Abstract
The underlying genetic etiology of late onset Alzheimer’s disease (LOAD) remains largely unknown, likely due to its polygenic architecture and a lack of sophisticated analytic methods to evaluate complex genotype-phenotype models. The aim of the current study was to overcome these limitations in a bi-multivariate fashion by linking intermediate magnetic resonance imaging (MRI) phenotypes with a genome-wide sample of common single nucleotide polymorphism (SNP) variants. We compared associations between 94 different brain regions of interest derived from structural MRI scans and 533,872 genome-wide SNPs using a novel multivariate statistical procedure, parallel-independent component analysis, in a large, national multi-center subject cohort. The study included 209 elderly healthy controls, 367 subjects with amnestic mild cognitive impairment and 181 with mild, early-stage LOAD, Caucasian adults, from the Alzheimer’s Disease Neuroimaging Initiative cohort. Imaging was performed on comparable 1.5T scanners at over 50 sites in the USA/Canada. Four primary “genetic components” were associated significantly with a single structural network including all regions involved neuropathologically in LOAD. Pathway analysis suggested that each component included several genes already known to contribute to LOAD risk (e.g. APOE4) or involved in pathologic processes contributing to the disorder, including inflammation, diabetes, obesity and cardiovascular disease. In addition significant novel genes identified included ZNF673, VPS13, SLC9A7, ATP5G2 and SHROOM2. Unlike conventional analyses, this multivariate approach identified distinct groups of genes that are plausibly linked in physiologic pathways, perhaps epistatically. Further, the study exemplifies the value of this novel approach to explore large-scale data sets involving high-dimensional gene and endophenotype data.
Keywords: epistasis, multivariate, ICA, genotype-phenotype, pathway, enrichment
Introduction
Late onset Alzheimer’s disease (LOAD), the commonest cause of late-life dementia (Bekris et al., 2010) has high heritability (Gatz et al., 2006a; Gatz et al., 2006b). However, its etiopathology, pathogenesis and major risk genes are only partly known, mainly due to its genetic complexity and heterogeneity. The “amyloid hypothesis” seems insufficient to fully explain LOAD etiology and alternative hypotheses continue to be advanced (Pimplikar et al., 2010).
To date only one gene of major effect, apolipoprotein E ε4 (APOE4), replicates as significantly influencing LOAD risk (Strittmatter et al., 1993), but does not account for all genetic variability, suggesting the interplay of multiple, mostly unidentified susceptibility loci of smaller effect size acting multiplicatively under a common disease variant model (Eccles and Tapper, 2010) and/or with environmental factors (Traynor and Singleton, 2010). Recent high-throughput genome wide association studies (GWAS) (van Es and van den Berg, 2009)(Grupe et al., 2007; Harold et al., 2009; Seshadri et al., 2010) have identified and replicated in addition to APOE4, other genes such as BIN1, CLU, ABCA7, CR1, PICALM, MS4A6A, CD33, MSA4E and CD2AP, all of which (apart from APOE) have modest effect sizes and cumulatively account for only 35% of the population attributable risk (Ku et al., 2011; Naj et al., 2011). However, if LOAD risk is mediated in part by common polymorphisms individually conferring low disease risk, acting in concert, typical univariate GWAS might not have enough power to consistently detect these effects unless they utilize very large sample sizes. This might be an inherent issue as obtaining such large sample sizes are usually quite difficult. Also more importantly univariate studies do not take into account the effect of multiple genes at once. This is important because major LOAD risk factors include obesity, cerebrovascular disease and diabetes, all disorders with significant genetic underpinnings (Profenno et al., 2010), suggesting causative genes might belong to common biological pathways shared by these conditions. To circumvent some of these issues, multivariate analyses have been suggested as an approach to identify important genetic factors in LOAD (Gandhi and Wood, 2010).
MRI captures robust phenotypic neuroanatomical LOAD biomarkers, most consistently implicating posterior cingulate and entorhinal cortices, hippocampus and other medial temporal structures (Jack et al., 2010a; Jack et al., 2010b; Smith, 2010; Villain et al., 2010) corresponding to sites of early, severe LOAD-related neuropathology. Imaging genetics attempts to bridge genetic variations with phenotypic trait markers, relating genotypic variations to underlying biological disease etiologies and increasing statistical power, thereby requiring smaller sample sizes (Potkin et al., 2009). However, such strategies require tools to simultaneously accommodate thousands of data points per feature set (e.g. ~105 voxels from imaging data and up to 106 SNPs from genetic data), posing a major statistical challenge. Often, large scale studies are performed in a univariate fashion that significantly limits either one or both feature sets. However, these techniques can curtail the usefulness of multidimensional data to identify potentially informative relationships. Conventional voxel-wise analyses are computationally time consuming on a genome-wide scale and ineffectively capture cumulative effects spread over multiple genes. Prior analyses (Biffi et al., 2010; Potkin et al., 2009; Shen et al., 2010) on the multi-site MRI/genetic ADNI dataset used massively univariate approaches: GWAS, that confirmed the risk status of APOE4 and identified TOMM40 (Shen et al., 2010) and hypothesis-driven analyses using pre-selected known affected brain regions plus GWAS, that reinforced the status of promising individual genes of interest (Biffi et al., 2010). However, no analyses have evaluated the premise that genetic determinants are not randomly distributed among relevant biological pathways but instead grouped together among specific biological processes, nor have they detected predicted groups of common, interactive risk polymorphisms.
Parallel independent component analysis (Para-ICA) a novel multivariate data-driven, hypothesis-free statistical technique, extends ICA to analyze multiple modalities simultaneously (Calhoun et al., 2009). Para-ICA identifies simultaneously clusters of associated, likely interacting genes related to either: (a) functional brain networks, (b) related structural brain regions, or (c) physiologic processes e.g. EEG patterns or other potential endophenotypes and shows their relationships (Calhoun et al., 2009). Beginning with two modalities (here, SNP’s and MRI-derived regional brain volume/thickness), we sought to discover underlying factors from both modalities and their connections. Similar to conventional ICA analyses, extracted structural MRI components are maximally independent within modality and loading coefficients represent variation among individuals. Networks or components extracted from genetic data are groups of interacting SNP loci, contributing with varying degrees to a genetic process affecting a downstream biological function, i.e. linear SNP combinations highly associated with related phenotypes. To date, this technique has been used mainly in schizophrenia and healthy controls to find genes responsible for brain structure and function using MRI and EEG patterns (Jagannathan et al., 2010; Liu et al., 2008; Meda et al., 2010). However, subject and SNP numbers in those studies were typically small.
Genetic and structural MRI data from the ADNI sample provide an ideal test bed to explore LOAD and to validate application of Para-ICA to larger datasets. The subject number (>800) and large genotypic dataset (>600,000 SNPs) allow for examination of feasibility of scaling up this technique where some valid results are published in this dataset from conventional, hypothesis-driven analyses (Biffi et al., 2010). Because many LOAD risk genes remain to be discovered, the technique can simultaneously be used to identify novel risk genes, as it identifies clusters of related, interacting SNPs.
We had the following goals: 1) to evaluate whether Para-ICA could be scaled up to deal with larger populations and many more SNPs than previously analyzed; 2) to identify new risk genes for LOAD and their corresponding endophenotypes and 3) to explore the different LOAD-mediating biological interactive pathways in which the identified risk genes might participate. We hypothesized that the method might identify previously unknown LOAD risk genes, as well as known candidate genes. We hypothesized that identified genes would group into LOAD-associated physiologic pathways and processes.
Materials and Methods
We evaluated associations between two data modalities, structural MRI (sMRI), (regional brain volumes and cortical thicknesses), and genome-wide genotypic data (SNPs), to reveal multivariate relationships between structural brain regions and SNP’s that differed between healthy controls, MCI and AD subjects.
Alzheimer’s Disease Neuroimaging Initiative (ADNI) Study
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.ucla.edu). ADNI results from efforts of many co-investigators from a broad range of academic institutions and private corporations, with subjects recruited from over 50 sites across the U.S. and Canada. For up-to-date information, see www.adni-info.org. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco.
Study Participants
Data derived from the ADNI database on 818 subjects included baseline 1.5 T MRI scans, Illumina SNP genotyping data, APOE genotype status and demographic information. We limited analyses to European-American ADNI subjects (classified initially into respective ethnic groups based on self-report and validated subsequently using genetic markers) to prevent confluence of population stratification effects on data, yielding a total of 209 HC (Mean/SD Age = 76.05/4.94; 113 Males) with no past history of neurological or psychiatric disorder, 367 subjects with MCI (Mean/SD Age = 74.95/7.37; 239 Males) and 181 subjects with clinically-assessed AD (Mean/SD Age = 75.57/7.48; 100 Males) for analysis.
Single Nucleotide Polymorphisms (SNP’s) - Genotype
Sample collection and Single nucleotide polymorphism (SNP) genotyping for more than 620,000 target SNPs across the whole genome was completed on all ADNI participants as described in (Saykin et al., 2010; Shen et al., 2010).
