Abstract
Large granular lymphocyte (LGL) leukemia characterized by clonal expansion of antigen-activated cytotoxic T cells (CTL). Patients frequently exhibit seroreactivity against a human T-cell leukemia virus (HTLV) epitope, BA21. Aplastic anemia, paroxysmal nocturnal hemoglobinuria and myelodysplastic syndrome are bone marrow failure diseases that can also be associated with similar aberrant CTL activation (LGL-BMF). We identified a BA21 peptide that was specifically reactive with LGL leukemia sera and found significantly elevated antibody reactivity against the same peptide in LGL-BMF sera. This finding of shared seroreactivity in LGL-BMF conditions and LGL leukemia suggests that these diseases might share a common pathogenesis.
Keywords: LGL leukemia, aplastic anemia, myelodysplastic syndrome, paroxysmal nocturnal hemoglobinuria, HTLV antibody
1. Introduction
The Bone Marrow Failure Disease Consortium (BMFDC) of the Rare Diseases Clinical Research Network was organized with National Institutes of Health support to study rare hematologic diseases, including LGL leukemia, myelodysplastic syndromes (MDS), aplastic anemia (AA), and paroxysmal nocturnal hemoglobinuria (PNH). Extensive data support the antigen activated nature of leukemia LGL cells [1]. T-LGL leukemia has been characterized as an accumulation of apoptosis resistant effector memory cytotoxic T lymphocytes (CTL) that constitutively express perforin and other markers of activated killer cells [1–3]. Like LGL leukemia, expansions of effector memory CTL have been noted in these other hematologic diseases [4–8]. However, T-LGL leukemia cells are typically CD3+CD8+CD57+DR+, with clonal rearrangement of the T cell receptor (TCR), whereas the CTL clones in these marrow failure diseases are less prominent and often oligoclonal [4–9]. It has been postulated that exposure to infectious agents might lead to CTL expansion in LGL leukemia, although the actual target recognized by these activated CTL has not been characterized [4, 10].
There are some reports of LGL leukemia developing in retrovirally-infected individuals [11–17]. Findings in LGL leukemia indicate that sera from 21% of patients are positive in an HTLV-1 and/or HTLV-2 ELISA, compared to 0.17% positive sera in normal donors. Subsequent Western blot analyses showed that this reactivity is sero-indeterminate and that most patients are not infected with a prototypical retrovirus [18–20].
It has been documented that non-infected patients with autoimmune disorders and chronic diseases of aging can demonstrate indeterminate retroviral serology [21, 22]. There are numerous critical differences in the pattern of reactivity for LGL leukemia patients and these other cross-reactive groups. According to a previous study, 84% of non-infected sero-indeterminate normal donors had antibodies to HTLV gag p19 protein only, 16% were reactive with HTLV gag p24 only, and 2.9% had dual gag p24 plus env p21e reactivity [18]. In contrast, only 4% of sero-indeterminate LGL leukemia patients were reactive to gag p19 only, while 82% reacted to gag p24, and 39% demonstrated dual gag p24/env p21e reactivity. Therefore, patients have a disease-directed antibody response against HTLV-1 that is markedly distinct from normal non-infected people.
We have demonstrated that HTLV env reactivity in LGL leukemia was directed at the BA21 region, overlapping the immunogenic p21e transmembrane [23]. Previous studies have indicated that 30% to 46% of sera from LGL leukemia patients were reactive to BA21. However, LGL-specific BA21 epitopes were not identified at that time. Results of an earlier study utilizing radio-immunoassays suggested that the amino terminus of BA21 (containing QEQCR) was more specifically reactive than the PPLE-containing region for patients infected with HTLV-1 [24].
Since the original descriptions of HTLV seroreactivity in LGL leukemia were drawn from clinical ELISAs designed to screen for HTLV-1/2 infection in blood donors, we used a combined microarray and ELISA platform to test antibody recognition. The reactive sequences were further tested on ELISA to identify a disease-specific BA21 epitope, i.e.; one that was consistently recognized by antibodies from LGL leukemia patients but not by serum antibodies from healthy donors. The selected LGL leukemia-specific epitope was located in the amino terminus of BA21. It was recognized by at least 40% of LGL leukemia sera but not by normal donor sera. We also determined the levels of BA21 IgG for participants in the BMFDC. We found that a substantial number of sera from participants with AA, MDS, and PNH also recognized the epitope. Reactivity with the LGL leukemia-specific BA21 epitope was associated with these BMF diseases. These data provide further support for the hypothesis that a variety of hematologic diseases associated with CTL expansion might result from a common pathogenetic mechanism.
2. Materials and methods
2.1 Sera
BA21 antibody testing was approved as part of a multi-institutional IRB protocol for the Rare Disease Clinical Research Network (RDCRN)-sponsored Bone Marrow Failure Diseases Consortium (BMFC). The consortium included four institutions: The Penn State Hershey Cancer Institute (Penn State College of Medicine, Hershey, PA USA), the Cleveland Clinic Taussig Cancer Center (Translational Hematology and Oncology Research, Cleveland, OH, USA), the H. Lee Moffitt Cancer Center (Malignant Hematology, Tampa, FL, USA), and the Jonsson Comprehensive Cancer Center (Department of Hematology/Oncology, University of California Los Angeles, Los Angeles, CA, USA).
Sera collected from consented bone marrow failure (BMF) patients at the time of enrolment (baseline samples) were included in this study. The BMF disorders in this study are aplastic anemia (AA), LGL leukemia, myelodysplastic syndrome (MDS) and paroxysmal nocturnal hemoglobinuria (PNH).
