Abstract
Background
Poly(ADP-ribose) polymerase-1 (PARP-1 catalyzes poly(ADP-ribosyl)ation to various proteins involved in many cellular processes, including DNA damage detection and repair, and cell proliferation and death. PARP-1 has been implicated in human carcinogenesis, but the association between the most-studied PARP-1 V762A polymorphism (rs1136410) and risk of various cancers was reported with inconclusive results.
Aims
To assess the association between the PARP-1 V762A polymorphism and cancer risk.
Methods
A meta-analysis of 21 studies with 12027 cancer patients and 14106 cancer-free controls was conducted to evaluate the strength of the association using odds ratio (OR) with 95% confidence interval (CI).
Results
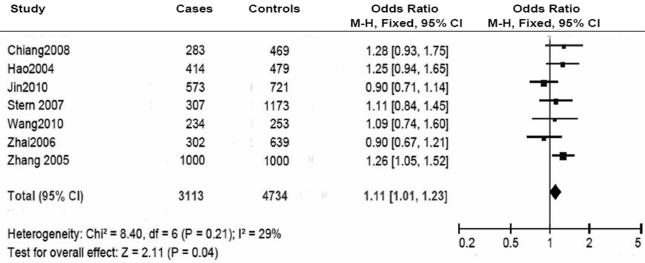
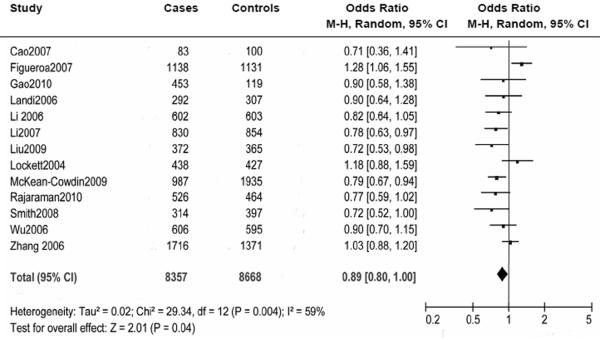
Overall, no significant association was found between the PARP-1 V762A polymorphism and cancer risk. In the stratified analyses, however, it was found that the variant A allele of the PARP-1 V762A polymorphism was associated with an increased risk of cancer among Asian populations (VA+AA vs.VV: OR = 1.11, 95% CI: 1.01-1.23; Pheterogeneity = 0.210) but a decreased risk of cancer (VA+AA vs.VV: OR =0.89, 95% CI: 0.80-1.00; Pheterogeneity = 0.004), among Caucasian populations, especially for glioma risk (OR = 0.79, 95% CI: 0.69-0.90; Pheterogeneity = 0.800).
Conclusions
This meta-analysis found evidence for an association of the PARP-1 V 762A polymorphism with increased risk of cancer among Asians but decreased risk of cancer among Caucasians, particularly of glioma. Further well designed studies with large sample sizes of different ethnic populations and different cancer types are warranted to confirm these findings.
Keywords: DNA repair, Case-control study, Meta-analysis, Polymorphism, Susceptibility
Introduction
Genomic DNA damage in mammalian cells can be caused by numerous physical and chemical agents, of which some are endogenous products, such as reactive oxygen species generated by normal cellular metabolism processes, while others are exogenous insults, such as ultraviolet right, ionizing radiation, and genotoxic chemicals. Such DNA damage, if left unrepaired or repaired incorrectly, may induce mutations and genomic instability that lead to cellular malignant transformation and tumorgenesis [Hoeijmakers 2001; Jackson and Bartek 2009]. Nevertheless, human cells have developed a set of complex DNA repair systems that safeguard the integrity of the genome to minimize the consequences of detrimental mutations [Hoeijmakers 2001]. At least four main repair pathways are involved in the removal of DNA damage: base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), and double-strand breaks repair (DSBR) [Decordier, et al. 2010; Hoeijmakers 2001]. Inherited defects in DNA damage response and repair pathways predispose to the development of malignancies [Hoeijmakers 2001]. Indeed, several human hereditary diseases, which are characterized by severe developmental problems and /or a predisposition to cancer, have been found to be linked to deficiencies in DNA repair [Friedberg 2008; Wiesmuller, et al. 2002].
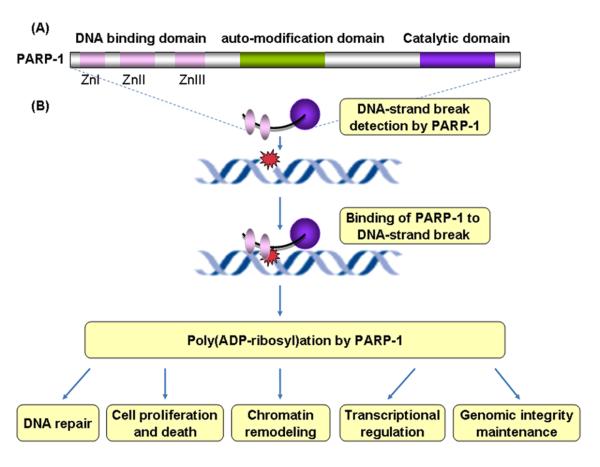
Poly(ADP-ribose) polymerase-1 (PARP-1), also known as ADPRT, is a major member of the PARP family that contains 18 distinct proteins in humans [Kim, et al. 2005]. Human PARP-1 protein (113KDa) is a nuclear protein comprised of three functional domains (Figure 1). The N-terminal DNA-binding domain (DBD) harbors two zinc-finger motifs that are important for the binding of PARP1 to DNA-strand breaks. A third zinc finger was found to be important for coupling damage-induced changes in the DBD to alterations in PARP-1 catalytic activity [Rouleau, et al.2010]. In the central auto-modification domain, specific glutamate and lysine residues serve as potential acceptor sites for poly(ADP-ribosyl) ation. This domain also comprises a BRCA1 C-terminus motif, via which PARP-1 mediates protein-protein interactions. The third functional domain is a C-terminal catalytic domain containing a ‘PARP-signature’ motif, a highly evolutionally conservative sequence that forms the active site [Ame, et al. 2004; Kim, et al. 2005; Rouleau, et al. 2010].
Figure 1.
Extensive studies of PARP-1 to date have implicated PARP-1 in a variety of cellular processes, all of which involve DNA-related transactions [Rouleau, et al. 2010] (Figure 1). Most importantly, PARP-1 functions as a DNA damage sensor in the DNA repair process [Kim, et al. 2005]. In general, in response to DNA damage generated either exogenously or endogenously, PARP-1 binds to sites of DNA damage through its DNA-binding domain, which activates the poly(ADP-ribosyl)ation of target proteins involved in numerous biological events. A growing number of evidence has shown that PARP-1 physically and functionally interacts with various proteins involved in multiple DNA repair pathways, including the BER, SSB, and DSBR pathways, and may recruit the repair proteins to sites of DNA damage (e.g., XRCC1 in BER, DNA-dependent protein kinase in DSBR repair), thereby plays a critical role in DNA damage repair [Burkle 2001; Kim, et al. 2005; Masson, et al. 1998; Ruscetti, et al. 1998]. In addition, PARP-1 is also involved in many other molecular and cellular processes, such as modulation of gene transcription, chromatin remodeling, maintaining telomeres and genomic stability, as well as cell proliferation and death [Gurung, et al. 2010; Masutani, et al. 2003; Masutani, et al. 2005; Tong, et al. 2001]. Accumulated evidence from the results of animal and cellular studies indicates that deficiency of PARP-1 leads to DNA repair defects, genomic instability, failure of induction of cell death, and modulation of gene transcription, thereby contributing to carcinogenesis [Masutani, et al. 2003; Masutani, et al. 2005].
The human PARP-1 gene is located on chromosome 1q41-42, consists of 23 exons, and spans 47.3 kb. To date, at least 439 single nucleotide polymorphisms (SNPs), including 17 non-synonymous SNPs (nsSNPs) in the PARP-1 gene, have been reported, of which only one coding nsSNP (rs1136410, V762A) is common [minor allele frequency (MAF) >0.05] (http://www.ncbi.nlm.nih.gov/SNP) and most frequently studied for its association with cancer risk. PARP-1 V762A is a base T to C transition at codon 762 in exon 17, which results in the substitution of alanine for valine in the catalytic domain of PARP-1, and PARP-1 V762A polymorphism was reported to be associated with an altered activity of PARP-1 [Lockett, et al. 2004; Wang, et al. 2007].
To date, a number of molecular epidemiological studies have been conducted to evaluate the effect of the PARP-1 V762A polymorphism on risk of many cancer types, including cancers of the brain [Liu, et al. 2009; McKean-Cowdin, et al. 2009; Rajaraman, et al. 2010; Yosunkaya, et al.2010], stomach [Miao, et al. 2006; Zhang, et al. 2009], breasts [Cao, et al. 2007; Haiman, et al. 2008; Smith, et al. 2008; Zhai, et al. 2006; Zhang, et al. 2006], colorectum [Brevik, et al. 2010; Stern, et al. 2009; Stern, et al. 2007], bladder [Figueroa, et al. 2007; Huang, et al. 2007; Wang, et al. 2010; Wu, et al. 2006], prostate [Gao, et al. 2010; Lockett, et al. 2004], head and neck [Li, et al. 2007], lung [Choi, et al. 2003; Landi, et al. 2006; Zhang, et al. 2005], esophagus [Hao, et al. 2004], and skin [Li, et al. 2006] as well as other types of cancer [Chiang, et al. 2008; Jin, et al. 2010 ; Rajaraman, et al. 2010; Shen, et al. 2006]. However, until now, those studies that investigated associations between the PARP-1 V762A polymorphism and cancer risk have yielded inconsistent results. Considering the potential important role of PARP-1 in carcinogenesis, we performed a meta-analysis to assess the association of the PARP-1 V762A polymorphism with overall cancer risk as well as the risk of specific type of cancers in different ethnic populations.
Materials and Methods
Literature search
For this meta-analysis, we searched for relevant studies by using PubMed software to search Medline (U.S. National Library of Medicine) to identify candidate studies. The latest search was performed on May 12, 2011. The following terms were jointly used for the search: ‘PARP-1’ or ‘ADPRT’, ‘polymorphism or variant’, ‘cancer’, ‘tumor’, or ‘carcinoma’. In addition, additional studies were also identified by a manual search for the reference cited in the retrieved studies and review articles. Search results were limited to studies published in English.
Inclusion and exclusion criteria
All human-associated studies were included, if they met the following criteria: (1) evaluation of the PARP-1 V762A polymorphism (rs1136410) and cancer risk, (2) case-control, nested case-control, or cohort study, (3) the study reported an odds ratio (OR) with 95% confidence interval (95% CI) or data for their calculation, and (4) the study was published in English, (5) Genotype distribution of control populations must be consistent with Hardy-Weinberg equilibrium (HWE). Exclusion criteria were: (1) duplicate data, (2) abstract, case report, comment, review and editorial, (3) no sufficient genotyping data were provided, (4) the outcome was benign tumors, precancerous lesions, and adenomas, (5) family-based study, (6) genotype distribution of control subjects was inconsistent with Hardy–Weinberg equilibrium (HWE). If studies had overlapped subjects, only the one with largest sample size was included in the final analysis.
Data extraction
Two investigators independently reviewed the articles and extracted the data from all eligible publications according to the criteria listed above. Disagreements were resolved by discussion between the two investigators. The following information was recorded for each study: first author, year of publication, country or region of origin, ethnicity, cancer type, number of cases and controls, number of cases and controls by genotype, source of control group (population-based or hospital-based), genotyping methods, minor allele frequency in controls. For studies including subjects of different ethnic descents, data were extracted separately for each ethnicity, which was categorized as Caucasian, Asian, and the mixed descendants (admixture of different ethnic groups).