Quality Control/Pre-processing
Prior to Para-ICA, genotyped SNP’s underwent two pre-processing stages. First, quality control parameters were employed to discard data unsuitable for further analysis. Samples (both subjects and SNP’s) were checked for missing data and those with missing call rates >5% were excluded. Remaining samples were imputed for missing values (<1%) by replacing data with the corresponding major genotype. Following this, all uninformative SNP’s (constant variance) were excluded. SNP’s were then checked for minor allele frequency (MAF); rare SNP variants with MAFs <0.01 were excluded. Highly correlated SNP’s (r>0.95) (in block sizes of 100kb) were removed. Finally, SNPs (in controls only) were checked for Hardy-Weinberg equilibrium set at a threshold of p<1E-7. QC SNP’s (N=533,872) were then carried over to the next processing stage. The above analyses were performed using custom scripts in Matlab 7.0 (www.mathworks.com).
The above pre-processed SNP’s were subjected to a univariate GWAS type case-control association analysis to identify those differing significantly across the three diagnostic groups. This 1) effectively restricted the core analysis to disease-related genetic data in the current sample, 2) reduced potential noise from interacting genes with little or no relationship to the disease model, providing hypothesis-free data-driven “enrichment,” and 3) improved accuracy and the linking coefficient of the Para-ICA algorithm (determined based on previous simulation results (Liu et al., 2008)). All SNP’s (using an additive model) were entered into a mass univariate ANOVA design with diagnostic group as the independent factor, using Matlab 7.0. SNP’s surviving a liberal p<0.05 uncorrected threshold were then advanced to the Para-ICA multivariate association analyses. As noted, at this stage, no multiple comparison correction was performed. All significant SNP’s with a p<0.05 uncorrected threshold (N=27,150) were carried forward to Para-ICA to determine genetic associations (including weak effects spread across multiple SNPs) with brain structures.
MRI Structural Imaging - Phenotype
All subjects underwent a high-resolution 3D structural MRI scan (MPRAGE) as detailed in http://www.adni-info.org. We utilized recently published ADNI imaging data, analyzed in Freesurfer V4.1.0 (http://surfer.nmr.mgh.harvard.edu/fswiki) (Shen et al., 2010), thus, brain structure preparation and analysis methods are described only briefly. An automated Freesurfer Bayesian segmentation and parcellation routine extracted and labeled cortical and subcortical tissue classes (Shen et al., 2010), yielding target region volumes, cortical thicknesses and total intracranial volumes for pre-defined brain structure regions-of-interest (ROIs). Freesurfer values for two independently collected MPRAGE scans per subject were averaged to yield a single volume/cortical thickness value. Table 1 lists all imaging phenotypes (N=94; bilateral volumes of interest and cortical thickness values). All values were normalized by Z-score transformation before entry into Para-ICA.
Table 1.
Details all Freesurfer-derived regions of interest used as the imaging phenotype
| Rank order | Region | Abs Z-Score for Network S1 |
|---|---|---|
| 1 | Left Superior Frontal ** | 2.82 |
| 2 | Right Superior Frontal ** | 2.80 |
| 3 | Left Entorhinal Cortex ** | 2.74 |
| 4 | Left Middle Temporal ** | 2.72 |
| 5 | Left Rostral Mid Frontal ** | 2.65 |
| 6 | Left Caudal Mid Frontal ** | 2.64 |
| 7 | Left Inferior Temporal ** | 2.58 |
| 8 | Right Middle Temporal ** | 2.58 |
| 9 | Right Hippocampus Volume ** | 2.57 |
| 10 | Right Entorhinal Cortex ** | 2.56 |
| 11 | Right Rostral Mid Frontal ** | 2.54 |
| 12 | Left Hippocampus Volume ** | 2.52 |
| 13 | Left Isthmus Cingulate ** | 2.51 |
| 14 | Right Caudal Mid Frontal ** | 2.47 |
| 15 | Right Isthmus Cingulate ** | 2.43 |
| 16 | Right Temporal Pole ** | 2.39 |
| 17 | Left Temporal Pole ** | 2.37 |
| 18 | Right Inferior Parietal ** | 2.31 |
| 19 | Right Inferior Temporal ** | 2.29 |
| 20 | Left Fusiform ** | 2.29 |
| 21 | Left Superior Temporal ** | 2.28 |
| 22 | Inferior Lateral Ventricle ** | 2.27 |
| 23 | Left Superior marginal ** | 2.24 |
| 24 | Right Superior marginal ** | 2.21 |
| 25 | Left Inferior Parietal ** | 2.21 |
| 26 | Right Post Cingulate ** | 2.17 |
| 27 | Right Precuneus ** | 2.16 |
| 28 | Right Superior Temporal ** | 2.15 |
| 29 | Right Amygdala Volume ** | 2.13 |
| 30 | Right Pars Operculum ** | 2.11 |
| 31 | Left Precentral ** | 2.11 |
| 32 | Left Pars Trianglaris ** | 2.10 |
| 33 | Left Amygdala Volume ** | 2.10 |
| 34 | Left BanksSTS ** | 2.08 |
| 35 | Right Pars Trianglaris ** | 2.08 |
| 36 | Left Post Cingulate ** | 2.07 |
| 37 | Left Lateral OrbFrontal ** | 2.07 |
| 38 | Left Precuneus ** | 2.06 |
| 39 | Right Parahippocampus ** | 2.01 |
| 40 | Right Fusiform ** | 2.00 |
| 41 | Right Lateral OrbFrontal | 1.97 |
| 42 | Left Pars Orb | 1.97 |
| 43 | Right Precentral | 1.97 |
| 44 | Left Pars Operculum | 1.96 |
| 45 | Left Parahippocampus | 1.95 |
| 46 | Right BanksSTS | 1.90 |
| 47 | Left Medial Orbito Frontal | 1.84 |
| 48 | Right Superior Parietal | 1.82 |
| 49 | Right Medial Orbito Frontal | 1.82 |
| 50 | Frontal Pole | 1.79 |
| 51 | Right Ventricle DC | 1.79 |
| 52 | Left Ventricle DC | 1.78 |
| 53 | Right Paracentral | 1.75 |
| 54 | 3rd Ventricle | 1.71 |
| 55 | Left Paracentral | 1.68 |
| 56 | Left Superior Parietal | 1.68 |
| 57 | Left Frontal Pole | 1.61 |
| 58 | Right Pars Orb | 1.56 |
| 59 | Right Frontal Pole | 1.52 |
| 60 | Right Pallidum Volume | 1.47 |
| 61 | Left Post central | 1.44 |
| 62 | Left Rostral Ant Cingulate | 1.41 |
| 63 | Right Post central | 1.38 |
| 64 | CC_Mid Anterior | 1.37 |
| 65 | Left Pallidum Volume | 1.32 |
| 66 | CC_Central | 1.32 |
| 67 | Right Putamen Volume | 1.24 |
| 68 | Left Accumbens Volume | 1.20 |
| 69 | CC_Mid Posterior | 1.18 |
| 70 | Right Thalamus Volume | 1.18 |
| 71 | CC_Posterior | 1.17 |
| 72 | Left Putamen Volume | 1.15 |
| 73 | Left Left ingual | 1.13 |
| 74 | Left Lateral Occipital | 1.11 |
| 75 | Right Lateral Occipital | 1.10 |
| 76 | Right Accumbens Volume | 1.06 |
| 77 | CC_Anterior | 1.05 |
| 78 | Right Caud Ant Cingulate | 1.03 |
| 79 | Right Left ingual | 0.95 |
| 80 | Left TransverseTemporal | 0.84 |
| 81 | Right Rostral Ant Cingulate | 0.75 |
| 82 | Left Caud Ant Cingulate | 0.69 |
| 83 | Left Thalamus Volume | 0.68 |
| 84 | Right Corpus Collosum | 0.66 |
| 85 | Left Corpus Collosum | 0.56 |
| 86 | Right Cuneus | 0.55 |
| 87 | Right TransverseTemporal | 0.51 |
| 88 | Left Pericalc | 0.46 |
| 89 | Left Caudate Volume | 0.31 |
| 90 | Right Caudate Volume | 0.31 |
| 91 | Left Cuneus | 0.30 |
| 92 | Right Cerebellar Cortex | 0.18 |
| 93 | Right Pericalc | 0.08 |
| 94 | Left Cerebellar Cortex | 0.05 |
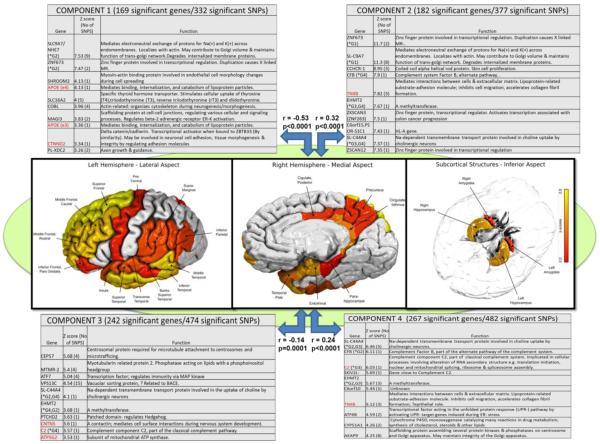
Denotes regions with significant loadings on imaging network S1, ranked in order of Z-score loads. S1 was the only imaging network that was significantly associated with any (in this case all four) genetic networks.
Genotype-Phenotype Associations (Para-ICA)
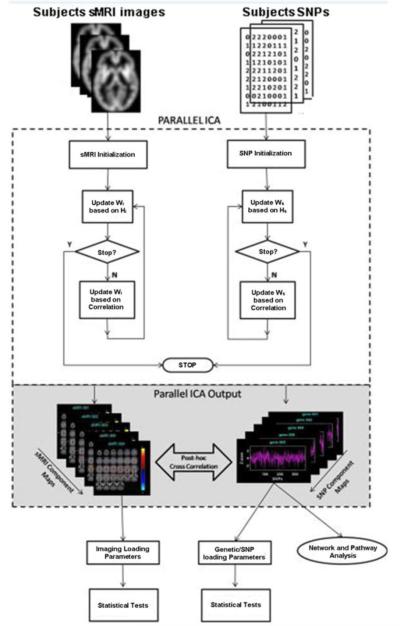
Para-ICA was implemented using the Fusion ICA Toolbox v2.0a; http://icatb.sourceforge.net in Matlab 7.0 to compute independent genetic/imaging networks and simultaneously identify and quantify association between the two modalities/features. This variant of ICA was designed for multimodality processing that extracts components using an entropy term based on information theory to maximize independence and enhances the interconnection by maximizing the linkage function in a joint estimation process (Calhoun et al., 2009; Liu et al., 2008). In addition, Para-ICA estimated loading parameters expressing the weight of the overall component for each subject. Overall correlation values between loading coefficients of the two sets of imaging and genetic component(s) were calculated component-wise for the aggregate sample to identify significantly associated feature sets. Comprehensive mathematical details of the algorithm and methodology are provided in (Liu et al., 2008). Data values from all three diagnostic groups were organized as a matrix of subjects by SNP’s/imaging-ROI values. These genotype and phenotype data matrices were input to the Para-ICA algorithm (see diagram in Figure 1). The number of independent estimated components for both SNP (12 components) and imaging data (8 components) was separately estimated using Akaike information criteria (AIC) (Calhoun et al., 2001). Resulting correlation values between the Para-ICA feature sets were appropriately corrected for multiple comparisons at this stage and a Bonferroni correction was applied based on 12×8=96 comparisons yielding a corrected p value threshold of 0.05/96 = 0.0005. Once significant feature set associations were identified, all contributing SNPs/imaging ROIs across each significant feature/network/component were thresholded at a supra level |Z|>2.0 to specifically identify dominant loadings for each individual network. SNP’s or regions surpassing this threshold were deemed to be contributing significantly to the overall signal of the corresponding component/network. Subsequently, loading coefficients of significantly associated components were tested in a case-control fashion to test if they differed significantly among diagnostic groups.
Figure 1.
Flow diagram illustrating the Para-ICA technique employed in this study.
Significant SNP’s from each component were then batch queried against the dbSNP database http://www.ncbi.nlm.nih.gov/projects/SNP/ to extract corresponding known gene information; genes from this query (derived for each component) were entered into the functional annotation tool, DAVID (http://david.abcc.ncifcrf.gov/) to identify enriched biological themes and visualize these genes on functional pathways e.g. KEGG and/or BioCarta. The Ingenuity pathway analysis tool (IPA; http://www.ingenuity.com/products/pathways_analysis.html) modeled and analyzed the complex biology and genetic interactions as canonical pathway models within the identified significant genetic network(s). SNPs associated with known genes were mapped to the Ingenuity Pathways Knowledgebase to delineate biological networks. Genes were also input to Funcassociate v2.0 (http://llama.mshri.on.ca/funcassociate/) to reveal significantly enriched functional attributes in each component compared against a gene ontology database. Finally, we performed standard chi-square association analyses on the top 10 Z-score-ranked genes from each Para-ICA-derived genetic network, to determine their relative association with the disease model.
Results
Initial data pre-processing with a univariate “GWAS like” analysis (p<0.05 uncorrected) revealed N=27,150 SNPs that differed significantly across groups. It confirmed SNPs from APOE (ε4; p=6.6E-16; ε3; p=3.6E-09) and TOMM40 (p=7.25E-08) as the top three candidate genes, whose genotypes differed significantly across diagnostic groups, as identified in prior GWAS of the same parent dataset (Potkin et al., 2009).
Genotype-Phenotype associations using Para-ICA
From the 12 SNP and 8 anatomic principal component networks identified by AIC, Para-ICA identified four different independent genetic networks significantly associated with a single structural network (following Bonferroni correction). These four networks were component/networks 1 (G1; consisting of 169 significant genes/332 SNPs), 2 (G2; 182 genes/377 SNPs), 3 (G3; 267 genes/482 SNPs) and 4 (G4; 169 genes/332 SNPs). G1 and G3 had significant loadings (Z>2) from APOE (ε4). All four networks were significantly associated with only one structural brain network (S1) that encompassed 40 different unilateral regions (a combination of both volumes and cortical thickness surpassing a Z=2 threshold). Key structural regions loading heavily in this component were entorhinal cortex and middle temporal cortex thicknesses and amygdala and hippocampus volumes. A complete list of significant regions is highlighted with double asterisks in Table 1 and illustrated in Figure 2. Significant genotype-phenotype correlations (between loading parameters of each modality or feature) were as follows: 1) G1-S1 (r=−0.53; p<0.0001) 2) G2-S1 (r=0.32; p<0.0001) 3) G4-S1 (r=0.24; p<0.0001) and G3-S1 (r=−0.14; p=0.0001). Figure 2 summarizes these data with the top 10 representative genes from each genetic network along with their corresponding biological functions. Supplementary figure 1 (Fig S1) shows correlation (scatter) plots for these associations. Testing loading coefficients of the above networks for between-group differences revealed that in addition to having significant associations they also significantly discriminated groups by baseline diagnosis (AD, MCI, or healthy control). Mean loading coefficients across each genetic/structural network are shown in supplementary Figure 2 (Fig S2).
Figure 2.
Significant genotype-phenotype associations identified using the hypothesis-free Para-ICA approach. Genetic factors combine effects from all risk loci, thus explaining more phenotypic variability than traditional analytic approaches. Figure shows the top ten genes, ranked by Z-scores, plus their known functions, from each of the four genetic networks that were significantly correlated with the MRI network. Numbers of significant genes and SNPs within each network is listed at top. * Indicates that the same gene appears among the top ten genes of another network. Genes in red text have been previously implicated in LOAD risk.
Pathway analysis of genetic networks (components)
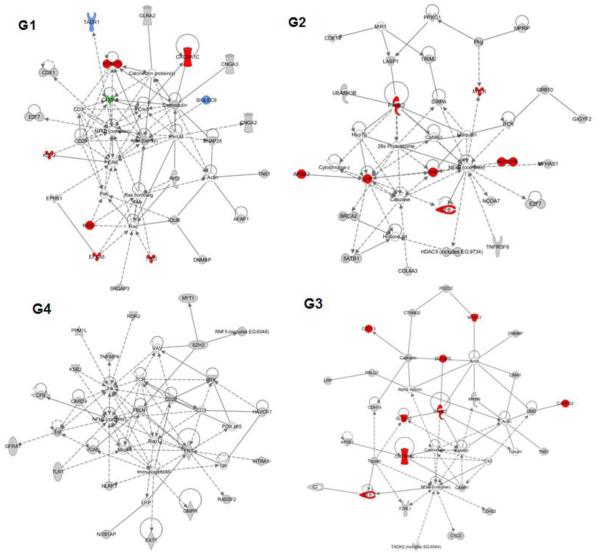
Ingenuity Pathway Analysis (IPA) software (http://www.ingenuity.com/) was used to detect, visualize, and explore relevant biological networks associated with each genetic component. Top networks for G1 were cellular assembly/organization, cell morphology and development. G2 was enriched with genes related to cardiovascular disease, neurologic disease and cardiac arteriopathy. Primary networks for G4 were cell cycle, cell death and inflammatory response. G3 had an over-representation of genes related to neurological and psychological disorders. All the above networks contained known Alzheimer’s-related proteins in their pathway interactions. Top dynamic networks from each genetic component are illustrated in Figure 3. Based on known gene functions, the top five IPA canonical pathways for each gene network (sorted in terms of genotype-phenotype linkage significance) were as follows:
Figure 3.
Representation of the top canonical pathways derived from Ingenuity pathway software for all four significant genetic networks. G1: Pathway enriched for cellular assembly/organization, cell morphology/development; G2: Pathway enriched for cardiac arteriopathy, cardiovascular and neurological disease; G4: Pathway enriched for cell cycle, cell death and inflammatory response; G3: Enriched for genetic, neurological and psychological disorders. Key: red = known AD implicated gene; blue = involved in long-term memory, green = involved in long-term potentiation of CA1 neurons.
G1: cAMP mediated signaling, sulfur metabolism, calcium signaling, vascular NO signaling and regulation of IL-2 expression in T lymphocytes. G2: Neuro-protective role of THOP1 in Alzheimer’s, NOS endothelial effects, Type 2 diabetes signaling, tyrosine metabolism, CYP450. G4: cAMP-mediated signaling, cardiac beta-adrenergic signaling, synaptic long-term potentiation, molecular cancer mechanisms, NOS endothelial effects. Significantly associated non-Neurologic disorders were type 2 diabetes (N=80), coronary artery disease (N=71), Crohn’s/inflammatory bowel disease (N=65). G3: Protein kinase A signaling, cardiac beta-adrenergic signaling, cAMP-mediated signaling, amino sugars metabolism, glycosaminoglycan degradation. Significantly associated non-neurologic disorders were Type 2 Diabetes (N=70), Crohn’s disease/IBD (N=57), coronary artery disease (N=57).
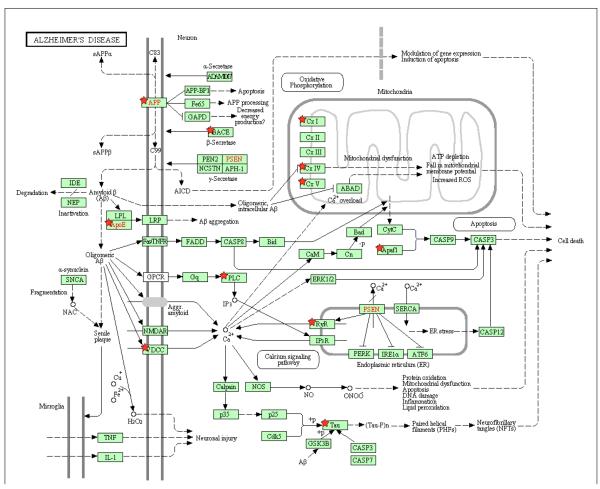
The DAVID functional annotation tool revealed that the significant genes from all four genetic networks are involved in multiple biological pathways including Alzheimer’s disease, adherens junction, arrhythmogenic right ventricular and dilated cardiomyopathy, axon guidance, calcium signaling, cell adhesion, ECM receptor interaction, focal adhesion and tight junction and smooth muscle contraction. Figure 4 illustrates (marked with red stars) the significant genes in our study directly related to Alzheimer’s disease on a KEGG pathway map derived from the above tool. Genes analyzed using the Funcassociate v2.0 toolkit revealed several (N=60) significantly over-represented attributes compared against the gene ontology database. The results presented in Table 2 are rank ordered based on adjusted p value along with the number of genes in the query, number of genes in the overall attribute and their odds ratios.
Figure 4.
Genes identified as part of the significant Para-ICA networks highlighted on a KEGG Alzheimer’s pathway diagram derived from the DAVID functional association tool. Genes identified using red stars occurred in 1 or more of our 4 gene networks. Genes indicated in red text are the primary genes identified to date responsible for the early form of AD (PSEN, APP) or account for a major proportion of LOAD risk (APOE).
Table 2.
Results derived from the Funcassociate web tool showing significantly enriched functional attributes, queried against the gene ontology database. Genes fed into the analysis comprised all significant genes from all four genetic networks identified in the study
| Rank | N | X | LOD | P | P-adj | Gene Ontology Attribute |
|---|---|---|---|---|---|---|
| 1 | 20 | 100 | 1.847 | 3.50E-28 | <0.001 | antigen processing and presentation of peptide antigen via MHC class I |
| 2 | 20 | 103 | 1.831 | 6.70E-28 | <0.001 | antigen processing and presentation of peptide antigen |
| 3 | 23 | 206 | 1.56 | 3.70E-26 | <0.001 | MHC class I protein complex |
| 4 | 23 | 335 | 1.327 | 2.90E-21 | <0.001 | MHC protein complex |
| 5 | 23 | 377 | 1.271 | 4.10E-20 | <0.001 | antigen processing and presentation |
| 6 | 121 | 2180 | 0.409 | 6.60E-17 | <0.001 | plasma membrane part |
| 7 | 28 | 1152 | 0.864 | 5.60E-14 | <0.001 | immune response |
| 8 | 28 | 1348 | 0.791 | 2.50E-12 | <0.001 | immune system process |
| 9 | 160 | 3765 | 0.289 | 5.90E-12 | <0.001 | plasma membrane |
| 10 | 298 | 9033 | 0.234 | 1.10E-11 | <0.001 | membrane |
| 11 | 254 | 7427 | 0.237 | 3.40E-11 | <0.001 | membrane part |
| 12 | 7 | 33 | 1.841 | 8.80E-11 | <0.001 | MHC class I receptor activity |
| 13 | 9 | 150 | 1.364 | 1.30E-09 | <0.001 | thiolester hydrolase activity |
| 14 | 8 | 115 | 1.43 | 3.40E-09 | <0.001 | ubiquitin thiolesterase activity |
| 15 | 8 | 118 | 1.418 | 4.20E-09 | <0.001 | ubiquitin-specific protease activity |
| 16 | 8 | 119 | 1.414 | 4.50E-09 | <0.001 | small conjugating protein-specific protease activity |
| 17 | 19 | 171 | 0.776 | 5.90E-09 | <0.001 | Rho protein signal transduction |
| 18 | 100 | 6558 | 0.316 | 3.30E-08 | 0.002 | intrinsic to membrane |
| 19 | 16 | 131 | 0.802 | 4.50E-08 | 0.002 | Rho guanyl-nucleotide exchange factor activity |
| 20 | 275 | 8319 | 0.183 | 8.10E-08 | 0.003 | protein binding |
| 21 | 35 | 558 | 0.475 | 1.10E-07 | 0.003 | cytoskeletal protein binding |
| 22 | 15 | 140 | 0.802 | 1.10E-07 | 0.003 | regulation of Rho protein signal transduction |
| 23 | 11 | 192 | 0.979 | 1.10E-07 | 0.003 | ATPase activity, coupled to transmembrane movement of substances |
| 24 | 11 | 193 | 0.976 | 1.20E-07 | 0.003 | ATPase activity, coupled to movement of substances |
| 25 | 521 | 19305 | 0.204 | 1.30E-07 | 0.003 | cell part |
| 26 | 521 | 19306 | 0.204 | 1.30E-07 | 0.003 | cell |
| 27 | 11 | 197 | 0.967 | 1.40E-07 | 0.003 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances |
| 28 | 15 | 143 | 0.792 | 1.50E-07 | 0.003 | Ras guanyl-nucleotide exchange factor activity |
| 29 | 31 | 2607 | 0.533 | 1.50E-07 | 0.003 | protein complex |
| 30 | 50 | 909 | 0.38 | 1.80E-07 | 0.003 | cell adhesion |
| 31 | 50 | 909 | 0.38 | 1.80E-07 | 0.003 | biological adhesion |
| 32 | 6 | 15 | 1.551 | 2.00E-07 | 0.003 | prostaglandin E receptor activity |
| 33 | 64 | 1301 | 0.328 | 2.40E-07 | 0.003 | intrinsic to plasma membrane |
| 34 | 96 | 6449 | 0.297 | 2.50E-07 | 0.004 | integral to membrane |
| 35 | 11 | 209 | 0.939 | 2.60E-07 | 0.004 | primary active transmembrane transporter activity |
| 36 | 11 | 209 | 0.939 | 2.60E-07 | 0.004 | P-P-bond-hydrolysis-driven transmembrane transporter activity |
| 37 | 63 | 1285 | 0.326 | 3.30E-07 | 0.005 | integral to plasma membrane |
| 38 | 3 | 6 | 2.561 | 4.30E-07 | 0.007 | sulfonylurea receptor activity |
| 39 | 27 | 390 | 0.511 | 6.50E-07 | 0.012 | proteinaceous extracellular matrix |
| 40 | 6 | 15 | 1.457 | 6.90E-07 | 0.012 | cell adhesion molecule binding |
| 41 | 32 | 1350 | 0.468 | 6.90E-07 | 0.012 | transmembrane transporter activity |
| 42 | 17 | 577 | 0.677 | 7.00E-07 | 0.012 | active transmembrane transporter activity |
| 43 | 3 | 4 | 2.603 | 7.90E-07 | 0.017 | cGMP-dependent protein kinase activity |
| 44 | 58 | 1262 | 0.329 | 7.90E-07 | 0.017 | calcium ion binding |
| 45 | 19 | 258 | 0.622 | 9.10E-07 | 0.018 | guanyl-nucleotide exchange factor activity |
| 46 | 6 | 19 | 1.398 | 1.00E-06 | 0.019 | prostaglandin receptor activity |
| 47 | 27 | 400 | 0.499 | 1.10E-06 | 0.019 | extracellular matrix |
| 48 | 41 | 3215 | 0.416 | 1.10E-06 | 0.019 | macromolecular complex |
| 49 | 34 | 589 | 0.433 | 1.20E-06 | 0.022 | metal ion transport |
| 50 | 8 | 246 | 1.082 | 1.20E-06 | 0.022 | cysteine-type peptidase activity |
| 51 | 136 | 3975 | 0.212 | 1.30E-06 | 0.023 | localization |
| 52 | 14 | 409 | 0.733 | 1.60E-06 | 0.027 | ATPase activity, coupled |
| 53 | 45 | 1158 | 0.366 | 1.60E-06 | 0.029 | ion transmembrane transporter activity |
| 54 | 8 | 258 | 1.06 | 1.80E-06 | 0.029 | ubiquitin-dependent protein catabolic process |
| 55 | 8 | 259 | 1.059 | 1.80E-06 | 0.029 | modification-dependent protein catabolic process |
| 56 | 8 | 259 | 1.059 | 1.80E-06 | 0.029 | modification-dependent macromolecule catabolic process |
| 57 | 8 | 260 | 1.057 | 1.90E-06 | 0.029 | proteolysis involved in cellular protein catabolic process |
| 58 | 8 | 262 | 1.053 | 2.00E-06 | 0.032 | cellular protein catabolic process |
| 59 | 15 | 152 | 0.688 | 2.50E-06 | 0.046 | calcium ion transport |
| 60 | 8 | 42 | 1.057 | 2.60E-06 | 0.047 | voltage-gated calcium channel activity |
Key: Rank: Position in the attribute list ranked by significant of association with the query, N: Number of genes that went into the query with this attribute, X: Number of genes overall in the ontology database with this attribute, LOD: logarithm (base 10) of the odds ratio, p-val: Single hypothesis p-value of the association between attribute and query (based on Fisher’s exact test), p-adj: Adjusted p-value; fraction of 1000 null-hypothesis simulations having attributes with this single hypothesis p-value or smaller
Supplementary Association analysis of top genes from Para-ICA
Association analysis (to illustrate their relative disease association) of allelic frequencies for the top ten genes (based on ranked Z-scores) from the four genetic components revealed that the genes SLC9A7, SHROOM2, ZNF673, APOE (ε3, ε4), VPS13C and ATP5G2 showed stronger effects of disease associations compared to other genes within the component. Table 3 details the allelic frequency and the associated statistical value for each of the top 10 genes from all four genetic networks; due to partial overlap (1 gene could appear in more than 1 component) totaling 32 unique genes. A weighting score was derived by normalizing the chi-square value of each gene to the chi-square of the gene most associated with clinical disease status within each component.
Table 3.
Results of association analysis performed on the top ten genes from each genetic network sorted in descending order of their weighting scores. Weighting scores represent the relative effect of each SNPs association with disease status within a particular component, ranging from 0-1. A & B refer to the different allelic frequencies
| Gene | Locati on |
Functio n |
rs # | AD (# Alleles) |
MCI (# Alleles) |
HC (# Alleles) |
- N square |
p- value |
Weighti ng Score |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | A | B | A | B | A | B | |||||||
| APOE3 | 19q13.2 | N/A | E3 | 162 | 20 0 |
269 | 46 5 |
39 | 27 9 |
88 | 1.00E- 16 |
1.000 | |
| APOE4 | 19q13.2 | N/A | E4 | 209 | 15 3 |
521 | 27 3 |
358 | 60 | 80 | 1.00E- 15 |
0.909 | |
|
SHROO M2 |
Xp22.3 | intron |
rs240594 0 |
209 | 15 3 |
503 | 23 1 |
294 | 12 4 |
16.58 | 0.0003 ** |
0.188 | |
| ZNF673 | Xp11.3 | intron |
rs787630 4 |
103 | 25 9 |
250 | 48 4 |
172 | 24 6 |
14.04 | 0.0009 ** |
0.160 | |
| SLC9A7 |
Xp11.2 3 |
intron |
rs188325 5 |
99 | 26 3 |
241 | 49 3 |
165 | 25 3 |
13 | 0.0015 ** |
0.148 | |
|
SLC16A 2 |
Xq13.2 | intron | rs479640 | 89 | 27 3 |
252 | 48 2 |
138 | 28 0 |
11.15 | 0.0038 | 0.127 | |
| MAGI3 | 1p13.2 | intron |
rs755295 4 |
154 | 20 8 |
373 | 36 1 |
226 | 19 2 |
10.97 | 0.0041 | 0.125 | |
| COBL | 7p12.2 | intron |
rs143749 0 |
195 | 16 7 |
374 | 36 0 |
255 | 16 3 |
10.9 | 0.0043 | 0.124 | |
| PLXDC2 |
10p12.3 1 |
intron |
rs235883 9 |
102 | 26 0 |
265 | 46 9 |
150 | 26 8 |
7.54 | 0.0229 | 0.086 | |
| CTNND2 | 5p15.2 | intron | rs757459 | 215 | 14 7 |
388 | 34 6 |
210 | 20 8 |
6.93 | 0.0311 | 0.079 | |
| G2 | |||||||||||||
| ZNF673 | Xp11.3 | intron |
rs787630 4 |
103 | 25 9 |
250 | 48 4 |
172 | 24 6 |
14.04 | 0.0009 ** |
1.000 | |
| SLC9A7 | Xp11.3 | intron |
rs188325 5 |
99 | 26 3 |
241 | 49 3 |
165 | 25 3 |
13 | 0.0015 ** |
0.926 | |
| TNXB | 6p21.3 | inron |
rs223968 9 |
133 | 22 9 |
213 | 52 1 |
114 | 30 4 |
9.47 | 0.0088 | 0.675 | |
|
ZSCAN1 2 |
6p22.1 |
Mis- sense |
rs136138 5 |
260 | 10 2 |
497 | 23 7 |
258 | 16 0 |
9.24 | 0.0098 | 0.658 | |
| EHMT2 | 6p21.3 | intron |
rs284445 7 |
146 | 21 6 |
236 | 49 8 |
131 | 28 7 |
8.9 | 0.0116 | 0.634 | |
|
ZKSCAN 3 |
6p22.1 | intron | rs213228 | 258 | 10 4 |
491 | 24 3 |
258 | 16 0 |
8.03 | 0.018 | 0.572 | |
|
SLC44A 4 |
6p21.3 | intron |
rs273642 8 |
139 | 22 3 |
227 | 50 7 |
127 | 29 1 |
7.41 | 0.0246 | 0.528 | |
| CFB | 6p21.3 | intron |
rs415165 7 |
223 | 13 9 |
509 | 22 5 |
286 | 13 2 |
6.96 | 0.0307 | 0.496 | |
| C6orf15 | 6p21.3 | intron |
rs313097 7 |
259 | 10 3 |
504 | 23 0 |
264 | 15 4 |
6.7 | 0.0349 | 0.477 | |
| CCHCR1 | 6p21.3 | intron | rs746647 | 256 | 10 6 |
490 | 24 4 |
261 | 15 7 |
6 | 0.0496 | 0.427 | |
|
G 3 |
|||||||||||||
| ATP5G2 |
12q13.1 3 |
intron |
rs180063 4 |
111 | 25 1 |
166 | 56 8 |
134 | 28 4 |
14.97 | 0.0006 ** |
1.000 | |
| VPS13C | 15q22.2 | intron |
rs198191 6 |
190 | 17 2 |
449 | 28 5 |
216 | 20 2 |
12.84 | 0.0016 ** |
0.858 | |
| ATF7 |
12q13.1 3 |
intron | rs784568 | 189 | 17 3 |
337 | 39 7 |
230 | 18 8 |
9.82 | 0.0073 | 0.656 | |
| CEP57 | 11q21 | intron |
rs301775 6 |
154 | 20 8 |
316 | 41 8 |
215 | 20 3 |
8.95 | 0.0113 | 0.598 | |
| EHMT2 | 6p21.3 | intron |
rs284445 7 |
146 | 21 6 |
236 | 49 8 |
131 | 28 7 |
8.9 | 0.0116 | 0.595 | |
| MTMR2 | 11q21 | intron | rS193364 | 225 | 13 7 |
437 | 29 7 |
219 | 19 9 |
8.66 | 0.0132 | 0.578 | |
| PTCHD2 | 1p36.2 2 | intron |
rs237913 5 |
160 | 20 2 |
259 | 47 5 |
165 | 25 3 |
8.32 | 0.0156 | 0.556 | |
| CNTN5 | 11q22.1 | intron |
rs108929 01 |
134 | 22 8 |
314 | 42 0 |
146 | 27 2 |
7.86 | 0.0196 | 0.525 | |
|
SLC44A 4 |
6p21.3 2 | intron |
rs273642 8 |
139 | 22 3 |
227 | 50 7 |
127 | 29 1 |
7.41 | 0.0246 | 0.495 | |
| C2 | 6p21.3 | intron |
rs273433 5 |
204 | 15 8 |
355 | 37 9 |
211 | 20 7 |
6.22 | 0.0445 | 0.415 | |
| G4 | |||||||||||||
| SKIV2L | 6p21.3 | intron | rs592229 | 227 | 13 5 |
388 | 34 6 |
239 | 17 9 |
9.69 | 0.0078 | 0.999 | |
| TNXB | 6P21.3 | intron |
rs223968 9 |
133 | 22 9 |
213 | 52 1 |
114 | 30 4 |
9.47 | 0.0088 | 0.976 | |
| EHMT2 | 6p21.3 | intron | rS159445 | 268 | 94 | 486 | 24 8 |
277 | 14 1 |
7.71 | 0.0211 | 0.795 | |
| AKAP9 | 7q21.2 | intron | rs756647 | 234 | 12 8 |
461 | 27 3 |
234 | 18 4 |
7.39 | 0.0248 | 0.762 | |
|
SLC44A 4 |
6p21.3 | intron |
rs224266 5 |
227 | 13 5 |
397 | 33 7 |
238 | 18 0 |
7.34 | 0.0254 | 0.757 | |
| CFB | 6p21.3 | intron |
rs415165 7 |
223 | 13 9 |
509 | 22 5 |
286 | 13 2 |
6.96 | 0.0307 | 0.718 | |
| ATF6B | 6p12.3 | intron |
rs222862 8 |
242 | 12 0 |
526 | 20 8 |
315 | 10 3 |
6.9 | 0.0316 | 0.711 | |
| C6orf10 | 6p21.3 | intron | rs485774 | 203 | 15 9 |
455 | 27 9 |
272 | 14 6 |
6.81 | 0.0331 | 0.702 | |
|
CYP51A 1 |
7q21.2 | intron |
rs230155 9 |
233 | 12 9 |
454 | 28 0 |
234 | 18 4 |
6.34 | 0.0419 | 0.654 | |
| C2 | 6p21.3 | intron |
rs273433 5 |
204 | 15 8 |
355 | 37 9 |
211 | 20 7 |
6.22 | 0.0445 | 0.641 | |
Genes that significantly discriminate groups after correction for multiple comparisons (Bonferroni-corrected across 40 chi-square tests).
Discussion
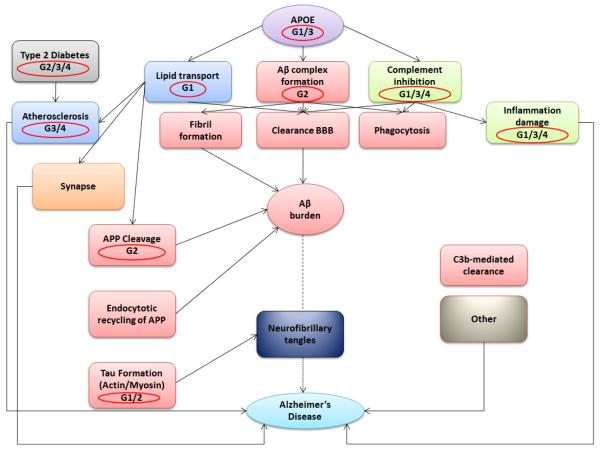
As hypothesized, we validated a scaled-up Para-ICA approach to reveal novel interactive genes and pathways for LOAD, thus highlighting one of the primary advantages of Para-ICA which is the use of modest sample sizes compared to conventional GWAS analyses to effectively capture genotype-phenotype relationships. Dominant loading coefficients were contained in all major regions affected by LOAD pathology in the single structural component significantly associated with four different SNP/genetic networks. Seven other structural components were unassociated with other gene components. The genetic components identified included SNPs from APOE4 plus multiple other risk genes (and putatively protective SNPs, e.g. APOE2) either previously identified in LOAD risk (e.g. ATF7 in G3 (Lin et al., 2006)) or involved in one or more biological processes thought to contribute to LOAD pathology. Figure 5 summarizes the involvement of these four genetic risk networks on a LOAD physiologic pathway diagram.
Figure 5.
A summary of different genetic components (circled in red) identified in the current study projected onto an Alzheimer’s disease functional interaction pathway (modified from Sleegers K et al., TIGS 2009)
The most significant association was between G1-S1. This genetic network had significant loading contributions from a total of 169 different genes (332 SNPs) and correlated negatively with brain network S1, implying increased genetic load is related to decreased brain volume/thickness within the network. This association was notable as the S1 had high loadings from APOE4 (in the top 10 gene loadings) and S1 included regions known to be affected early and severely in LOAD, including entorhinal, middle temporal and prefrontal cortices and hippocampus. More importantly, this genetic network had high loadings from several other genes (SLC9A7/NHE7, ZNF673, SHROOM2) previously unidentified in LOAD pathology. Given that they were part of the same independent genetic component as APOE, this finding both confirms APOE’s established role as an important LOAD risk gene and suggests that these additional SNPs may interact with APOE to influence disease risk, supporting APOE’s role as a LOAD risk factor rather than a direct cause (Guerreiro et al., 2010). The protein encoded by SLC9A7 mediates Na+/H+ exchange across cell surface plasma membranes (Kagami et al., 2008) cycling between the cell surface and intracellular trans-Golgi network and recycling endosomes, which are vital to APP processing (Marks and Berg, 2010). SLC9A7 co-localizes with actin, implicated in tau formation (Gallo, 2007). LOAD lymphoblasts show abnormalities modulated by sodium/hydrogen exchanger blockers (Urcelay et al., 2001). Overall, this genetic network was enriched with genes dominant in cell signaling pathways. Other strongly contributing genes are involved in lipid transport and tau formation (via actin/myosin binding). ZNF673 is associated with X-linked mental retardation, (Lugtenberg et al., 2006; Ramaswamy et al., 2010) and is close (~0.2 MB) to SCL9A7 on Xp11.3.
The second most significant association in genotype/phenotype correlation was G2-S1. This correlation was positive. G2 comprised 182 unique genes (377 SNPs). Some top-ranked genes from this component overlapped with those from G1, including ZNF673 and SLC9A7. These genes had a significant differential distribution among diagnostic groups, suggesting a role of actin localization and transcriptional regulation in LOAD. Other top genes from this network involved in important AD-related processes included the complement system, involved in amyloid-beta formation and inflammatory damage (van Es and van den Berg, 2009).
Network G4 correlated positively with S1. G4 contained several genes associated with risk for non-neurologic disorders, including diabetes and cardiovascular disease, both LOAD risk factors (Profenno et al., 2010). Top genes from this network, previously unidentified in the context of LOAD, belonged to the complement factor/inhibition pathway related to amyloid-beta clearance (35) or are associated with major histocompatibility class III. Additionally, AKAP9, a top 10 gene in this network, maintains neuronal Golgi integrity and is involved in LOAD pathogenesis (Stieber et al., 1996). Regarding association analysis, no top 10 gene from this network was significantly differentially distributed in the disease groups, suggesting that G4 comprises multiple SNPs of low effect acting together through diverse biological risk pathways, especially inflammation, (see Eikelenboom et al. 2006) to significantly affect LOAD-related neuropathology.
The final genotype-phenotype association was a negative correlation between G3 and S1. G3 included ATP5G2, a subunit of mitochondrial ATP-synthase, which was over-represented in the disease group. Mitochondrial ATP-synthase in entorhinal cortex is a target of oxidative stress in LOAD (Terni et al., 2010) and part of LOAD apoptosis pathways. Several other G3 genes included dominant signaling from CNTN5, recently associated with multiple AD MRI characteristics (Biffi et al., 2010), CEP57, a microtubular/centrosomal localizer (Meunier et al., 2009), MTMR2, an endosomal regulator (Lee et al., 2010), and ATF7, associated with LOAD in Lin et al. (Lin et al., 2006). The loading of previously identified LOAD genes and associated pathobiological pathways further supports the relevance of this genetic network.
Analyzing significant genes from all four components using DAVID and visualizing related processes on KEGG pathways revealed that genes grouped in multiple LOAD-relevant biological processes (see Figure 4). Additional prominent processes not shown in figure included cellular communication, cardiovascular diseases, signal transduction, calcium signaling, cell adhesion and neuronal developmental processes (e.g. axon guidance). Many such processes are implicated in LOAD pathology (e.g. neuronal calcium signaling (Kostiuk et al., 2010; LaFerla, 2002; Mattson and Chan, 2003)). Semaphorin 3A, an axon-guiding membrane protein, accumulates in hippocampus in AD (Koncina et al., 2007).
Major themes deriving from the top 32 Z score-defined genes in the 4 SNP components suggest several major pathophysiological LOAD pathways, especially when such genes co-occurred within a component. From G1, APOE may relate to LOAD risk through pathways not directly linked to amyloid-beta, including actin-related mechanisms. Actin cytoskeletal changes as a path to tau formation (Gallo, 2007) are implicated across all components by SCLC987/NHE7 (Kagami et al., 2008; Ohgaki et al., 2008), SHROOM2 and COBL (Dominguez, 2009) and microtubule-related genes including MTMR2, CEP57 and CTNND2 (Bamburg and Bloom, 2009; Meunier et al., 2009). Three such genes were present in component 1. Immune function, especially the complement system, related to amyloid-beta clearance (Guerreiro et al., 2010; Kolev et al., 2009) and expressed in cerebrovascular smooth muscle (Walker et al., 2008), is suggested by ATF7, CFB, C2, SKIV2L, C6orf10 and C6orf15 (Li et al., 2006; Veerhuis, 2010). These genes support the known role of the complement system in LOAD pathogenesis (van Es and van den Berg, 2009), while adding new gene candidates, e.g. C2. Complement is present in dystrophic LOAD neurites, involved in immune response and linked to synaptic pruning (Hollingworth et al., 2010). Five immune related/complement genes are present in G4.
CTNND2/Delta Catenin/NPRAP is associated with GSK3-beta, hence BAP and tau (Bareiss et al., 2010). CNTN5 encodes contactin5; other contactins participate in LOAD risk, (Biffi et al., 2010; Osterfield et al., 2008). The prominence of SCLC987/NHE7(and MTMR2) suggests the importance of the trans-Golgi network and recycling endosome (Lee et al., 2010). Endosomal processing of APP involving SorLA is of importance in LOAD (Lin et al., 2005; Marks and Berg, 2010; Ohgaki et al., 2008). VPS proteins are related to this process (He et al., 2005; Marks and Berg, 2010), although VPS13C has yet to be implicated. VPS13C is associated with maintenance of plasma glucose levels (Saxena et al., 2010); the related VPS26 is linked with BACE/memapsin2 (He et al., 2005). CL44A4 is involved in choline uptake (Jurgensen and Ferreira, 2010). MTMR2 has relevance to excitatory synapses (Lee et al., 2010). ZKSCAN3/ZNF263 is associated with vascular endothelial growth factor (Yang et al., 2008).
The above data suggest involvement of multiple genes influencing varied, complex pathways that might interact mutually to contribute to LOAD. Output from Para-ICA lends itself readily to functional pathway analysis and ultimately systems biology. We also identified novel putative LOAD risk genes, confirmed via testing allelic frequency distributions among disgnostic groups in standard case-control association analyses. It is notable that while none of these genes survived a standard GWAS study, they have high impact when their effect is evaluated in the context of other SNPs. In addition, the SNP components detected several genes previously unknown in the context of LOAD risk, having high Z scores, exceeding those for APOE. Several of these (e.g. SLC9A7, ZNF673, VPS13) were: (a) identified by multiple (up to 17) SNPs, (b) mediate processes plausibly associated with LOAD risk from pathway analyses and prior publications, (c) had SNPs differentially distributed among diagnostic groups and (d) are prominently expressed in brain. These results suggest validity of these novel loci as candidate LOAD risk genes.
Examining loading coefficients of the gene and structural networks revealed a stepped response pattern (see Fig S2), with MCI values falling between those of healthy control and AD, except for in G3, where they were elevated in MCI compared to AD, suggesting that this gene component may act to either protect against or hasten regional brain deterioration in MCI to influence progression rate to AD.
Our study has limitations. Although the Para-ICA method is data driven, we restricted the genetic dataset to a disease-related subset. This focused analysis might fail to uncover genes affecting LOAD pathology via other interactive pathways that may not straightforwardly show group differences. However, since we employed a liberal statistical threshold to limit the genetic dataset to disease-related genes, we were able to include numerous SNPs discarded by conventional univariate studies. Our AD/MCI-focused gene set analysis may not have detected other genetic associations to brain structure. The analysis was carried out only in European-Americans, by far the most numerous ethnicity in the dataset. Future studies could include larger mixed populations. Also, since para-ICA identified multivariate relationships at the gene network level (comprised of linear combinations of SNPs), the directionality and effect magnitude of individual SNPs is not immediately transparent. Our supplementary association analysis to derive the top SNPs might be slightly biased, as they were already pre-selected at a liberal cut-off to be included in the multivariate analysis. Given these limitations and the novelty of our study, our results require further validation and replication in more diverse and larger independent datasets.
In conclusion, we met our major study goals by 1) confirming the feasibility of a hypothesis-blind, multivariate approach to corroborate LOAD genes associated with known pathologic mechanisms and to discover new putative disease-relevant genes that interact but fail individually to reach genome-wide significance. These data thus extend existing GWAS and hypothesis-driven analyses on the same ADNI data set (Biffi et al., 2010; Saykin et al., 2010; Shen et al., 2010). 2) The Para-ICA approach identifies genes in relatively modest sized samples that are plausibly linked collectively in known physiologic pathways, perhaps epistatically and suggests itself as a novel method for exploring other large-scale data sets involving gene and endophenotype information such as BSNIP or COGS (Calkins et al., 2007; Thaker, 2008), in psychotic disorders where the neuropathology and genetic basis are less well-defined than LOAD. Finally, 3) we identified plausible new biological pathways associated with AD neuropathology. Possible therapies resulting from our findings might include agents targeted to the complement and/or immune systems.
Supplementary Material
Acknowledgements
The study was supported by the following grants and research support to Dr. Andrew Saykin from Eli Lilly and Company, Siemens AG, Welch Allyn Inc., the NIH (R01 CA101318 [PI], R01 AG19771 [PI], RC2 AG036535 [Core Leader], P30 AG10133-18S1 [Core Leader], and U01 AG032984 [Site PI and Chair, Genetics Working Group]), the Indiana Economic Development Corporation (IEDC #87884), and the Foundation for the NIH, Dr. Vince Calhoun NIH ROIEB005846. We would also like to thank Mrs. Joanna Mounce for assistance with the biological pathways and network analyses used in these studies.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California,San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
Dr. Andrew Saykin receives research support from Eli Lilly and Company, Siemens AG, Welch Allyn Inc.
References
- Bamburg JR, Bloom GS. Cytoskeletal pathologies of Alzheimer disease. Cell Motil Cytoskeleton. 2009;66:635–649. doi: 10.1002/cm.20388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareiss S, Kim K, Lu Q. Delta-catenin/NPRAP: A new member of the glycogen synthase kinase-3beta signaling complex that promotes beta-catenin turnover in neurons. J Neurosci Res. 2010;88:2350–2363. doi: 10.1002/jnr.22414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekris LM, Yu CE, Bird TD, Tsuang DW. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol. 2010;23:213–227. doi: 10.1177/0891988710383571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, Salat D, Rosand J. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Liu J, Adali T. A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. Neuroimage. 2009;45:S163–172. doi: 10.1016/j.neuroimage.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. Actin filament nucleation and elongation factors--structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles D, Tapper W. The influence of common polymorphisms on breast cancer. Cancer Treat Res. 2010;155:15–32. doi: 10.1007/978-1-4419-6033-7_2. [DOI] [PubMed] [Google Scholar]
- Gallo G. Tau is actin up in Alzheimer’s disease. Nat Cell Biol. 2007;9:133–134. doi: 10.1038/ncb0207-133. [DOI] [PubMed] [Google Scholar]
- Gandhi S, Wood NW. Genome-wide association studies: the key to unlocking neurodegeneration? Nat Neurosci. 2010;13:789–794. doi: 10.1038/nn.2584. [DOI] [PubMed] [Google Scholar]
- Gatz M, Mortimer JA, Fratiglioni L, Johansson B, Berg S, Reynolds CA, Pedersen NL. Potentially modifiable risk factors for dementia in identical twins. Alzheimers Dement. 2006a;2:110–117. doi: 10.1016/j.jalz.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006b;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E, Karnoub M, Nowotny P, Tacey K, Catanese J, Sninsky J, Brayne C, Rubinsztein D, Gill M, Lawlor B, Lovestone S, Holmans P, O’Donovan M, Morris JC, Thal L, Goate A, Owen MJ, Williams J. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- Guerreiro RJ, Gustafson DR, Hardy J. The genetic architecture of Alzheimer’s disease: beyond APP, PSENs and APOE. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Li F, Chang WP, Tang J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE) J Biol Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Jones L, Owen MJ, Williams J. Alzheimer’s disease genetics: current knowledge and future challenges. Int J Geriatr Psychiatry. 2010 doi: 10.1002/gps.2628. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr., Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM, Schuff N, Krueger G, Killiany RJ, Decarli CS, Dale AM, Carmichael OW, Tosun D, Weiner MW. Update on the magnetic resonance imaging core of the Alzheimer’s disease neuroimaging initiative. Alzheimers Dement. 2010a;6:212–220. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr., Wiste HJ, Vemuri P, Weigand SD, Senjem ML, Zeng G, Bernstein MA, Gunter JL, Pankratz VS, Aisen PS, Weiner MW, Petersen RC, Shaw LM, Trojanowski JQ, Knopman DS. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain. 2010b;133:3336–3348. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan K, Calhoun VD, Gelernter J, Stevens MC, Liu J, Bolognani F, Windemuth A, Ruano G, Assaf M, Pearlson GD. Genetic associations of brain structural networks in schizophrenia: a preliminary study. Biol Psychiatry. 2010;68:657–666. doi: 10.1016/j.biopsych.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgensen S, Ferreira ST. Nicotinic receptors, amyloid-beta, and synaptic failure in Alzheimer’s disease. J Mol Neurosci. 2010;40:221–229. doi: 10.1007/s12031-009-9237-0. [DOI] [PubMed] [Google Scholar]
- Kagami T, Chen S, Memar P, Choi M, Foster LJ, Numata M. Identification and biochemical characterization of the SLC9A7 interactome. Mol Membr Biol. 2008;25:436–447. doi: 10.1080/09687680802263046. [DOI] [PubMed] [Google Scholar]
- Kolev MV, Ruseva MM, Harris CL, Morgan BP, Donev RM. Implication of complement system and its regulators in Alzheimer’s disease. Curr Neuropharmacol. 2009;7:1–8. doi: 10.2174/157015909787602805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncina E, Roth L, Gonthier B, Bagnard D. Role of semaphorins during axon growth and guidance. Adv Exp Med Biol. 2007;621:50–64. doi: 10.1007/978-0-387-76715-4_4. [DOI] [PubMed] [Google Scholar]
- Kostiuk OP, Korol T, Korol SV, Romanenko SV, Pinchenko VO, Kostiuk PH. [Alteration of calcium signaling as one of the mechanisms of Alzheimer’s disease and diabetic polyneuropathy] Fiziol Zh. 2010;56:130–138. [PubMed] [Google Scholar]
- Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of genome-wide association studies: where are we now? J Hum Genet. 2011;55:195–206. doi: 10.1038/jhg.2010.19. [DOI] [PubMed] [Google Scholar]
- LaFerla FM. Calcium dyshomeostasis and intracellular signalling in Alzheimer’s disease. Nat Rev Neurosci. 2002;3:862–872. doi: 10.1038/nrn960. [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim Y, Han K, Kim H, Kim E. The phosphoinositide 3-phosphatase MTMR2 interacts with PSD-95 and maintains excitatory synapses by modulating endosomal traffic. J Neurosci. 2010;30:5508–5518. doi: 10.1523/JNEUROSCI.4283-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Partain J, Nixon RR, Allen CN, Irwin RP, Jakobs PM, Litt M, Hershberger RE. Mutations of presenilin genes in dilated cardiomyopathy and heart failure. Am J Hum Genet. 2006;79:1030–1039. doi: 10.1086/509900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PI, Martin ER, Browning-Large CA, Schmechel DE, Welsh-Bohmer KA, Doraiswamy PM, Gilbert JR, Haines JL, Pericak-Vance MA. Parsing the genetic heterogeneity of chromosome 12q susceptibility genes for Alzheimer disease by family-based association analysis. Neurogenetics. 2006;7:157–165. doi: 10.1007/s10048-006-0047-z. [DOI] [PubMed] [Google Scholar]
- Lin PJ, Williams WP, Luu Y, Molday RS, Orlowski J, Numata M. Secretory carrier membrane proteins interact and regulate trafficking of the organellar (Na+,K+)/H+ exchanger NHE7. J Cell Sci. 2005;118:1885–1897. doi: 10.1242/jcs.02315. [DOI] [PubMed] [Google Scholar]
- Liu J, Demirci O, Calhoun VD. A Parallel Independent Component Analysis Approach to Investigate Genomic Influence on Brain Function. IEEE Signal Process Lett. 2008;15:413–416. doi: 10.1109/LSP.2008.922513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg D, Yntema HG, Banning MJ, Oudakker AR, Firth HV, Willatt L, Raynaud M, Kleefstra T, Fryns JP, Ropers HH, Chelly J, Moraine C, Gecz J, van Reeuwijk J, Nabuurs SB, de Vries BB, Hamel BC, de Brouwer AP, van Bokhoven H. ZNF674: a new kruppel-associated box-containing zinc-finger gene involved in nonsyndromic X-linked mental retardation. Am J Hum Genet. 2006;78:265–278. doi: 10.1086/500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks N, Berg MJ. BACE and gamma-secretase characterization and their sorting as therapeutic targets to reduce amyloidogenesis. Neurochem Res. 2010;35:181–210. doi: 10.1007/s11064-009-0054-1. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Chan SL. Neuronal and glial calcium signaling in Alzheimer’s disease. Cell Calcium. 2003;34:385–397. doi: 10.1016/s0143-4160(03)00128-3. [DOI] [PubMed] [Google Scholar]
- Meda SA, Jagannathan K, Gelernter J, Calhoun VD, Liu J, Stevens MC, Pearlson GD. A pilot multivariate parallel ICA study to investigate differential linkage between neural networks and genetic profiles in schizophrenia. Neuroimage. 2010;53:1007–1015. doi: 10.1016/j.neuroimage.2009.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier S, Navarro MG, Bossard C, Laurell H, Touriol C, Lacazette E, Prats H. Pivotal role of translokin/CEP57 in the unconventional secretion versus nuclear translocation of FGF2. Traffic. 2009;10:1765–1772. doi: 10.1111/j.1600-0854.2009.00985.x. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki R, Fukura N, Matsushita M, Mitsui K, Kanazawa H. Cell surface levels of organellar Na+/H+ exchanger isoform 6 are regulated by interaction with RACK1. J Biol Chem. 2008;283:4417–4429. doi: 10.1074/jbc.M705146200. [DOI] [PubMed] [Google Scholar]
- Osterfield M, Egelund R, Young LM, Flanagan JG. Interaction of amyloid precursor protein with contactins and NgCAM in the retinotectal system. Development. 2008;135:1189–1199. doi: 10.1242/dev.007401. [DOI] [PubMed] [Google Scholar]
- Pimplikar SW, Nixon RA, Robakis NK, Shen J, Tsai LH. Amyloid-independent mechanisms in Alzheimer’s disease pathogenesis. J Neurosci. 2010;30:14946–14954. doi: 10.1523/JNEUROSCI.4305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, Weiner M, Macciardi F. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Profenno LA, Porsteinsson AP, Faraone SV. Meta-analysis of Alzheimer’s disease risk with obesity, diabetes, and related disorders. Biol Psychiatry. 2010;67:505–512. doi: 10.1016/j.biopsych.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Ramaswamy V, Castillo M, Bolduc FV. Developmental disability: duplication of zinc finger transcription factors 673 and 674. Pediatr Neurol. 2010;43:209–212. doi: 10.1016/j.pediatrneurol.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, Lyssenko V, Bouatia-Naji N, Dupuis J, Jackson AU, Kao WH, Li M, Glazer NL, Manning AK, Luan J, Stringham HM, Prokopenko I, Johnson T, Grarup N, Boesgaard TW, Lecoeur C, Shrader P, O’Connell J, Ingelsson E, Couper DJ, Rice K, Song K, Andreasen CH, Dina C, Kottgen A, Le Bacquer O, Pattou F, Taneera J, Steinthorsdottir V, Rybin D, Ardlie K, Sampson M, Qi L, van Hoek M, Weedon MN, Aulchenko YS, Voight BF, Grallert H, Balkau B, Bergman RN, Bielinski SJ, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Buchanan TA, Bumpstead SJ, Cavalcanti-Proenca C, Charpentier G, Chen YD, Chines PS, Collins FS, Cornelis M, G JC, Delplanque J, Doney A, Egan JM, Erdos MR, Firmann M, Forouhi NG, Fox CS, Goodarzi MO, Graessler J, Hingorani A, Isomaa B, Jorgensen T, Kivimaki M, Kovacs P, Krohn K, Kumari M, Lauritzen T, Levy-Marchal C, Mayor V, McAteer JB, Meyre D, Mitchell BD, Mohlke KL, Morken MA, Narisu N, Palmer CN, Pakyz R, Pascoe L, Payne F, Pearson D, Rathmann W, Sandbaek A, Sayer AA, Scott LJ, Sharp SJ, Sijbrands E, Singleton A, Siscovick DS, Smith NL, Sparso T, Swift AJ, Syddall H, Thorleifsson G, Tonjes A, Tuomi T, Tuomilehto J, Valle TT, Waeber G, Walley A, Waterworth DM, Zeggini E, Zhao JH, Illig T, Wichmann HE, Wilson JF, van Duijn C, Hu FB, Morris AD, Frayling TM, Hattersley AT, Thorsteinsdottir U, Stefansson K, Nilsson P, Syvanen AC, Shuldiner AR, Walker M, Bornstein SR, Schwarz P, Williams GH, Nathan DM, Kuusisto J, Laakso M, Cooper C, Marmot M, Ferrucci L, Mooser V, Stumvoll M, Loos RJ, Altshuler D, Psaty BM, Rotter JI, Boerwinkle E, Hansen T, Pedersen O, Florez JC, McCarthy MI, Boehnke M, Barroso I, Sladek R, Froguel P, Meigs JB, Groop L, Wareham NJ, Watanabe RM. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, Risacher SL, Nho K, Huentelman MJ, Craig DW, Thompson PM, Stein JL, Moore JH, Farrer LA, Green RC, Bertram L, Jack CR, Jr., Weiner MW. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT, Jr., Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, Huentelman MJ, Craig DW, Dechairo BM, Potkin SG, Jack CR, Jr., Weiner MW, Saykin AJ. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD. Neuroimaging through the course of Alzheimer’s disease. J Alzheimers Dis. 2010;19:273–290. doi: 10.3233/JAD-2010-1217. [DOI] [PubMed] [Google Scholar]
- Stieber A, Mourelatos Z, Gonatas NK. In Alzheimer’s disease the Golgi apparatus of a population of neurons without neurofibrillary tangles is fragmented and atrophic. Am J Pathol. 1996;148:415–426. [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terni B, Boada J, Portero-Otin M, Pamplona R, Ferrer I. Mitochondrial ATP-synthase in the entorhinal cortex is a target of oxidative stress at stages I/II of Alzheimer’s disease pathology. Brain Pathol. 2010;20:222–233. doi: 10.1111/j.1750-3639.2009.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor BJ, Singleton AB. Nature versus nurture: death of a dogma, and the road ahead. Neuron. 2010;68:196–200. doi: 10.1016/j.neuron.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcelay E, Ibarreta D, Parrilla R, Ayuso MS, Martin-Requero A. Enhanced proliferation of lymphoblasts from patients with Alzheimer dementia associated with calmodulin-dependent activation of the na+/H+ exchanger. Neurobiol Dis. 2001;8:289–298. doi: 10.1006/nbdi.2000.0381. [DOI] [PubMed] [Google Scholar]
- van Es MA, van den Berg LH. Alzheimer’s disease beyond APOE. Nat Genet. 2009;41:1047–1048. doi: 10.1038/ng1009-1047. [DOI] [PubMed] [Google Scholar]
- Veerhuis R. Histological and Direct Evidence for the Role of Complement in the Neuroinflammation of AD. Curr Alzheimer Res. 2010 doi: 10.2174/156720511794604589. [DOI] [PubMed] [Google Scholar]
- Villain N, Chetelat G, Desgranges B, Eustache F. [Neuroimaging in Alzheimer’s disease: a synthesis and a contribution to the understanding of physiopathological mechanisms] Biol Aujourdhui. 2010;204:145–158. doi: 10.1051/jbio/2010010. [DOI] [PubMed] [Google Scholar]
- Walker DG, Dalsing-Hernandez JE, Lue LF. Human postmortem brain-derived cerebrovascular smooth muscle cells express all genes of the classical complement pathway: a potential mechanism for vascular damage in cerebral amyloid angiopathy and Alzheimer’s disease. Microvasc Res. 2008;75:411–419. doi: 10.1016/j.mvr.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang L, Wu Q, Boyd DD. Unbiased screening for transcriptional targets of ZKSCAN3 identifies integrin beta 4 and vascular endothelial growth factor as downstream targets. J Biol Chem. 2008;283:35295–35304. doi: 10.1074/jbc.M806965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.