Additional LGL leukemia in this study sera were collected under the same inclusion/exclusion criteria for the LGL Leukemia Registry and Tissue Bank located at Penn State Hershey Cancer Institute (Hershey, PA). Sera from age and gender-matched healthy anonymous donors were collected during the same period by Florida Blood Services (St Petersburg, FL) and by the Hershey Medical Center Blood Bank (Hershey, PA). Inclusion and exclusion criteria for all participants including normal donors are detailed in a supplemental document (Supplementary Data).
Pooled normal control sera were purchased from Sigma (St Louis, MO). HTLV-1/2 infected control sera were purchased from Zeptometrix (Buffalo, NY) and from SeraCare (Milford, MA). Inclusion criteria included confirmed infection with HTLV-1 or HTLV-2. Gender, age and health status were not available for these samples. Screening for all individual donors/specimens was performed as indicated in Table 1.
Table 1. Donor and Specimen Screening.
Screening parameters are listed.
| DONOR & SPECIMEN SCREENING | ||
|---|---|---|
| Universal Screening (For All Normal Control & BMF Participants) | NORM CTL | BMF PTS |
| Demographics including date of birth | X | X |
| History and Physical Exam | ||
| CBC/Differential/Reticulocyte count | ||
| Serum iron/total iron binding capacity/ferritin | ||
| RBC folate/B12 serology | ||
| Pathogen Screening (HIV/HTLV/HBV/HCV/Chagas/Syphillis) | ||
| Standard of Care, General (to establish diagnosis) | NORM CTL | BMF PTS |
|---|---|---|
| Symptoms and Treatment | NA | X |
| Comprehensive metabolic panel | ||
| Cytogenetics if bone marrow aspiration if done. | ||
| Bone marrow biopsy/aspiration* | ||
| HLA TYPING (KIR-Ligand) if not already known | ||
| Quality of life Questionnaire |
| Standard of Care, Disease Specific (to establish diagnosis) | NORM CTL | AA | LGL | MDS | PNH |
|---|---|---|---|---|---|
| Factor V Leiden, Protein S, Protein C, Prothrombin 2010A | NA | X | |||
| PNH clones by flow cytometry (CD55 & CD59 on granulocytes) | X | X | |||
| DEB testing (Fanconianaemia) in patients < 21 years old | X | ||||
| TCR-gamma rearrangement | X | X | |||
| Serum Protein electrophoresis | X | ||||
| Rheumatoid factor | X | ||||
| Low grade lymphoma panel | X | ||||
| Bone marrow flow cytometry of blast gate** | X | ||||
| Parvovirus B-19 IgM and IgG serologies | |||||
| Chest x-ray; CT scan if chest x-ray inconclusive |
: Biopsy/aspiration was performed only if clinically indicated.
: Flow cytometry was done only if bone marrow cells were already collected.
NORM CTL: normal control donors/specimens, BMF PTS: Bone marrow failure participants, NA, not applicable, AA: aplastic anemia, LGL: LGL leukemia, MDS: myelodysplastic syndrome, PNH: paroxysmal nocturnal haemoglobinuria
2.2 Epitope prediction analyses
Online databases were mined for epitope predictions for the BA21 sequence. Databases used were LEPD (Linear Epitope Prediction Database, http://140.121.196.30/LEPD_Antigenicity.php) [25] and the Immune Epitope Database (IEDB, http://tools.immuneepitope.org/main/html/bcell_tools.html). Reinforced merging (LEPD) was combined with Kolaskar & Tongaonkar Antigenicity[26] plus BEpiPred [27] (both at IEDB) algorithms.
2.3 Biologics
Peptides for array experiments were synthesized by Invitrogen EvoQuest Laboratory Services (Carlsbad, CA). Recombinant BA21 peptides for enzyme-linked immunosorbent assays (ELISAs) were purchased from Biosynthesis Inc. (Lewisville, TX). Anti-human IgG antibodies and recombinant human IgGs were obtained from Sigma.
2.4 Reagents
ELISA reagents were purchased from Sigma and from Fisher Scientific (Pittsburgh, PA). Dynal 4HB ELISA plates were used for all experiments. For peptide arrays, epoxy slides and buffers for printing and blocking were purchased from ArrayIt (Sunnyvale, CA). Cover slips for array slides were purchased from Fisher Scientific. Fluorescent dye was obtained from Invitrogen (Carlsbad, CA).
2.5 BA21 epitope screening array
The entire BA21 region was split into overlapping peptides, based on the published work of Sokol, et al (Table 2, BA21.1 – BA21.4) [28]. Anti-human IgG was used as a positive control. A sequence derived from duck hepatitis B virus core protein, which is non-reactive with human sera on arrays, was used as a negative control (CLT.NR).
Table 2.
Linear B-cell Epitope Prediction for BA21.Peptides and their sequences are shown, along with regions of each peptide identified to have the following antigenic properties: beta-turns, hydropathicity, exposed surfaces, charged (polar) regions, bulky regions (high molecular weight), hydrophobic regions, along with regions containing loops and other flexible structures. An antigenicity score was assigned to the regions with greatest antigenicity (High Antigenicity). Shaded regions represent the peptides with the greatest predicted overall reactivity with the darkest shading denoting the most reactive region of BA21.
| PEPTIDE ID | PEPTIDE SEQUENCE |
|---|---|
| BA21.1 | QEQCRFPNITNSHVSIL
|
| BA21.2 | QEQCRFPNITNSHVSILQER
|
| BA21.3 | NSHVSILQERPPLENRVLTGW
|
| BA21.4 | PPLENRVLTGWGLNWDLGLSQW
|
| (−) DHBV Core | EEAEEIPLGDLFKHQEERI
|
| Q | E | Q | C | R | F | P | N | I | T | N | S | H | V | S | I | L | Q | E | R | P | P | L | E | N | R | V | L | T | G | W | G | L | N | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EPITOPE FEATURE | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 |
| BETA-TURN | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||
| HYDROPATHICITY | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||
| SURFACE | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||
| POLARITY | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||
| MOLECULAR WT | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||
| HYDROPHOBICITY | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||
| FLEXIBILITY | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||||||
| LOW ANTIGENICITY | ||||||||||||||||||||||||||||||||||
| HIGH ANTIGENICITY | X | X | X | X | X | X | X |
The peptides and anti-IgG were suspended in protein printing buffer and applied to ArrayIt SuperEpoxy slides using a MicroGrid II robotic arrayer (BioRobotics, Woburn, MA). Slides were printed and prepared following manufacturer recommendations (http://arrayit.com/Products/Microarray_Slides/Microarray_Slides_SuperEpoxy_1/microarray_slides_superepoxy_1_and_2_protein.html). Binding sera were visualized on an Axon GenePix 4000B scanner (Molecular Devices, Sunnyvale CA) and GenePix Pro 4.0 software (Molecular Devices). The signal data was transferred to GeneSpring software (Agilent Technologies, Santa Clara CA) for analysis. IgG recognition was confirmed by BA21 peptide ELISA.
2.6 BA21 peptide ELISA design and controls
Peptides BA21.1 and BA21.4 (shown on Table 2) as well as the entire BA21 sequence (BA21) were tested. Peptides were applied to 96-well Immulon 2 plates (Dynal) at previously determined optimal concentrations. Commercially available pooled normal donor AB sera and 5 each HTLV-1/-2 infected donor sera were included in all BA21 ELISAs as calibrators and controls. IgG concentrations were standardized as described in the Supplementary Data. At least two ELISAs were performed for each test. For each individual, mean values represent IgG values from two plates with an inter-plate standard deviation equal to or less than 10% of the mean IgG value.
2.7 Statistical analyses
There are no data on BA21 seroreactivity in healthy non-infected donor sera. For this reason, a 99% confidence interval was employed to identify the upper limits of reactivity for the normal donor sera and this cut-off value was then applied to each LGL-BMF group to determine which samples had elevated anti-BA21 IgG. The determination of significant differences in anti-BA21 IgG levels for each group compared to the normal donors was calculated by unpaired two-sided T tests. When smaller data sets were analyzed, the results of the T tests were compared to significance outcomes determined by Mann-Whitney Wilcoxon (MWW) test, using α = 0.01 to derive the Z-critical value. The resulting MWW p value is significant if it is close to 1 or 0.
3. Results
3.1 Prediction of BA21 B cell epitopes
Antigenicity was predicted for three regions of BA21, namely an amino terminus containing EQCR (BA21.1, BA21.2), a region near the center of BA21 containing PPLE (BA21.3, BA21.4), and a region at the carboxyl terminus containing WGLN (BA21.4) (Table 2). Of these, PPLE-containing sequences were predicted to be the most immunogenic. We screened peptides containing the predicted epitopes using microarray and ELISA methods.
3.2 Array and ELISA screening for LGL leukemia-specific BA21 epitopes
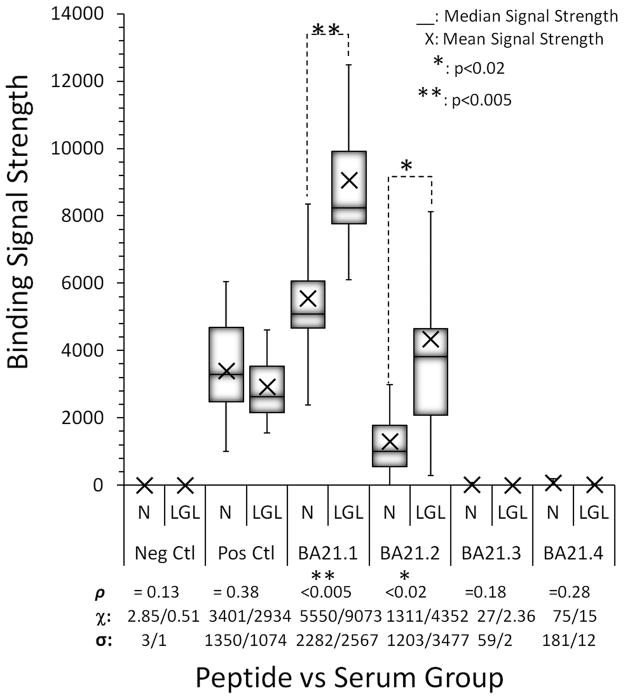
A peptide array was used to perform the first screening of the BA21 epitopes for 20 patients and 20 normal controls (Fig. 1). The array was constructed with overlapping BA21 peptides BA21.1 – BA21.4, including the peptides that were analyzed in the prediction analyses. A peptide from the duck hepatitis B virus core reacted with none of the sera and was used as a non-reactive control. As shown in Figure 1, the positive control anti-IgG1 antibody captured a portion of the total IgG from both groups. In contrast the BA21 epitopes hybridized to all IgG subtypes. The result for this positive control was also used to confirm that both groups produce equivalent amounts of IgG1. For BA21.1, median values in Normal and Leukemia groups were 5088 and 8247 respectively; the distributions in the two groups differed significantly using the Mann-Whitney Wilcoxon test with the Z-Critical value set for 99% certainty (α=0.01, U = 13, n1 = n2 = 11, P < 0.005 two-tailed). The p value from the Mann-Whitney Wilcoxon test was 0.89 in favor of the LGL leukemia group. Similarly for BA21.2, median values in Normal and Leukemia groups were 1005 and 3822 respectively; and the distributions also differed significantly using the same test (α=0.01, U = 22, n1 = n2 = 11, P < 0.02 two-tailed). For this peptide, the MWW p value was 0.82 in favor of LGL leukemia. None of the other groups were significantly different according to the outcomes of unpaired two-tailed T tests and Mann-Whitney Wilcoxon tests.
Fig 1.
Peptide Array Epitope Mapping. Overlapping peptides were tested for seroreactivity against LGL leukemia and normal donor sera (Peptide vs. Serum Group). The signal strength of the sera was interpreted as reactivity. Neg Ctl: duck hepatitis B virus core peptide. Pos Ctl: Anti-IgG1. ρ: probability, χ: mean value, σ: standard deviation.
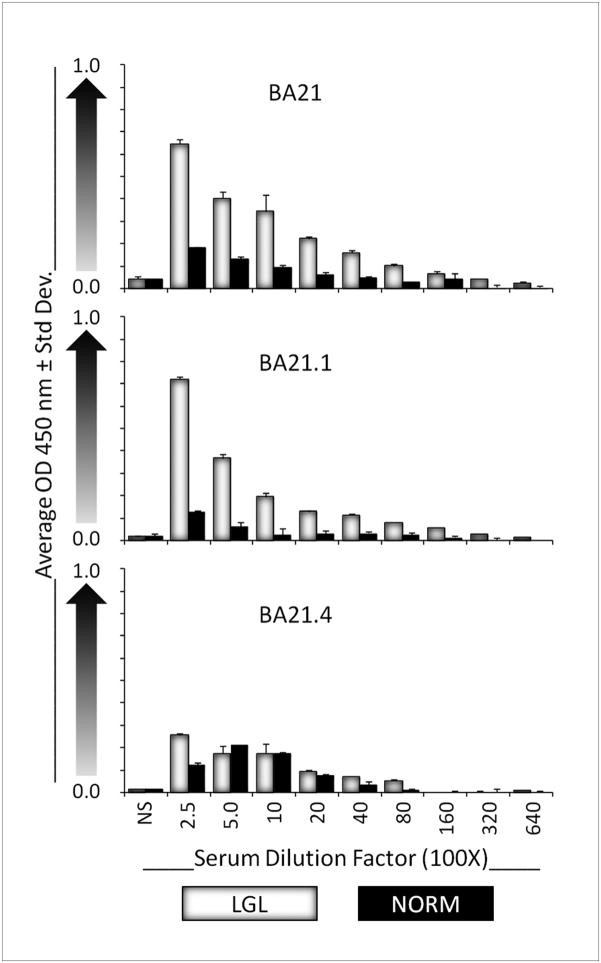
Peptides derived from the amino terminus of BA21 elicited significantly stronger signal intensities from the LGL leukemia sera than from normal sera. For the shortest BA21 EQCR-containing peptide BA21.1, the mean signal strength for LGL leukemia sera was 9073 (SD: 2567; range: 6094 – 15531). For the normal sera it was only 5550 (SD: 2282; range: 2377–10800). The difference between the LGL: Normal signals was significant with p<0.005. The slightly longer version of this peptide, BA21.2, also elicited a significantly greater signal from the leukemia sera than from the normal sera (p<0.02). For the PPLE-containing peptides, there was no significant difference in binding signal strength for normal versus leukemia sera (BA21.3, p=0.18; BA21.4, p=0.28). We validated the array findings by testing an additional 5 leukemia sera and 12 normal donor sera on ELISA (Fig. 2). In our ELISA optimization testing, we learned that BA21.2 was more reactive with normal sera than was BA21.1 (data not shown). Therefore we focused our attention on BA21.1.
Fig 2.
BA21 Epitope ELISAs. Serum IgG levels are proportional to the Average OD 450nm (Y axis). Sera were serially diluted in PBS 250X to 64,000X. NS: no sera. LGL: LGL leukemia sera. Norm: Normal donor sera.
3.3 Testing the LGL leukemia-specific BA21 epitope
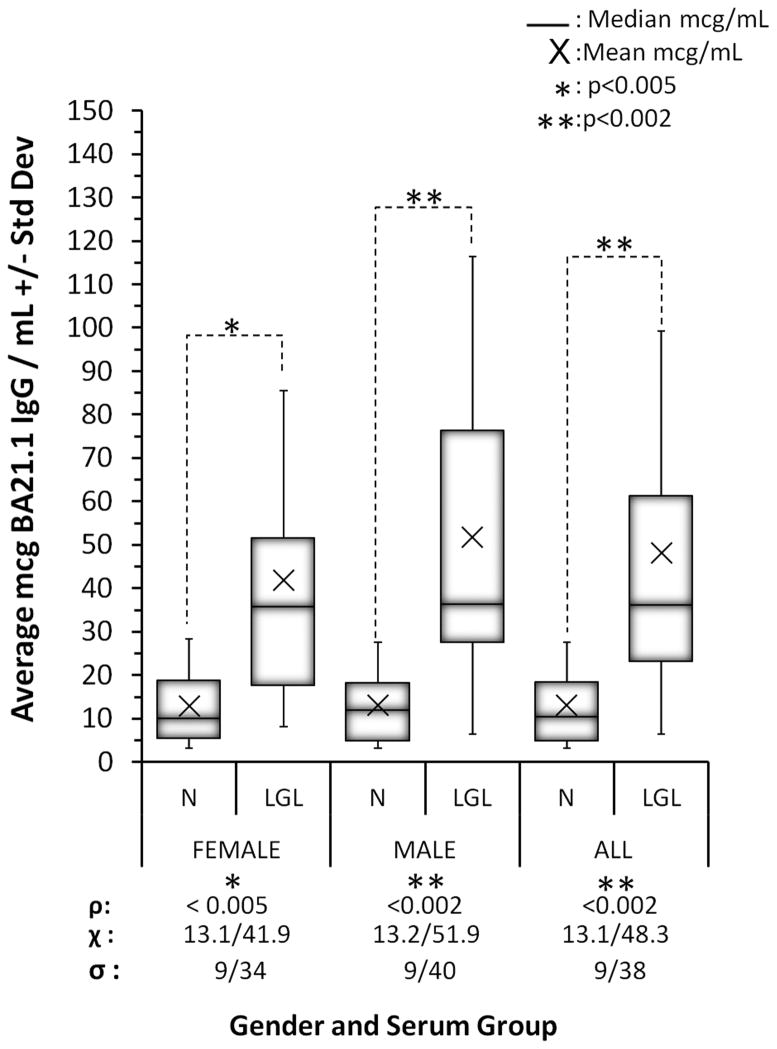
The BA21.1 epitope was tested on 42 age and gender-matched specimens from LGL leukemia patients and from normal control donors. Peptide BA21.1 was significantly more reactive with LGL leukemia sera than with normal sera (p<0.002) (Fig. 3). From the 99% confidence interval of the normal serum values it was determined that the upper value for normal sera was 37.4 μg IgG/mL. None of the 42 normal sera (0%) and 20 of the 42 leukemia sera (48%) had significantly greater than normal reactivity. The BA21.1 peptide was up to two-fold more specific for LGL leukemia than was the entire BA21 antigen [23, 28]. To confirm the significance of the unpaired two-tailed T tests, we applied Mann-Whitney Wilcoxon testing to the three groups. Results of this test suggested that the distributions for normal and leukemia sera differed significantly in favor of LGL leukemia, but with no significant gender difference within the leukemia or normal group. For male versus female LGL leukemia, the MWW p value was 0.52 in favor of males (α=0.01, U = 179, n Female = 15, n Male = 27, P = 0.42 two-tailed). For male versus female normal donors, the MWW p value was 0.51 in favor of males (α=0.01, U = 198, n Female = 15, n Male = 27, P = 0.98 two-tailed).
Fig 3.
LGL Leukemia Serum Reactivity to BA21.1. ELISA data is shown. Females: n=15, average age was 49.5 for female LGL leukemia patients and 49.2 for normal female donors. Males: n=27, average age was 62.8 for male LGL leukemia patients and 60 for normal male donors. ρ: probability, χ: mean value, σ: standard deviation.
3.4 BA21 antibody reactivity in bone marrow failure diseases
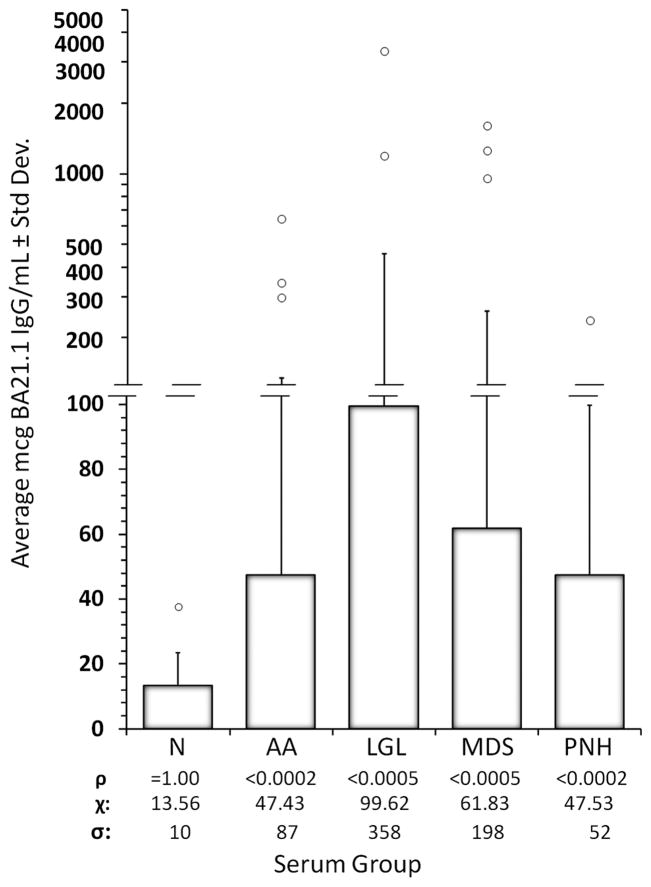
Next, 102 LGL leukemia sera, 230 BMF sera and 237 normal donor sera were analyzed for reactivity to BA21.1. We found significantly elevated antibody reactivity against BA21.1 in all patient groups (Fig. 4). The 99% confidence interval for normal sera was 38.90 μg BA21.1 IgG/mL sera. Only 1 of 237 (0.42%) normal sera were above this level, while 41 of 102 (40%) LGL leukemia sera were reactive. Likewise, a considerable percentage of sera from all of the BMF groups exceeded the 99% confidence interval value against BA21.1: 28 of 83 (34%) AA, 39 of 126 (31%) MDS, and 9 of 21 (43%) PNH.
Fig 4.
BA21 IgG levels in LGL Leukemia and BMF Diseases. ELISA data is shown. N: normal donors, AA: aplastic anemia, LGL: large granular lymphocyte leukemia, MDS: myelodysplastic syndrome, PNH: paroxysmal nocturnal hemoglobinuria (all described in text). Open circles: individual values > 99% CI for each group. ρ: probability, χ: mean value, σ: standard deviation (also shown as bars).
3.5 BA21 reactivity and transfusion status
Regarding transfusion status, 24% of LGL leukemia patients with transfusion data received a transfusion prior to serum collection. In contrast, 62% of AA, 51% of MDS, and 60% of PNH patients with transfusion data reported receiving at least one transfusion prior to baseline serum collection. However, there was no correlation of transfusion status with BA21 reactivity.
3.6 BA21 reactivity and immune-modulating therapy
Treatment with growth factors and immune suppressive drugs were analyzed for an association with elevated versus normal BA21 IgG levels. Differences in BA21 IgG could not be analyzed for some therapies because few participants were being treated at baseline (time of registration). Of the variables with sufficient data, none were significantly associated with normal or elevated BA21 IgG, including intravenous immunoglobulin (IVIG), epoetin alpha, anti-thymoglobulin, cyclosporine, prednisone and methotrexate. There was also no significant association of BA21 reactivity status with a history of autoimmune disease. Detailed information on these baseline data which include clinical lab values, demographics, TCR clonotype, medical history and treatment history are provided as supplemental data (supplemental data S1 and S2).
3.7 Elevated BA21.1 IgG as a feature of LGL leukemia and BMF diseases
Pooled risk determinations for BMF patients versus normal donors revealed that individuals with elevated BA21.1 IgG were 128 times (OR = 128.4:1) more likely to have LGL leukemia, AA, MDS or PNH than to be in a normal condition (Table 3). The specificity of the BA21.1 IgG test to any BMF condition was 99%. For LGL leukemia alone, the Odds Ratio based on relative risk was 158:1 (not shown). 35% of all LGL-BMF disease sera tested in this study demonstrated abnormally high BA21.1 antibody levels, independent of any association to over 140 baseline parameters.
Table 3.
BA21.1 IgG Specificity and Risks.Specificity, sensitivity and risks associated with elevated BA21.1 IgG were calculated for sera from patients with LGL leukemia and BMF diseases, and for sera from normal controls. BA21 IgG: BA21.1 HI: Specimens with BA21.1 IgG levels greater than the 99% confidence interval limit of normal sera. BA21.1 NRM: Specimens with normal levels of BA21.1 IgG. LGL-BMF: Patients with LGL leukemia and BMF diseases. Normal: Healthy participants.
| BA21 IgG SPECIFICITY & RISKS | ||||
|---|---|---|---|---|
| BA21 IgG | LGL-BMF | NORMAL | SUM | PARAMETER |
| Count of BA21.1 HI | 117 | 1 | 118 | TOTAL # BA21.1 HI |
| Count of BA21.1 NRM | 215 | 236 | 451 | TOTAL # BA21.1 NRM |
| Total Count | 332 | 237 | 569 | TOTAL # Tested |
| Sensitivity of BA21.1 Serology: LGL-BMF | 35.2% | |||
| Specificity of BA21.1 Serology: LGL-BMF | 99.6% | |||
| CALCULATION | VALUE | DESCRIPTION |
|---|---|---|
| Positive Predictive Value | 99% | Ability to predict LGL-BMF disease specimen |
| Negative Predictive Value | 52% | Ability to rule out normal specimen |
| False Positive Rate | 0.42% | Percent of normal specimens that would have elevated BA21.1 IgG results on this test |
| Likelihood Ratio | 83.5:1 | Odds that a LGL-BMF specimen vs a normal specimen will produce elevated BA21.1 IgG |
| Overall Odds Ratio | 128.4:1 | Odds that elevated BA21.1 IgG will be found in someone with a LGL-BMF condition vs someone with a normal condition |
4. Discussion
Our results show that sera from patients with AA, MDS, and PNH have high titer antibodies reactive to the LGL leukemia-specific BA21 epitope. This epitope, which is derived from the amino terminus of BA21 and which contains a QEQC signal motif, was designated as BA21.1. A previous study showed no difference in BA21 reactivity for patients with acute myelogenous leukemia, chronic myelogenous leukemia, acute lymphoblastic leukemia or non-Hodgkin’s lymphoma, as compared to normal controls [23]. Therefore, BA21 reactivity may be specific for these hematologic diseases.
Previous studies have shown that sera from 30% to 46% of LGL leukemia patients are reactive to the BA21 protein, which is located in the transmembrane region of the HTLV-1 envelope [23, 28]. In this study, we used a microarray format to identify the most disease-specific reactive regions of BA21, and then used a custom ELISA to validate the reactivity of the new epitopes. The mean concentration of BA21.1 IgG for LGL leukemia sera (99.6 μg/mL) was more than 7-fold greater than the mean concentration in normal sera (13.6 μg/mL). AA and PNH sera were both 3.5 fold more reactive than normal sera, and each had a mean IgG concentration of 47.5 μg/mL. Finally, the mean specific BA21.1 IgG concentration for MDS sera was 61.8 μg/mL which was more than 4.5-fold greater than normal. Taken together, over 35% of patients with LGL-BMF conditions were reactive to BA21.1, at levels more than three standard deviations above normal. The percent of LGL leukemia sera reactive for this epitope (40%) is similar to previous reports. The reason for this percentage is still unknown. It is possible that this 40% represents a subset of LGL leukemia patients who may have different immune responses to the antigen(s) represented by the BA21 epitopes tested in this study. Perhaps an additional explanation is that BA21 is an imperfect representation of an uncharacterized LGL leukemia viral antigen, therefore reactivity is still much less than 100%. Of interest were the numbers of BMF patients who also responded to the same BA21 epitope, while reactivity with normal sera was significantly less. This suggests that a specific immunogenetic link may exist for a persistent subset of these patients. Elevated BA21.1 IgG was associated with an overall 128 times greater likelihood of having any LGL-BMF disease and a 158-fold greater chance of having LGL leukemia.
The common seroreactivity to BA21 in LGL leukemia and these other hematologic disorders suggests a shared pathogenesis. Therefore we were interested in interrogating the RDCRN BMFC database to determine if there were demographic or medical factors associated with BA21 seropositivity. Multiple transfusions increase the probability of producing cross-reactive HLA antibodies, which could result in elevated serum proteins [29], while IVIG therapy inhibits the production of HLA antibodies[30]. An important negative finding in this respect was the lack of association of BA21 antibody levels with transfusion history or with IVIG. Although the analyses of available TCR data (in supplemental data S2) did not point to a single clonotype associated with BA21 reactivity, other epitopes that may be important include epitopes with related structural conformation found in other retroviruses. We are currently studying these to determine if a common naturally-occurring antigenic motif is important to LGL leukemia.
5. Conclusions
This is the first report showing that BMF patients and LGL leukemia patients have similarly elevated levels of BA21 antibody. The significant odds ratios suggest that elevated BA21.1 IgG is associated with LGL leukemia and with LGL-BMF diseases. The antigen (s) responsible for eliciting BA21 seroreactivity remains unknown, and the actual relationship of seroreactivity to disease development has not been defined. Even so, these findings are useful because they establish a relationship between disease-specific BA21 IgG, LGL leukemia and other hematologic diseases. Overall, these data suggest that there might be a common pathogenesis for LGL leukemia, AA, MDS and PNH.
Supplementary Material
Acknowledgments
We would like to acknowledge the provision of assistance and materials in the development of the peptide array, provided by the Penn State College of Medicine Functional Genomics Core Facility, specifically Robert M. Brucklacher, BA BS and Bill Freeman, PhD (Facility Director).
Grant Support
The project described was supported by Grant Number U54RR019397,” Bone Marrow Failure Clinical Research Center” to J.M., Cleveland Clinic Award #5401 to T.P.L., “Longitudinal Studies in Bone Marrow Failure Diseases”, and a pilot project award #5409, “HTLV-2 Seroreactivity in LGL Leukemia” to T.P.L., all from the NIH Rare Diseases Clinical Research Network. Additional support for this work was obtained from NIH funding. NCI Grants CA133525 and CA 94872 were awarded to T.P.L. NCI Grant CA112112 was awarded to P.E.B. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH Rare Diseases Clinical Research Network.
Footnotes
Contribution: S.B.N. contributed to the overall concept, designed and performed the research, performed databases extractions, analyzed the data, made the figures and wrote the paper. D.J. K. contributed to the array concept and design, performed array experiments, analyzed the data, made the figures and wrote the paper. M.C. designed and performed the cytokine experiments, provided specimen data, and contributed to the writing. R.B.I. analyzed data, provided critical input on the results, and contributed to the writing. K.T.B. performed array experiments, database extraction, and managed the BMF and LGL Leukemia Registry specimens. N.R.J. performed ELISA experiments and analyzed data. L.S. contributed to the overall concept and research design, contributed vital new reagents, and analyzed data. E.S. and J.L. contributed statistical analyses. D.C. contributed statistical analyses and performed database extraction. P.K.EB, R.P. and A.F.L. contributed critical input on the analyses, contributed to the writing and provided BMF specimens and specimen data. J. P.M. contributed to the overall concept and research design, provided critical input for the results, contributed to the writing and provided BMF specimens and clinical data. T.P.L. contributed to the overall research concept and design, analyzed data, provided critical input on the data analyses and results, provided BMF specimens and clinical data, provided LGL Leukemia Registry specimens and clinical data and wrote the paper.
Conflict of interest
The authors declare no competing financial interests.
The following Supplementary Information File accompanies this study:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dearden C. Large granular lymphocytic leukaemia pathogenesis and management. Br J Haematol. 2011;152:273–83. doi: 10.1111/j.1365-2141.2010.08494.x. [DOI] [PubMed] [Google Scholar]
- 2.Melenhorst JJ, Sorbara L, Kirby M, et al. Large granular lymphocyte leukaemia is characterized by a clonal T-cell receptor rearrangement in both memory and effector CD8(+) lymphocyte populations. Br J Haematol. 2001;112:189–94. doi: 10.1046/j.1365-2141.2001.02509.x. [DOI] [PubMed] [Google Scholar]
- 3.Kothapalli R, Bailey RD, Kusmartseva I, et al. Constitutive expression of cytotoxic proteases and down-regulation of protease inhibitors in LGL leukemia. Int J Oncol. 2003;22:33–9. [PubMed] [Google Scholar]
- 4.Maciejewski JP, O’Keefe C, Gondek L, et al. Immune-mediated bone marrow failure syndromes of progenitor and stem cells: molecular analysis of cytotoxic T cell clones. Folia Histochem Cytobiol. 2007;45:5–14. [PubMed] [Google Scholar]
- 5.Plasilova M, Risitano A, Maciejewski JP. Application of the molecular analysis of the T-cell receptor repertoire in the study of immune-mediated hematologic diseases. Hematology. 2003;8:173–81. doi: 10.1080/1024533031000107505. [DOI] [PubMed] [Google Scholar]
- 6.Karadimitris A, Manavalan JS, Thaler HT, et al. Abnormal T-cell repertoire is consistent with immune process underlying the pathogenesis of paroxysmal nocturnal hemoglobinuria. Blood. 2000;96:2613–20. [PubMed] [Google Scholar]
- 7.Zeng W, Nakao S, Takamatsu H, et al. Characterization of T-cell repertoire of the bone marrow in immune-mediated aplastic anemia: evidence for the involvement of antigen-driven T-cell response in cyclosporine-dependent aplastic anemia. Blood. 1999;93:3008–16. [PubMed] [Google Scholar]
- 8.Epling-Burnette PK, Painter JS, Rollison DE, et al. Prevalence and clinical association of clonal T-cell expansions in Myelodysplastic Syndrome. Leukemia. 2007;21:659–67. doi: 10.1038/sj.leu.2404590. [DOI] [PubMed] [Google Scholar]
- 9.Loughran TP., Jr Clonal diseases of large granular lymphocytes. Blood. 1993;82:1–14. [PubMed] [Google Scholar]
- 10.Rose MG, Berliner N. T-cell large granular lymphocyte leukemia and related disorders. Oncologist. 2004;9:247–58. doi: 10.1634/theoncologist.9-3-247. [DOI] [PubMed] [Google Scholar]
- 11.Horiuchi T, Hirokawa M, Satoh K, et al. Clonal expansion of gammadelta-T lymphocytes in an HTLV-I carrier, associated with chronic neutropenia and rheumatoid arthritis. Ann Hematol. 1999;78:101–4. doi: 10.1007/s002770050483. [DOI] [PubMed] [Google Scholar]
- 12.Loughran TP, Jr, Coyle T, Sherman MP, et al. Detection of human T-cell leukemia/lymphoma virus, type II, in a patient with large granular lymphocyte leukemia. Blood. 1992;80:1116–9. [PubMed] [Google Scholar]
- 13.Marlton P, Taylor K, Elliott S, et al. Monoclonal large granular lymphocyte proliferation in SLE with HTLV-I seroreactivity. Aust N Z J Med. 1992;22:54–5. doi: 10.1111/j.1445-5994.1992.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 14.Martin MP, Biggar RJ, Hamlin-Green G, et al. Large granular lymphocytosis in a patient infected with HTLV-II. AIDS Res Hum Retroviruses. 1993;9:715–9. doi: 10.1089/aid.1993.9.715. [DOI] [PubMed] [Google Scholar]
- 15.Nakano-Akamatsu S, Takahashi R, Sekioka Y, et al. CD20− and CD56-positive T-cell large granular lymphocyte leukemia in a human T-cell leukemia virus type 1 carrier. Int J Hematol. 2007;86:348–51. doi: 10.1532/IJH97.07076. [DOI] [PubMed] [Google Scholar]
- 16.Pulik M, Lionnet F, Genet P, et al. CD3+ CD8+ CD56− clonal large granular lymphocyte leukaemia and HIV infection. Br J Haematol. 1997;98:444–5. doi: 10.1046/j.1365-2141.1997.1913009.x. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto Y, Kawachi Y, Uchida T, et al. Adult T-cell leukaemia/lymphoma featuring a large granular lymphocyte leukaemia morphologically. Br J Haematol. 1994;86:383–5. doi: 10.1111/j.1365-2141.1994.tb04745.x. [DOI] [PubMed] [Google Scholar]
- 18.Loughran TP, Jr, Sherman MP, Ruscetti FW, et al. Prototypical HTLV-I/II infection is rare in LGL leukemia. Leuk Res. 1994;18:423–9. doi: 10.1016/0145-2126(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 19.Duong YT, Jia H, Lust JA, et al. Short communication: Absence of evidence of HTLV-3 and HTLV-4 in patients with large granular lymphocyte (LGL) leukemia. AIDS Res Hum Retroviruses. 2008;24:1503–5. doi: 10.1089/aid.2008.0128. [DOI] [PubMed] [Google Scholar]
- 20.Thomas A, Perzova R, Abbott L, et al. LGL leukemia and HTLV. AIDS Res Hum Retroviruses. 2010;26:33–40. doi: 10.1089/aid.2009.0124. [DOI] [PubMed] [Google Scholar]
- 21.Wang F, Xu RH, Han B, et al. High incidence of hepatitis B virus infection in B-cell subtype non-Hodgkin lymphoma compared with other cancers. Cancer. 2007;109:1360–4. doi: 10.1002/cncr.22549. [DOI] [PubMed] [Google Scholar]
- 22.Busch MP, Switzer WM, Murphy EL, et al. Absence of evidence of infection with divergent primate T-lymphotropic viruses in United States blood donors who have seroindeterminate HTLV test results. Transfusion. 2000;40:443–449. doi: 10.1046/j.1537-2995.2000.40040443.x. [DOI] [PubMed] [Google Scholar]
- 23.Loughran TP, Jr, Hadlock KG, Perzova R, et al. Epitope mapping of HTLV envelope seroreactivity in LGL leukaemia. Br J Haematol. 1998;101:318–24. doi: 10.1046/j.1365-2141.1998.00691.x. [DOI] [PubMed] [Google Scholar]
- 24.Palker TJ, Tanner ME, Scearce RM, et al. Mapping of immunogenic regions of human T cell leukemia virus type I (HTLV-I) gp46 and gp21 envelope glycoproteins with env-encoded synthetic peptides and a monoclonal antibody to gp46. J Immunol. 1989;142:971–8. [PubMed] [Google Scholar]
- 25.Chang HT, Pai TW, Fan TC, et al. A reinforced merging methodology for mapping unique peptide motifs in members of protein families. BMC Bioinformatics. 2006;7:38. doi: 10.1186/1471-2105-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–4. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 27.Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokol L, Agrawal D, Loughran TP., Jr Characterization of HTLV envelope seroreactivity in large granular lymphocyte leukemia. Leuk Res. 2005;29:381–7. doi: 10.1016/j.leukres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Turner D. The human leucocyte antigen (HLA) system. Vox Sang. 2004;87 (Suppl1):87–90. doi: 10.1111/j.1741-6892.2004.00438.x. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan KM. Immunomodulation in allogeneic marrow transplantation: use of intravenous immune globulin to suppress acute graft-versus-host disease. Clin Exp Immunol. 1996;104 (Suppl 1):43–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.