Statistical analysis
Genotypic frequency for the PARP-1 V762A polymorphism was tested for deviation from HWE in control subjects using the chi-square goodness of fit, and a P value < 0.05 was considered significant. Odds ratios (OR) and corresponding 95% confidence interval (95% CI) were used to estimate the association between the PARP-1 V762A polymorphism and cancer risk. We estimated the risk for variant homozygous AA and heterogeneous VA genotypes, compared with the wild-type homozygous VV genotype, and then for the (VA + AA) versus (vs.) VV and AA versus (VA + VV), assuming recessive and dominant effect models, respectively. The statistical heterogeneity among studies was assessed with the Q-test and I2 statistics [Lau, et al. 1997]. The heterogeneity across studies was considered significant when a P value < 0.1 for the Q statistic [Lau, et al. 1997]. If there was no significant heterogeneity, the fixed-effects model was used to estimate the summary OR and 95% CI; otherwise, the random-effects model was used [DerSimonian and Laird 1986]. To explore potential sources of heterogeneity across studies, we did stratification and meta-regression analyses. Stratified analyses were conducted by ethnicity (Caucasian, Asian, and mixed), type of cancers and specific type of cancers in different ethnic populations (if one cancer type contains less than three studies, it was merged into the ‘other cancers’ group), source of controls (hospital-based and population-based), and genotyping methods. Publication bias was evaluated with the funnel plot and Begg’s and Egger’s tests [Begg and Mazumdar 1994; Egger, et al. 1997]. A P value < 0.05 was used as an indication for the presence of potential publication bias. Sensitivity analyses were conducted by removing one study at a time to assess the influence of individual studies on the pooled ORs. All analyses were conducted using Review Manager (v.5.0; Oxford, England) and Stata software (version 8.2; StataCorp LP, College Station, TX, USA). All the P values were two-sided.
Results
Characteristics of studies
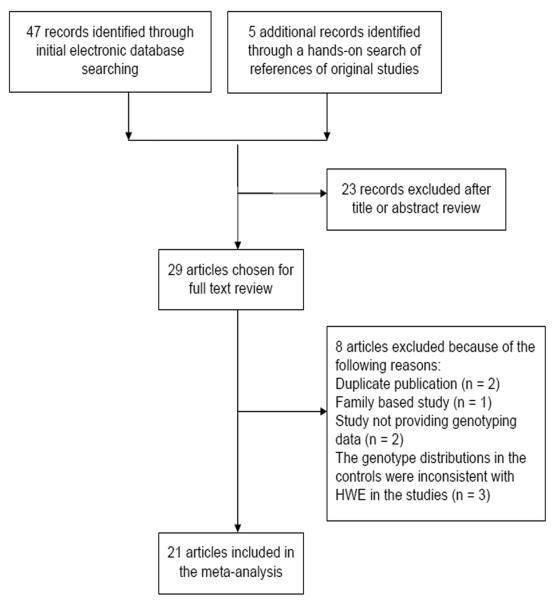
The flowchart of study selection for this meta-analysis is presented in Figure 2. A total of 52 published records up to May 12, 2011 were identified, of which 29 were considered potentially eligible for inclusion in this meta-analysis and were retrieved in full texts. Among these, eight studies were excluded: three studies had the controls with genotype distributions inconsistent with HWE (P < 0.050) [Miao, et al. 2006; Yosunkaya, et al. 2010; Zhang, et al. 2009], two were duplicate publications [Huang, et al. 2007; Stern, et al. 2009], one was a family-based study [Brevik, et al. 2010], and two provided no genotyping data [Choi, et al. 2003; Haiman, et al. 2008]. Finally, 21 articles were selected in the meta-analysis. In addition, the studies investigating multiple types of cancers [Rajaraman, et al. 2010] or multiple ethnicities[Lockett, et al. 2004; Smith, et al. 2008] were separated into multiple studies in the subgroup analysis. One study [Huang, et al. 2007; Liu, et al. 2009] that only provided the total number of variant genotypes (VA + AA and VV) was included in the analysis for the dominant model but not for other genetic models. Main characteristics of the included studies are presented in Table 1. Overall, 21 publications including 12027 cases and 14106 controls were available for this meta-analysis. All studies were case-control studies, including two studies for glioma, one study for multiple types of cancer (adult meningioma, glioma, and acoustic neuroma), four studies for breast cancer, three studies for bladder cancer, and eleven studies for other types of cancers such as lung cancer, gastric cancer, melanoma, prostate cancer. Among 21 studies included in this meta-analysis, 11 studies were conducted in Caucasians, seven studies in Asians, three studies in the mixed ethnic populations (including Caucasians and African-Americans). The distributions of genotypes in the controls were all in agreement with HWE.
Figure 2.
Table 1.
Characteristics of studies included in the meta-an analysis
| First author | Year | Country | Ethnicitya | Types of cancerb | OR (95%CI)c (VA +AA vs. VV) |
Sample size (Cases /Controls) |
Source of controls | Genotyping methodg |
|---|---|---|---|---|---|---|---|---|
| Lockett | 2004 | America | Mixed | Prostate Cancer | 1.27 (1.11 - 1.46)d | 488 / 524 | hospital-based | Mass ARRAY |
| Hao | 2004 | China | Asian | ESCC | 1.25 (1.07 - 1.46) d | 414 / 479 | hospital-based | PCR-RFLP |
| Zhang | 2005 | China | Asian | Lung Cancer | 1.68 (1.27 - 2.23) d | 1000 / 1000 | hospital-based | PCR-RFLP |
| Zhang | 2006 | America | Caucasian | Breast Cancer | 1.03 (0.96 - 1.11) d | 1716 / 1371 | population-based | TaqMan |
| Zhai | 2006 | China | Asian | Breast Cancer | 0.87 (0.64 - 1.19) e | 302 / 639 | hospital-based | PCR-RFLP |
| Li | 2006 | America | Caucasian | Cutaneous Melanoma | 0.94 (0.68 - 1.30) d | 602 / 603 | hospital-based | PCR-RFLP |
| Shen | 2006 | America | Multiple | NHL | 0.87 (0.66 - 1.15) d | 455 / 535 | population-based | TaqMan |
| Wu | 2006 | America | Caucasian | Bladder Cancer | 0.89 (0.68 - 1.15) d | 606 / 595 | hospital-based | TaqMan |
| Landi | 2006 | Multiple regions | Caucasian | Lung Cancer | 0.90 (0.75 - 1.08) d | 292 / 307 | hospital-based | APEX |
| Li | 2007 | America | Caucasian | SCCHN | 0.79 (0.63 - 1.00) e | 830 / 854 | hospital-based | PCR-RFLP |
| Cao | 2007 | France | Caucasian | Breast Cancer | 0.71 (0.48 - 1.06) d | 83 / 100 | hospital-based | Sequence |
| Figueroa | 2007 | Spain | Caucasian | Bladder Cancer | 1.24 (1.02 - 1.51) d | 1138 / 1131 | hospital-based | TaqMan |
| Stern | 2007 | Singapore | Asian | Colorectal Cancer | 1.1 (0.8 - 1.4) e | 307 / 1173 | population-based | TaqMan |
| Chiang | 2008 | Taiwan | Asian | Thyroid Carcinoma | 1.28 (1.04 - 1.56) d | 283 / 469 | hospital-based | TaqMan |
| Smith | 2008 | America | Multiple | Breast Cancer | 0.79 (0.66 - 0.96) d | 366 / 469 | hospital-based | MassARRAY |
| McKean-Cowdin | 2009 | America | Caucasian | Glioblastoma | 0.80 (0.67 - 0.95) e | 987 / 1935 | hospital-based and population-based |
MassARRAY |
| Liu | 2009 | America | Caucasian | Glioma | 0.71 (0.52 - 0.97) e | 372 / 365 | population-based | MassARRAY |
| Jin | 2010 | Korea | Asian | NHL | 0.90 (071 - 1.04) e Glioma: 0.86 (0.63 - 1.17) e |
573 / 721 | population-based | PCR-HRM |
| Rajaraman | 2010 | America | Caucasian | Multiple Cancers | Meningioma: 0.67 (0.42 - 1.07) e Acoustic neuroma: 0.53 (0.28 - 1.00) e |
526 / 464 | hospital-based | TaqMan |
| Wang | 2010 | China | Asian | Bladder Cancer | 1.08 (0.89 - 1.04) d | 234 / 253 | hospital-based | PCR-RFLP |
| Gao | 2010 | America | Caucasian | Prostate Cancer | 0.90 (0.82 - 0.99) d | 453 / 119 | hospital-based | Sequence |
Ethnicitya: Mixed: mixed ethnicities.
Cancer types: ESCC: Esophageal Squamous Cell Carcinoma; NHL: Non-Hodgkin lymphoma; SCCHN: Squamous Cell Carcinoma of the Head; and Neck; Multiple cancers: Meningioma, Glioma, and Acoustic neuroma.
VA + AA vs. VV: Dominant model.
Unadjusted odds ratio (OR) and 95% Confidence Interval (CI).
Adjusted OR (95 CI %).
Genotyping method: MassARRAY: Genotyping was performed using the Sequenom MassARRAY iPLEX™ platform2. MassARRAY Workstation version 3.3 software was used to process and analyze iPLEX SpectroCHIP bioarrays; APEX: polymorphism was analyzed together for a given sample by a microarray technique based on the arrayed primer extension principle; PCR-RFLP: Polymerase Chain Reaction-restriction Fragment Length Polymorphism; PCR-HRM: PCR cycling and high resolution melting (HRM) analysis was performed on the Rotor-Gene 6000™.
Quantitative synthesis
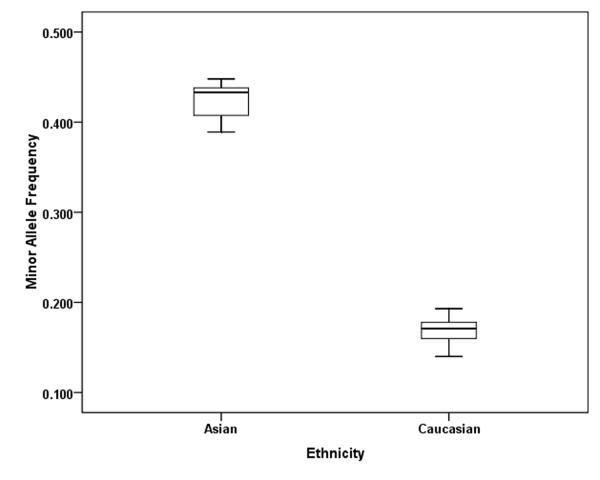
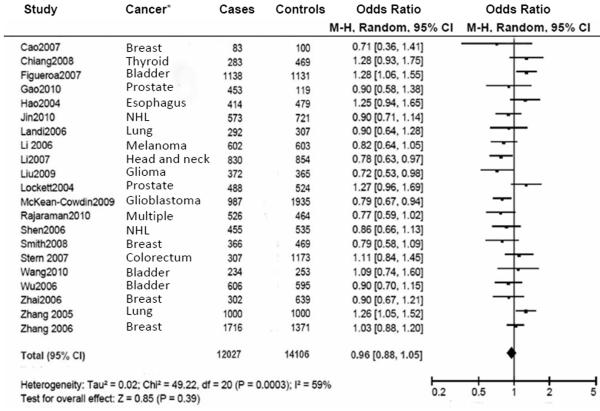
We included 12027 cancer patients and 14106 control subjects in this meta-analysis. We found that the frequency of the variant allele (A) of the PARP-1 V762A polymorphism was significantly higher in Asians (0.423, 95% CI: 0.403 - 0.443) than in Caucasians (0.166, 95% CI: 0.154-0.178) (Figure 3). Overall, no significant association was found between the PARP-1 V762A polymorphism and cancer susceptibility (Table 2, Figure 4). Individuals carrying the A allele did not have an altered cancer risk, compared with individuals with the wild-type VV homozygous genotype (VA vs. VV: OR = 0.97, 95% CI: 0. 90-1.06, Pheterogeneity = 0.010; AA vs. VV: OR = 0.99, 95% CI: 0.84-1.17; Pheterogeneity = 0.010). Similarly, no significant association with the risk of cancer was found in either a dominant model (VA+AA vs. VV: OR = 0.96, 95% CI: 0.88-1.05, Pheterogeneity = 0.0003) or a recessive model (AA vs. VA+VV: OR =1.03, 95% CI: 0.93 -1.14, Pheterogeneity = 0.100) (Table 2).
Figure 3.
Table 2.
Meta-analysis for the association between the PARP-1 V762/A polymorphism and cancer risk
| Variables | No. of subjects Cases / Controls |
na | VA vs. VV | AA vs. VV | VA +AA vs. VVb | AA vs. VA +VVc | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| OR (95% CI) | P d | OR (95% CI) | P d | OR (95% CI) | P d | OR (95% CI) | P d | |||
| Total | 12027 / 14106 | 21 | 0.97 (0.90-1.06) | 0.010 | 0.99 (0.84-1.17) | 0.010 | 0.96 (0.88-1.05) | 0.0003 | 1.03 (0.93-1.14) | 0.100 |
| Ethnicities | ||||||||||
| Asian | 3113 / 4734 | 7 | 1.09 (0.98-1.21) | 0.560 | 1.16 (0.95-1.41) | 0.070 | 1.11 (1.01-1.23) | 0.210 | 1.12 (0.99-1.26) | 0.230 |
| Caucasian | 8357 / 8668 | 13 | 0.91 (0.81-1.02) | 0.009 | 0.84 (0.70-1.02) | 0.280 | 0.89 (0.80-1.00) | 0.004 | 0.86 (0.71-1.04) | 0.350 |
| Mixede | 557 / 704 | 3 | 0.93 (0.71-1.21) | 0.250 | 0.66 (0.26-1.69) | - | 0.91 (0.70-1.18) | 0.240 | 1.12 (0.99-1.26) | - |
| Types of cancers | ||||||||||
| Glioma | 1699 / 2764 | 3 | 0.81 (0.70-0.95) | 0.670 | 0.69 (0.45-1.07) | 0.610 | 0.79 (0.69-0.90) | 0.800 | 0.73 (0.48-1.13) | 0.580 |
| Breast cancer | 2467 / 2579 | 4 | 0.96 (0.85-1.10) | 0.330 | 0.90 (0.68-1.18) | 0.850 | 0.96 (0.84-1.08) | 0.370 | 0.92 (0.71-1.19) | 0.860 |
| Bladder cancer | 1978 / 1979 | 3 | 1.10 (0.84-1.44) | 0.060 | 0.99 (0.70-1.40) | 0.850 | 1.09 (0.86-1.39) | 0.080 | 0.96 (0.69-1.33) | 0.820 |
| Other cancers f | 5883 / 7712 | 13 | 0.98 (0.91-1.07) | 0.100 | 1.05 (0.83-1.34) | 0.006 | 0.98 (0.86-1.11) | 0.004 | 1.04 (0.86-1.27) | 0.040 |
| Sources of controls | ||||||||||
| Population-based | 3423 / 4165 | 5 | 1.00 (0.89-1.12) | 0.600 | 0.94 (0.77-1.14) | 0.520 | 0.95 (0.86-1.05) | 0.190 | 0.95 (0.79-1.13) | 0.670 |
| Hospital-based | 7617 / 8006 | 15 | 0.99 (0.89-1.10) | 0.010 | 1.03 (0.83-1.29) | 0.010 | 0.99 (0.88-1.12) | 0.001 | 1.03 (085-1.24) | 0.060 |
| Otherg | 987 / 1935 | 1 | - | - | - | - | - | - | - | - |
| Genotyping methods | ||||||||||
| PCR-RFLP | 3382 / 3828 | 6 | 0.98 (0.83-1.16) | 0.050 | 1.06 (0.79-1.43) | 0.020 | 1.00 (0.82-1.21) | 0.005 | 1.05 (0.84-1.33) | 0.080 |
| Other h | 8645 / 10278 | 15 | 0.96 (0.83-1.11) | 0.150 | 0.97 (0.88-1.07) | 0.030 | 0.95 (0.85-1.05) | 0.004 | 0.96 (0.84-1.11) | 0.300 |
Number of comparisons.
VA + AA vs VV: Dominant model
AA vs VV + VA: Recessive model
P value of Q-test for heterogeneity test. The random-effects model was used when P-value for heterogeneity test < 0.1; otherwise, the fixed-effects model was used.
Mixed: one comparison between the AA genotype and the VV genotype, and one comparison for the recessive model (AA vs.VA /VV).
Other cancers: each type of cancer which contains less than three studies was merged into the ‘other cancers’ group.
Other: Controls of this study (McKean-Cowdin, et al.) were from both hospital-based controls and population-based controls.
Other The genotyping methods included TaqMan, MassARRAY assays, Sequence, ant the others.
Figure 4.
In the stratified analysis by ethnicity, a significantly increased cancer risk was found among Asians in the dominant model (VA+AA vs. VV: OR = 1.11, 95% CI: 1.01-1.23, Pheterogeneity = 0.210) (Table 2, Figure 5). In contrast, the PARP-1 V762A polymorphism was significantly associated with a decreased cancer risk among Caucasians in the dominant model (VA+AA vs. VV: OR =0.89, 95% CI: 0.80-1.00, Pheterogeneity = 0.004) (Table 2, Figure 6). No significant associations were detected in all genetic models in the mixed ethnicity population group. No significant associations were found when the data were stratified by the source of control subjects and by genotyping methods (Table 2).
Figure 5.
Figure 6.
Further stratified analyses by specific type of cancers and ethnicity were performed. In Caucasians, the VA heterozygous genotype was found to be associated with significantly decreased risk of glioma compared with the common VV homozygous genotype (VA vs. AA: OR = 0.81, 95% CI: 0.70-0.95, Pheterogeneity = 0.670). A significant main effect was also observed in the dominant model for glioma (VA+AA vs. VV: OR = 0.79, 95% CI: 0.69-0.90, Pheterogeneity = 0.800). However, no significant association was found in ether breast cancer or other cancers (each type of cancers having less than three studies was merged into the ‘other cancers’ group) (Table 3). We were not able to further evaluate the effect of the PARP-1 V762A polymorphism on cancer risk by specific type of cancers in Asian and in the mixed ethnicity populations because only one studies of specific type of cancers were included in these two groups.
Table 3.
Meta-analysis for the association between the PARP-1 V762/A polymorphism and the risk of different types of cancers in Caucasians
| Variables | No. of subjects Cases / Controls |
na | VA vs. VV | AA vs. VV | VA +AA vs. VVb | AA vs. VA +VVc | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| OR (95% CI) | P d | OR (95% CI) | P d | OR (95% CI) | P d | OR (95% CI) | P d | |||
| Types of cancers | 15 | |||||||||
| Glioma | 1699 / 2764 | 3 | 0.81 (0.70-0.95) | 0.670 | 0.69 (0.45-1.07) | 0.610 | 0.79 (0.69-0.90) | 0.800 | 0.73 (0.48-1.13) | 0.580 |
| Breast cancer | 2113 / 1868 | 3 | 0.86 (0.63-1.17) | 0.090 | 0.91 (0.63-1.32) | 0.660 | 0.95 (0.83-1.09) | 0.110 | 0.92 (0.64-1.32) | 0.670 |
| Other cancerse | 4545 / 4964 | 9 | 0.93 (0.79-1.08) | 0.020 | 0.86 (0.66-1.11) | 0.170 | 0.91 (0.78-1.07) | 0.008 | 0.87 (0.67-1.13) | 0.200 |
Number of comparisons.
VA /AA vs VV: Dominant model
AA vs VV /VA: Recessive model
P value of Q-test for heterogeneity test. The random-effects model was used when P-value for heterogeneity test < 0.1; otherwise, the fixed-effects model was used.
Other cancers: each type of cancer which contains less than three studies was merged into the ‘other cancers’ group.
Evaluation of heterogeneity
In the present study, heterogeneity across studies was observed in overall comparisons. Therefore, we assessed the source of heterogeneity for the dominant model (VA+AA vs. VV) by ethnicity, type of cancers, source of controls, and genotyping methods. We found that ethnicity (P = 0.044) and type of cancers (P = 0.038), but not source of controls (P = 0.903) and genotyping techniques (P = 0.146), contributed to substantial heterogeneity.
Sensitivity analysis
In the sensitivity analysis, the influence of each study on the pooled OR was examined by repeating the meta-analysis while omitting each study, one at a time. Such a leave-one-out sensitivity analysis indicated that no single study influenced the pooled ORs qualitatively (data not shown), which indicted the stability of our overall result.
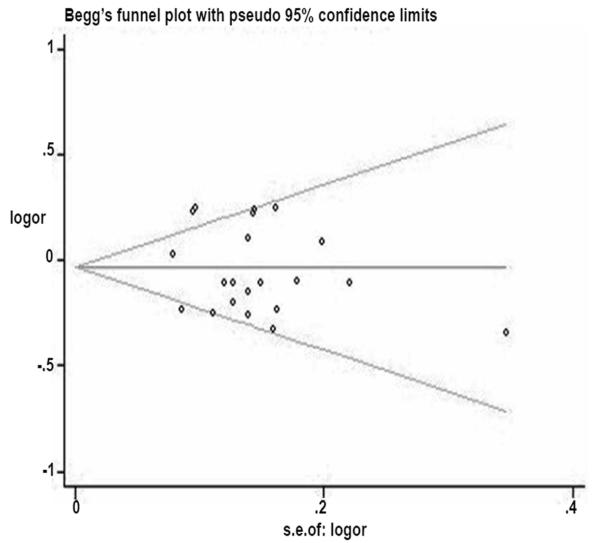
Publication bias
Begger’s funnel plot and Egger’s test were performed to evaluate the publication bias of the publication bias of literature on cancer. The shape of the funnel plot seemed asymmetrical (Figure 7), which was confirmed by an Egger’s test (t = −0.74, P = 0.465 for VA+AA vs. VV).
Figure 7.
Discussion
Carcinogenesis is a multi-step process involving aberrations in a variety of cellular processes including DNA damage detection and repair, genome maintenance, cell-cycle control, proliferation, differentiation, and cell death [Kim, et al. 2005]. PARP-1 catalyzes poly(ADP-ribosyl)ation, which was functionally involved in all of these processes, suggesting a possible connection between PARP-1 function and development of cancer [Kim, et al. 2005; Masutani, et al. 2003]. A body of evidence shows that PARP-1 plays a critical role in human carcinogenesis [Bhatia, et al. 1990a; Bhatia, et al. 1990b; Bieche, et al. 1996; Lyn, et al. 1993; Masutani, et al. 2005; Prasad, et al. 1990; Shiokawa, et al. 2005]. It has been found that PARP-1 knockout mice (PARP-1–/–) treated with either alkylating agents or γ-irradiation displayed high genomic instability, high frequencies of chromosome aberrations, and shortened telomere compared with the wild-type mice [d’Adda di Fagagna, et al. 1999; de Murcia, et al. 1997; Samper, et al. 2001]. PARP-1–/–mice also showed an increased susceptibility to chemically-induced tumorigenesis in the liver, colon, and lung [Nozaki, et al. 2003; Tsutsumi, et al. 2001], although in certain situations, PARP-1 may also facilitate the growth of tumor cells [Martin-Oliva, et al. 2004] . For example, Martin-Olva et al. found that PARP-1–/– mice displayed a strikingly reduced susceptibility to skin carcinogenesis [Martin-Oliva, et al. 2004]. Dysregulation of PARP-1 expression has been found in a variety of human cancers, such as breast cancer, head and neck cancer, melanoma, and colorectal cancer [Goncalves, et al. 2010; Nosho, et al. 2006; Staibano, et al. 2005]. Furthermore, reduced PARP-1 activity in human peripheral blood lymphocytes has been observed in patients with cancers of the breasts, colon, lung, larynx, and prostate as well as glioma [Barton, et al. 2009; Lockett, et al. 2004; Pero, et al. 1990; Rajaee-Behbahani, et al. 2002]. Therefore, it is biologically plausible that the PARP-1 V762A polymorphism residing in the catalytic domain of the PARP-1 enzyme, which causes an amino acid change from Val to Ala, may alter the enzymatic activity of PARP-1 and thereby contribute to cancer susceptibility.
In the present meta-analysis with 12027 cancer cases and 14106 controls, no significant association between the PARP-1 V762A polymorphism and overall risk of cancer was found. However, in the subgroup analyses by ethnicity, the variant 762A allele was found to be associated with an increased risk of cancer among Asian populations but a decreased risk of cancer, particularly glioma, among Caucasian populations,. These findings indicate that the PARP-1 V762A polymorphism may play a role in cancer development, at least in some ethnic groups or some specific cancer types. However, it is still unclear whether the same polymorphism may have the same effect across different types of cancer, though evidence has shown that a few SNPs, such as rs401681 in CLPTM1L, are associated with risk of many cancer types including basal cell carcinoma and cancers of the lung, bladder, prostate, and cervix [Rafnar, et al. 2009]. Therefore, our results should be interpreted cautiously. Nevertheless, our findings are interesting and worthy of further investigation.
If significant heterogeneity is present, pooled summary estimates from such meta-analyses are hard to interpret. In our meta-analysis, obvious heterogeneity across studies was observed in the overall comparison and some subgroup analyses. We then used the meta-regression analysis to explore the sources of heterogeneity. We found that ethnicity and type of cancers might contribute to the potential heterogeneity, suggesting that the PARP-1 V762A polymorphism may modulate cancer risk differently depending on ethnicity and type of cancers. Indeed, in the stratified analysis by ethnicity, the effect of the variant A allele of the PAPR-1 V762A polymorphism appeared to be associated with cancer risk more likely in Asians than in Caucasians. When stratifying by type of cancers, we found that the variant A allele of this polymorphism was associated with a decreased risk for glioma in Caucasians but not for other types of cancer in the dominant model, and no heterogeneity was found across the studies.
In our meta-analysis, we found that the minor allele frequencies of the PARP-1 V762A polymorphism exhibited striking racial differences. The A allele frequency of Asians was significantly higher (42.3%) than that of Caucasians (16.6%). Similarly, in the HapMap database, the frequency of the minor A allele among Asians (45%) is also higher than that of European (15%) (http://hapmap.ncbi.nlm.nih.gov/). To date, no association study between the PARP-1 V762A polymorphism and glioma risk has been reported in Asians. Therefore, further studies are needed to explore the role of the PARP-1 V762A polymorphism in the development of different type of cancers including glioma in different ethnic populations.
Some potential limitations in this meta-analysis should be considered. Firstly, only papers published in English were included in this meta-analysis, and this could have introduced bias in our results. Secondly, obvious heterogeneity across studies, which might result from different ethnicities and types of cancer, existed in overall comparisons and also some subgroup analyses. Although various genotyping techniques were used in those studies used in our meta-analysis, including polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), TaqMan, and MassARRAY assays, it did not contribute to the observed heterogeneity across studies (P = 0.146 ). Thirdly, the small sample sizes in some subgroup analyses may have limited statistical power. Furthermore, due to limited studies included in this meta-analysis, we were unable to perform further subgroup analyses, such as by specific type of cancers in Asians. Fourthly, our results were based on unadjusted OR estimates, because not all studies included in this meta-analysis provided adjusted ORs or the ORs were not adjusted by the same potential confounders, such as age, sex, ethnicity, and other environmental factors. A more precise analysis should be conducted if individual data were available. Fifthly, despite the confirmatory findings from this meta-analysis, we can not rule out the possibility that the current significant results might be due to chance, because many factors, such as those we mentioned above and potential publication bias that may exist but was undetectable in this meta-analysis, could have an impact on our analysis. Therefore, these results should be interpreted with caution and confirmed from additional analysis with more published studies in the future. Finally, evaluation of the potential gene-gene and gene-environment interaction effects on the risk of cancer was limited because of lacking the original data of the reviewed studies.
In conclusion, our meta-analysis with a total of 12027 cases and 14106 controls suggests that the variant A allele of PARP-1 V763A polymorphism may increase the risk of cancer in Asian populations but decrease the risk of cancer, especially for the risk of glioma, in Caucasian populations. Further well designed studies with large sample sizes of different ethnic populations and different cancer types are warranted. Furthermore, the effects of gene-gene and gene-environment interactions on the risk of cancer should also be taken into account.
Acknowledgments
This work was supported in part by the National Institutes of Health grants R01 CA131274 and R01 ES011740 (Q. Wei). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest statement: None declared.
Reference
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26(8):882–93. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Barton VN, Donson AM, Kleinschmidt-DeMasters BK, Gore L, Liu AK, Foreman NK. PARP1 expression in pediatric central nervous system tumors. Pediatr Blood Cancer. 2009;53(7):1227–30. doi: 10.1002/pbc.22141. [DOI] [PubMed] [Google Scholar]
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101. [PubMed] [Google Scholar]
- Bhatia K, Huppi K, Cherney B, Raffeld M, Smulson M, Magrath I. Relative predispositional effect of a PADPRP marker allele in B-cell and some non B-cell malignancies. Curr Top Microbiol Immunol. 1990a;166:347–57. doi: 10.1007/978-3-642-75889-8_43. [DOI] [PubMed] [Google Scholar]
- Bhatia KG, Cherney BW, Huppi K, Magrath IT, Cossman J, Sausville E, Barriga F, Johnson B, Gause B, Bonney G. A deletion linked to a poly(ADP-ribose) polymerase gene on chromosome 13q33-qter occurs frequently in the normal black population as well as in multiple tumor DNA. Cancer Res. 1990b;50(17):5406–13. others. [PubMed] [Google Scholar]
- Bieche I, de Murcia G, Lidereau R. Poly(ADP-ribose) polymerase gene expression status and genomic instability in human breast cancer. Clin Cancer Res. 1996;2(7):1163–7. [PubMed] [Google Scholar]
- Brevik A, Joshi AD, Corral R, Onland-Moret NC, Siegmund KD, Le Marchand L, Baron JA, Martinez ME, Haile RW, Ahnen DJ. Polymorphisms in base excision repair genes as colorectal cancer risk factors and modifiers of the effect of diets high in red meat. Cancer Epidemiol Biomarkers Prev. 2010;19(12):3167–73. doi: 10.1158/1055-9965.EPI-10-0606. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkle A. PARP-1: a regulator of genomic stability linked with mammalian longevity. Chembiochem. 2001;2(10):725–8. doi: 10.1002/1439-7633(20011001)2:10<725::AID-CBIC725>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Cao WH, Wang X, Frappart L, Rigal D, Wang ZQ, Shen Y, Tong WM. Analysis of genetic variants of the poly(ADP-ribose) polymerase-1 gene in breast cancer in French patients. Mutat Res. 2007;632(1-2):20–8. doi: 10.1016/j.mrgentox.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Chiang FY, Wu CW, Hsiao PJ, Kuo WR, Lee KW, Lin JC, Liao YC, Juo SH. Association between polymorphisms in DNA base excision repair genes XRCC1, APE1, and ADPRT and differentiated thyroid carcinoma. Clin Cancer Res. 2008;14(18):5919–24. doi: 10.1158/1078-0432.CCR-08-0906. [DOI] [PubMed] [Google Scholar]
- Choi JE, Park SH, Jeon HS, Kim KM, Lee GY, Park RW, Kam S, Kim IS, Kim CH, Jheon SH. No association between haplotypes of three variants (codon 81, 284, and 762) in poly(ADP-ribose) polymerase gene and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2003;12(9):947–9. others. [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Hande MP, Tong WM, Lansdorp PM, Wang ZQ, Jackson SP. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat Genet. 1999;23(1):76–80. doi: 10.1038/12680. [DOI] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94(14):7303–7. doi: 10.1073/pnas.94.14.7303. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decordier I, Loock KV, Kirsch-Volders M. Phenotyping for DNA repair capacity. Mutat Res. 2010;705(2):107–29. doi: 10.1016/j.mrrev.2010.05.002. [DOI] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Egger M, Smith G Davey, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa JD, Malats N, Real FX, Silverman D, Kogevinas M, Chanock S, Welch R, Dosemeci M, Tardon A, Serra C. Genetic variation in the base excision repair pathway and bladder cancer risk. Hum Genet. 2007;121(2):233–42. doi: 10.1007/s00439-006-0294-y. others. [DOI] [PubMed] [Google Scholar]
- Friedberg EC. A brief history of the DNA repair field. Cell Res. 2008;18(1):3–7. doi: 10.1038/cr.2007.113. [DOI] [PubMed] [Google Scholar]
- Gao R, Price DK, Dahut WL, Reed E, Figg WD. Genetic polymorphisms in XRCC1 associated with radiation therapy in prostate cancer. Cancer Biol Ther. 2010;10(1):13–8. doi: 10.4161/cbt.10.1.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves A, Finetti P, Sabatier R, Gilabert M, Adelaide J, Borg JP, Chaffanet M, Viens P, Birnbaum D, Bertucci F. Poly(ADP-ribose) polymerase-1 mRNA expression in human breast cancer: a meta-analysis. Breast Cancer Res Treat. 2010;127(1):273–81. doi: 10.1007/s10549-010-1199-y. [DOI] [PubMed] [Google Scholar]
- Gurung RL, Balakrishnan L, Bhattacharjee RN, Manikandan J, Swaminathan S, Hande MP. Inhibition of poly (ADP-Ribose) polymerase-1 in telomerase deficient mouse embryonic fibroblasts increases arsenite-induced genome instability. Genome Integr. 2010;1(1):5. doi: 10.1186/2041-9414-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Hsu C, de Bakker PI, Frasco M, Sheng X, Van Den Berg D, Casagrande JT, Kolonel LN, Le Marchand L, Hankinson SE. Comprehensive association testing of common genetic variation in DNA repair pathway genes in relationship with breast cancer risk in multiple populations. Hum Mol Genet. 2008;17(6):825–34. doi: 10.1093/hmg/ddm354. others. [DOI] [PubMed] [Google Scholar]
- Hao B, Wang H, Zhou K, Li Y, Chen X, Zhou G, Zhu Y, Miao X, Tan W, Wei Q. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64(12):4378–84. doi: 10.1158/0008-5472.CAN-04-0372. others. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Huang M, Dinney CP, Lin X, Lin J, Grossman HB, Wu X. High-order interactions among genetic variants in DNA base excision repair pathway genes and smoking in bladder cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2007;16(1):84–91. doi: 10.1158/1055-9965.EPI-06-0712. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin XM, Kim HN, Lee IK, Park KS, Kim HJ, Choi JS, Juhng SW, Choi C. PARP-1 Val762Ala polymorphism is associated with reduced risk of non-Hodgkin lymphoma in Korean males. BMC Med Genet. 2010;11:38. doi: 10.1186/1471-2350-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19(17):1951–67. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Landi S, Gemignani F, Canzian F, Gaborieau V, Barale R, Landi D, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P. DNA repair and cell cycle control genes and the risk of young-onset lung cancer. Cancer Res. 2006;66(22):11062–9. doi: 10.1158/0008-5472.CAN-06-1039. others. [DOI] [PubMed] [Google Scholar]
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–6. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- Li C, Hu Z, Lu J, Liu Z, Wang LE, El-Naggar AK, Sturgis EM, Spitz MR, Wei Q. Genetic polymorphisms in DNA base-excision repair genes ADPRT, XRCC1, and APE1 and the risk of squamous cell carcinoma of the head and neck. Cancer. 2007;110(4):867–75. doi: 10.1002/cncr.22861. [DOI] [PubMed] [Google Scholar]
- Li C, Liu Z, Wang LE, Strom SS, Lee JE, Gershenwald JE, Ross MI, Mansfield PF, Cormier JN, Prieto VG. Genetic variants of the ADPRT, XRCC1 and APE1 genes and risk of cutaneous melanoma. Carcinogenesis. 2006;27(9):1894–901. doi: 10.1093/carcin/bgl042. others. [DOI] [PubMed] [Google Scholar]
- Liu Y, Scheurer ME, El-Zein R, Cao Y, Do KA, Gilbert M, Aldape KD, Wei Q, Etzel C, Bondy ML. Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol Biomarkers Prev. 2009;18(1):204–14. doi: 10.1158/1055-9965.EPI-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockett KL, Hall MC, Xu J, Zheng SL, Berwick M, Chuang SC, Clark PE, Cramer SD, Lohman K, Hu JJ. The ADPRT V762A genetic variant contributes to prostate cancer susceptibility and deficient enzyme function. Cancer Res. 2004;64(17):6344–8. doi: 10.1158/0008-5472.CAN-04-0338. [DOI] [PubMed] [Google Scholar]
- Lyn D, Cherney BW, Lalande M, Berenson JR, Lichtenstein A, Lupold S, Bhatia KG, Smulson M. A duplicated region is responsible for the poly(ADP-ribose) polymerase polymorphism, on chromosome 13, associated with a predisposition to cancer. Am J Hum Genet. 1993;52(1):124–34. [PMC free article] [PubMed] [Google Scholar]
- Martin-Oliva D, O’Valle F, Munoz-Gamez JA, Valenzuela MT, Nunez MI, Aguilar M, de Almodovar JM Ruiz, del Moral R Garcia, Oliver FJ. Crosstalk between PARP-1 and NF-kappaB modulates the promotion of skin neoplasia. Oncogene. 2004;23(31):5275–83. doi: 10.1038/sj.onc.1207696. [DOI] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18(6):3563–71. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani M, Nakagama H, Sugimura T. Poly(ADP-ribose) and carcinogenesis. Genes Chromosomes Cancer. 2003;38(4):339–48. doi: 10.1002/gcc.10250. [DOI] [PubMed] [Google Scholar]
- Masutani M, Nakagama H, Sugimura T. Poly(ADP-ribosyl)ation in relation to cancer and autoimmune disease. Cell Mol Life Sci. 2005;62(7-8):769–83. doi: 10.1007/s00018-004-4509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean-Cowdin R, Barnholtz-Sloan J, Inskip PD, Ruder AM, Butler M, Rajaraman P, Razavi P, Patoka J, