D184S) (lower panels) were diluted in buffer D and loaded onto a glycerol gradient in buffer C supplemented with 5 mM ATP and 5 mM GuHCl (where indicated). Sedimentation analysis was performed (Beckman SW60 rotor; 15–45% (v/v) glycerol gradient; 46,000 rpm; 16 h; 4°C). The position of Hsp104 and Hsp104 (D184S) in the gradients was visualized by SDS-PAGE and Coomassie Blue staining. b Kinetics of the partial trypsin digestion of 4 μg of Hsp104 (upper panel) or Hsp104 (D184S) (lower panel) in the presence of 2 mM ATP and presence or absence of 5 mM GuHCl (as indicated). Reactions (10 μl) were assembled and incubated at 30°C with 0.07 μg of trypsin. At the indicated time points, reactions were stopped by the addition of Laemmli buffer. Proteolysis products were analyzed by SDS-PAGE and Coomassie Blue staining
Partial trypsin digestion was used to further examine the protein conformation, since it was previously reported that changes in a protein’s digestion pattern occur in response to protein oligomerization (Rudyak et al. 2001). Accordingly, we observed that in the presence of ATP, the addition of GuHCl inhibits the proteolytic cleavage of Hsp104 (Fig. 4b). Not only was the disappearance of major protein bands restricted but also the patterns of digestion products were also different in the presence of GuHCl. This suggests that in the presence of GuHCl, the structure of Hsp104 changes in such a way that the protein becomes more resistant to trypsin digestion. Under the same experimental conditions, the presence of GuHCl had no effect on either the pattern or the rate of trypsin cleavage for Hsp104 (D184S) (Fig. 4b). It is worth noting that digestion of the Hsp104 (D184S) mutant by trypsin was substantially slower compared to wild-type Hsp104.
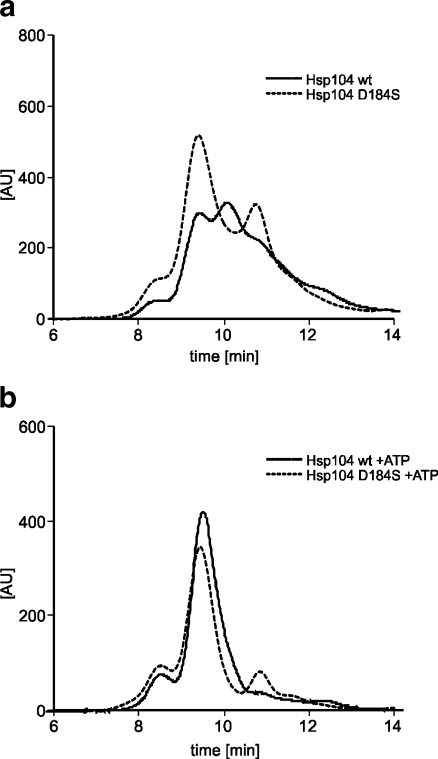
We additionally performed size-exclusion chromatography experiments to compare the elution profiles of Hsp104 and Hsp104 (D184S) in the presence and absence of ATP. In the absence of ATP, nearly all of the Hsp104 (D184S) was eluted in the position characteristic for its hexameric form as opposed to wild-type Hsp104 which was eluted in at least three unresolved peaks corresponding to different oligomeric species of Hsp104 (Fig. 5). In the presence of ATP, both proteins formed hexamers (Fig. 5). The difference in the hexameric status of both Hsp104 and its mutant observed in sedimentation and size-exclusion chromatography is most probably due to the experimental conditions, which differ substantially in time length of experiment, temperature, and glycerol content of the buffer. This shows that the sedimentation experiment allows better characterization of the influence of GuHCl on the oligomerization of Hsp104. All combined results suggest that mutation D184S results in an Hsp104 protein possessing a more stable oligomeric form not influenced by guanidine.
Fig. 5.
The D184S mutation of Hsp104 influences the oligomeric state of the protein in the absence of ATP. Size exclusion chromatography of Hsp104 wt and Hsp104 (D184S) mutant proteins was performed on a PROTEIN KW-804 column (Shodex packed column for HPLC) using a Waters 600 LCD HPLC Pump. Elution profiles of Hsp104 wt and Hsp104 (D184S) (20 μg each) were recorded in the absence (a) or presence of 5 mM ATP (b) in the running buffer (50 mM Tris, pH 7.5, 20 mM MgCl2, 150 mM KCl, 10% (v/v) glycerol) at a flow rate of 1 ml/min. The protein content of each fraction (tyrosine fluorescence) was measured using a Waters 474 Scanning Fluorescence Detector, at excitation wavelength of 280 nm, emission 335 nm, and a bandwidth of 40 nm
Taken together, this data suggests that the aspartic acid residue in position 184 in Hsp104 is involved in the interaction with guanidine, which presence severely inhibits both ATPase and disaggregation activities and promotes hexamerization of wild-type Hsp104. Mutation D184S results in a protein possessing lower ATPase and disaggregation activities. However, these activities are only weakly influenced by GuHCl. The oligomeric state of Hsp104 (D184S) is shifted towards hexamers, and the presence of GuHCl does not further influence its oligomeric status.
GuHCl efficiently stimulates ClpB’s ATPase activity but inhibits its disaggregating activity
Since it has been shown that the Hsp104 mutation D184S confers resistance to guanidine and is located in a highly conserved region of Hsp100 chaperone family members (Lee et al. 2003), we tested whether GuHCl would also affect another member of this family, the bacterial chaperone ClpB. To our surprise, an efficient stimulation of ClpB ATPase activity was observed in a GuHCl titration experiment (Fig. 6a), as opposed to inhibition of the analogous Hsp104 activity. Addition of 5 mM GuHCl resulted in a fivefold stimulation of the ATP hydrolysis rate of ClpB, with 50% of the maximal stimulation achieved at 1.5 mM GuHCl (Fig. 6a). As it has already been reported that the presence of casein increases the ATPase activity of ClpB (Woo et al. 1992), we tested the influence of both casein and GuHCl on the rate of ATP hydrolysis. Casein alone stimulates the ClpB ATPase activity 11-fold, and in the presence of both effectors, the ATPase rate increases over 35-fold (Supplementary Fig. S1). Therefore, we conclude that casein and GuHCl act independently in stimulating the ClpB ATPase activity.
Fig. 6.
The ClpB (D178S) mutant is not influenced by GuHCl in comparison to wild-type ClpB. a The ATPase activity of ClpB in the presence of increasing concentrations of GuHCl was measured spectrophotometrically in a coupled enzymatic assay. The concentration of ClpB was 4 μM. b Urea-denatured firefly luciferase (50 nM) was incubated with the KJE-ClpB chaperone system (0.5 μM ClpB, 1 μM DnaK, 0.2 μM DnaJ, 0.1 μM GrpE) at 24°C in the absence (filled circles) or presence of 5 mM GuHCl (filled squares) or with the KJE chaperone system alone in the absence of GuHCl (filled diamonds). Aliquots were withdrawn at the indicated time points, and the enzymatic activity of refolded luciferase was measured in a scintillation counter. c Urea-denatured firefly luciferase (50 nM) refolding was measured as in (b) the presence of increasing concentrations of GuHCl at a fixed concentration of ClpB (0.5 μM). The rate of reaction (increase in luciferase enzymatic activity) was calculated from the slope of the linear part of each curve and plotted as a function of GuHCl concentration. d The ATPase activity ClpB (D178S) in the presence of increasing concentrations of GuHCl was measured spectrophotometrically in a coupled enzymatic assay. The concentration of ClpB (D178S) was 4 μM. e Urea-denatured firefly luciferase (50 nM) was incubated with the KJE-ClpB (D178S) chaperone system (0.5 μM ClpB (D178S), 1 μM DnaK, 0.2 μM DnaJ, 0.1 μM GrpE) at 24°C in the absence (open circles) or presence of 5 mM GuHCl (open squares) or with the KJE chaperone system alone in the absence of GuHCl (filled diamonds). Aliquots were withdrawn at the indicated time points, and the enzymatic activity of refolded luciferase was measured in a scintillation counter. Experiments presented in the figure are representative of data obtained from separate protein preparations
We have previously shown that the DnaK, DnaJ, and GrpE (KJE)-ClpB chaperone system disaggregates GuHCl-denatured substrates (Ziętkiewicz et al. 2004). Therefore, we did not expect low concentrations of GuHCl to completely inhibit ClpB’s disaggregating activity as it does in the case of Hsp104-dependent disaggregation. However, since we observed a strong stimulatory effect of GuHCl on the ATPase activity of ClpB, we decided to test how different concentrations of GuHCl affect the KJE-ClpB disaggregation system. We followed the rate and efficiency of the disaggregation reaction using firefly luciferase. Although substantial inhibition of the disaggregation rate was observed in the presence of 5 mM GuHCl, a similar level of total disaggregation was achieved as in the absence of GuHCl (Fig. 6b). The results of a GuHCl titration experiment show that the addition of GuHCl significantly reduces the luciferase refolding rate (Fig. 6c). Similar GuHCl-dependent inhibition of the KJE-ClpB chaperone system was also observed when refolding the substrate, GFP (Fig. 7B and Supplementary Fig. S2A). The inhibition of GFP disaggregation by GuHCl was also shown to be irrespective of the concentration of ClpB used in the assay (Supplementary Fig. S2B).
Fig. 7.
GuHCl inhibits the Hsp100-dependent step in protein disaggregation for both yeast and bacterial Hsp70-Hsp100 disaggregating chaperones. A Heat-aggregated GFP (9 μg) was incubated with Hsp104 and Ssa1p/Ydj1p chaperones (2, 3, and 1 μM, respectively) in the absence (solid line) or presence of 5 mM GuHCl (dotted line). GFP reactivation was monitored in real time in a spectrofluorometer (excitation and emission wavelengths, 395 and 510 nm, respectively). B Heat-aggregated GFP (9 μg) was incubated with the KJE-ClpB chaperone system (0.5 μM ClpB, 1 μM DnaK, 0.2 μM DnaJ, 0.1 μM GrpE) in the absence (solid line) or presence of 5 mM GuHCl (dotted line). GFP reactivation was monitored as in A. C Sedimentation analysis of heat-aggregated GFP. (a) Heat-aggregated GFP (9 μg) was incubated with the bacterial Hsp70 system (KJE; 4.4 μM DnaK, 0.34 μM DnaJ, 0.45 μM GrpE) (top panel) or with both ClpB and the KJE system (KJEB; 0.5 μM ClpB, 4.4 μM DnaK, 0.34 μM DnaJ, 0.45 μM GrpE) in the absence (middle panel) or presence of 5 mM GuHCl (bottom panel) for 15 min at 24°C. The reactions were then stopped by the addition of EDTA to a final concentration of 40 mM, and the status of GFP molecules in reaction mixtures was investigated by sedimentation analysis (Beckman SW60 rotor; 15–45% (v/v) glycerol gradient; 40,000 rpm; 2 h; 4°C). The position of GFP in the gradients was visualized by SDS-PAGE followed by immunoblotting for GFP. (b) Heat-aggregated GFP was incubated with the yeast Hsp70 system (SY; 7.5 μM Ssa1p, 2.9 μM Ydj1p) (top) or with both Hsp104 and the Hsp70 system (SYHsp104; 2 μM Hsp104, 7.5 μM Ssa1p, 2.9 μM Ydj1p) in the absence (middle) or presence of 5 mM GuHCl (bottom) for 45 min at 24°C. The reactions were stopped, sedimented, and analyzed as in (a). (c) Control sedimentations; native (GFPN) and heat-aggregated GFP (GFPD) were subjected to glycerol gradient centrifugation and visualized as in (a)
Our in vitro experiments indicated that the presence of GuHCl influences the rate but not the total amount of ClpB-mediated disaggregation. To test possible in vivo effects, we monitored E. coli thermotolerance in the presence of GuHCl. No phenotype was observed. However, when we monitored the removal of aggregates formed in bacterial cells following heat shock, we did observe a mild in vivo effect on the disappearance of temperature-induced intracellular aggregates (Supplementary Fig. S3). This delay is consistent with our in vitro results showing an impaired rate of ClpB-dependent disaggregation.
Since the aspartic acid in position 184 in Hsp104 was identified as crucial for thermotolerance development and interaction with GuHCl, and is found in a highly conserved region, we decided to introduce an analogous mutation in ClpB. The resultant protein, ClpB (D178S), was purified and compared with the wild-type ClpB. In the absence of GuHCl, the ATPase activity of this mutant is similar to that of wild-type ClpB (Fig. 6d). Yet, in contrast to wild-type ClpB, no increase in the ATP hydrolysis rate was observed in the presence of GuHCl (Fig. 6d). Next, we analyzed the ability of ClpB (D178S) to disaggregate aggregated luciferase. Like the analogous Hsp104 (D184S) mutant, the disaggregating activity of ClpB (D178S) is lower than that of wild-type ClpB (compare Fig. 6b, e). Moreover, its disaggregating activity is not influenced by the presence of GuHCl (Fig. 6e). In our sedimentation experiments, both ClpB and its D178 mutant were found in the fractions characteristic for a monomer, and GuHCl did not influence their sedimentation profiles (Supplementary Fig. S4).
However, partial trypsin digestion showed that addition of GuHCl to the sample containing ClpB and ATP inhibited the proteolytic cleavage of ClpB (Supplementary Fig. S5). Under the same experimental conditions, the presence of GuHCl had no effect on the rate of trypsin cleavage for ClpB (D178S). Similarly to Hsp104 (D184S), the ClpB (D178S) mutant was digested substantially slower by trypsin as compared to wild-type protein (Supplementary Fig. S5). These experiments show that GuHCl does not influence an oligomeric status of ClpB as efficiently as it does in case of Hsp104. However, the partial trypsin digestion approach suggests that GuHCl does have some effect on wild-type ClpB but not the D178S mutant.
GuHCl inhibits the Hsp100-dependent step of disaggregation
The process of protein disaggregation is not solely dependent on Hsp100 chaperones in that it requires the specific cooperative action of Hsp100 and Hsp70 chaperones (Glover and Lindquist 1998; Krzewska et al. 2001b). Therefore, we made an effort to test more directly whether it is indeed only Hsp100, and not Hsp70, which is inhibited by GuHCl. We first analyzed the influence of GuHCl on Hsp104-dependent disaggregation of a substrate other than firefly luciferase, namely GFP protein. GFP has been successfully used to analyze the disaggregation activity of the bacterial ClpB-DnaK, DnaJ, and GrpE chaperone system (Ziętkiewicz et al. 2004). As expected, the addition of yeast chaperones Hsp104, Ssa1p, and Ydj1p to aggregated GFP resulted in its reactivation, monitored in real time as an increase in GFP fluorescence (Fig. 7A). The disaggregation process was efficiently blocked by the presence of 5 mM GuHCl in the reaction mixture (Fig. 7A). As stated above, inhibition of the GFP disaggregation rate by the bacterial chaperones was also observed in the presence of 5 mM GuHCl (Fig. 7B and Supplementary Fig. S2A). Because of these observations using two different substrates in two different systems, we continued to test whether GuHCl specifically influences the Hsp100 or the Hsp70 component of the bacterial and yeast chaperone disaggregating systems.
We took advantage of the fact that the Hsp70 system works upstream of Hsp100 (Ziętkiewicz et al. 2004; Weibezahn et al. 2004) and is able to disentangle single polypeptides from aggregates on its own (Ziętkiewicz et al. 2006). In the absence of Hsp100, the released polypeptides do not fold properly into their native structures and re-aggregate, forming a new class of aggregates characterized by a smaller size and a significantly lower sedimentation coefficient (Ziętkiewicz et al. 2006). These observations allowed us to investigate the influence of GuHCl on the specific disaggregation steps. Sedimentation conditions (40,000 rpm, SW 60 Beckman rotor; 2 h; 4°C) were chosen in such a way that following centrifugation, native GFP remained at the top of the gradient (fractions 1–3; Fig. 7C, subpanel c); the large aggregates of GFP formed a pellet at the bottom of the tube (Fig. 7C, subpanel c), but GFP aggregates remodeled by the KJE chaperone system sedimented to the middle of the gradient (fractions 5–9; Fig. 7C, subpanel a). The yeast Hsp70 chaperone system (Ssa1p/Ydj1p) was also capable of efficiently remodeling GFP aggregates, and the reaction produced analogous, small aggregates sedimenting in the middle of the gradient (fractions 4–12; Fig. 7C, subpanel b). When bacterial (KJE) or yeast (Ssa1p/Ydj1p) Hsp70 chaperone systems were supplemented with ClpB or Hsp104, respectively, GFP was found at the top of the gradients in the position characteristic for native GFP (Fig. 7C). These top fractions exhibited green fluorescence (result not shown), indicating that the action of the Hsp70-Hsp100 chaperones resulted in disaggregation and efficient reactivation of GFP. When the complete chaperone reactions, bacterial KJE-ClpB and yeast Ssa1p/Ydj1p-Hsp104, were supplemented with 5 mM GuHCl, the formation of fluorescent GFP refolded by the bacterial system was significantly inhibited (Fig. 7C, subpanel a) and essentially blocked in the case of the yeast system (Fig. 7C, subpanel b). However, it is important to note that under these conditions, the initial large aggregates of GFP, which typically form a pellet at the bottom of the tube, were efficiently converted by bacterial chaperones to small aggregates that sediment in the middle of the gradient (Fig. 7C, subpanel a). Similar, albeit slightly less efficient conversion, was also observed for yeast chaperones (Fig. 7C, subpanel b). These observations suggest that the activity of the bacterial (KJE) and yeast (Ssa1p/Ydj1p) Hsp70 chaperone systems was not affected by the presence of GuHCl. Since the hydrolysis of ATP was shown to be required for Hsp70-dependent remodeling of aggregates (Ziętkiewicz et al. 2004), this observation is also consistent with the lack of influence of GuHCl on the ATPase activity of both the KJE and Ssa1p/Ydj1p systems (Supplementary Table S1). Therefore, we conclude that GuHCl affects the Hsp100-dependent step in the disaggregation process of both bacterial and yeast chaperones.
Discussion
In this study, we analyzed the influence of guanidine hydrochloride on the abilities of two homologous bi-chaperone systems, bacterial KJE-ClpB and yeast Ssa1/Ydj1-Hsp104, to disaggregate protein substrates. Our results clearly show that guanidine inhibits the disaggregating activity of both bi-chaperone systems, albeit to different degrees. Disaggregation in the yeast system is blocked completely, whereas the effect is less pronounced in the bacterial system. We observed a two- to three-fold inhibition in the rate of disaggregation mediated by bacterial chaperones, but the efficiency was unaffected. Our attempts to observe the effect of GuHCl on E. coli thermotolerance were unsuccessful (results not shown), although we did observe a somewhat minor in vivo effect on the disappearance of temperature-induced intracellular aggregates. This delay in removal of aggregated proteins is consistent with our in vitro results showing an impaired rate of disaggregation. The inhibitory effect of GuHCl on bacterial chaperones has not been described previously either in vivo or in vitro.
For both Hsp104 and ClpB, the change of the conserved aspartic acid (D184 and D178, respectively) to serine resulted in the formation of protein with lower disaggregating activity, yet in both cases, the activities of the respective mutant proteins did not change in response to GuHCl, unlike their wild-type counterparts. Analysis of the structural models of Hsp104 and ClpB (Lee et al. 2003, 2010; Ziętkiewicz et al. 2010) shows that D184 and D178, respectively, are located in the middle of NBD1 domain, in close proximity to the first ATP binding site and not at the interface between adjacent subunits. Therefore, it is unlikely that the observed effect is due to the direct interaction of GuHCl with the amino acids at the subunit interface, which in turn influences the oligomerization of the chaperones. Collective data suggest that D184 in Hsp104 is involved in the interaction with positively charged guanidine. Lack of negative charge at this position in the D184S mutant results in the lack of interaction between Hsp104 and guanidine. This mutant possesses substantially lower ATPase and disaggregating activities in vitro, yet in vivo, the observed phenotype is almost normal. This suggests that a certain robustness in the functioning of the Hsp104 protein in the chaperone network exists. It is worth mentioning that both Hsp104 and Hsp104 (D184S) were expressed in yeast cells at the same level (Supplementary Fig. S6), and three independent Hsp104 (D184S) purifications resulted in a similarly active chaperone protein. An analogous robustness in ClpB functions also exists in E. coli since the observed two- to three-fold guanidine-dependent inhibition of ClpB activity in the presence of GuHCl does not result in an observable phenotypic effect.
Our results clearly assign the guanidine-dependent inhibition of the Hsp70/Hsp100 bi-chaperone system to the Hsp100-dependent disaggregation step. This is based on the observations that in both bacterial and yeast systems, neither Hsp70’s ATPase activity nor its ability to remodel protein substrate aggregates was disturbed by the presence of GuHCl. Thus, at which point(s) in the Hsp100-dependent step does GuHCl influence disaggregation? And what is the difference(s) between the two studied systems? Guanidine affects the ATPase activities of the bacterial and yeast Hsp100 chaperones oppositely, stimulating ClpB and inhibiting Hsp104. Yet, in both cases, disaggregation activity is inhibited. These results suggest that there is no simple correlation between the rate of ATP hydrolysis by Hsp100 chaperones and their ability to disaggregate proteins. Recently, it was shown that restrained ATP hydrolysis promoted substrate remodeling activity of ClpB and Hsp104, most probably by inducing the balance between substrate holding and unfolding (Doyle et al. 2007).
In contrast to its opposite effect on ATPase activities, GuHCl seems to have a stimulatory effect on the oligomerization of both ClpB and Hsp104. The effect is much more pronounced for Hsp104 since stable hexameric structures were easily detected by us and others (Grimminger et al. 2004) in sedimentation analysis. The effect for ClpB was observed only using trypsin digestion approach suggesting the transient stabilization of the hexameric structure. Interestingly, inhibition of the disaggregation reaction correlated with the increased stability of the oligomeric chaperone structures. One can speculate that during the disaggregation process, Hsp100 oligomerization and deoligomerization events must take place, and thus, the stabilization of Hsp100 oligomers does not necessarily lead to an increase in the overall efficiency (or rate) of the disaggregation reaction. On the other hand, it has to be remembered that results obtained for ClpX show that a stable hexameric variant of this chaperone is fully active in a translocation assay (Martin et al. 2005).
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 549 kb)
Acknowledgments
We thank Dr. Dariusz Wyrzykowski for his help with ITC experiments. We thank Drs. Szymon Ziętkiewicz, Debbie Ang, and Wojciech Sawula for discussions and critical reading of the manuscript. This work was supported by the Polish Ministry of Science and Higher Education grant No. N301 507338. E.M. and N.L. were supported by TEAM/2009/-3/5 program.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Bösl B, Grimminger V, Walter S. The molecular chaperone Hsp104—a molecular machine for protein disaggregation. J Struct Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Cyr D, Douglas MG. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. Asymmetric deceleration of Clpb and Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40:1375–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/S0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Grimminger V, Richter K, Imhof A, Buchner J, Walter S. The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J Biol Chem. 2004;279:7378–7383. doi: 10.1074/jbc.M312403200. [DOI] [PubMed] [Google Scholar]
- Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- Jung G, Jones G, Masison DC. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc Natl Acad Sci USA. 2002;99:9936–9941. doi: 10.1073/pnas.152333299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Wada C, Yoshioka S, Yura T. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock sigma factor (sigma 32) J Bacteriol. 1991;173:4247–4253. doi: 10.1128/jb.173.14.4247-4253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzewska J, Konopa G, Liberek K. Importance of two ATP-binding sites for oligomerization, ATPase activity and chaperone function of mitochondrial Hsp78 protein. J Mol Biol. 2001;314:901–910. doi: 10.1006/jmbi.2001.5190. [DOI] [PubMed] [Google Scholar]
- Krzewska J, Langer T, Liberek K. Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett. 2001;489:92–96. doi: 10.1016/S0014-5793(00)02423-6. [DOI] [PubMed] [Google Scholar]
- Laskowska E, Kuczynska-Wisnik D, Skórko-Glonek J, Taylor A. Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol Microbiol. 1996;22:555–571. doi: 10.1046/j.1365-2958.1996.1231493.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FTF. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/S0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- Lee S, Sielaff B, Lee J, Tsai FTF. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci USA. 2010;107:8135–8140. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Lewandowska A, Ziętkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker T, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rüdiger S, Röder D, Langen H, Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Strub C, Rist W, Weibezahn J, Bukau B. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- Norby JG. Coupled assay of Na+, K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Lindquist S. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J Biol Chem. 1994;269:4480–4487. [PubMed] [Google Scholar]
- Rudyak SG, Brenowitz M, Shrader TE. Mg2+-linked oligomerization modulates the catalytic activity of the Lon (La) protease from Mycobacterium smegmatis. Biochemistry. 2001;40:9317–9323. doi: 10.1021/bi0102508. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Lindquist S. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Ware DM, Queitsh C, Kowal AS, Lindquist SL. Subunit interactions influence the biochemical and biological properties of Hsp104. Proc Natl Acad Sci USA. 2001;98:914–919. doi: 10.1073/pnas.031568098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-V. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/S1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Squires CL, Pendersen S, Ross B, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite MF, Mundy CR, Cox BS. Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. AAA proteins. Lords of the ring. J Cell Biol. 2000;150:F13–F19. doi: 10.1083/jcb.150.1.F13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Motohashi K, Yoshida M. Roles of the two ATP binding sites of ClpB from Thermus thermophilus. J Biol Chem. 2002;277:5804–5809. doi: 10.1074/jbc.M109349200. [DOI] [PubMed] [Google Scholar]
- Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Bukau B, Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J Biol Chem. 2003;278:32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FTF, Mogk A, Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Woo KM, Kim KI, Goldberg AL, Ha DB, Chung CH. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J Biol Chem. 1992;267:20429–20434. [PubMed] [Google Scholar]
- Ziegelhoffer T, Lopes-Buesa P, Craig EA. The dissociation of ATP from hsp70 of Saccharomyces cerevisiae is stimulated by both Ydj1p and peptide substrates. J Biol Chem. 1995;270:10412–10419. doi: 10.1074/jbc.270.18.10412. [DOI] [PubMed] [Google Scholar]
- Ziętkiewicz S, Krzewska J, Liberek K. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem. 2004;279:44376–44383. doi: 10.1074/jbc.M402405200. [DOI] [PubMed] [Google Scholar]
- Ziętkiewicz S, Lewandowska A, Stocki P, Liberek K. Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. J Biol Chem. 2006;281:7022–7029. doi: 10.1074/jbc.M507893200. [DOI] [PubMed] [Google Scholar]
- Ziętkiewicz S, Ślusarz MJ, Ślusarz R, Liberek K, Rodziewicz-Motowidło S. Conformational stability of the full-atom hexameric model of the ClpB chaperone from Escherichia coli. Biopolymers. 2010;93:47–60. doi: 10.1002/bip.21294. [DOI] [PubMed] [Google Scholar]
- Zylicz M, Ang D, Liberek K, Georgopoulos C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 549 kb)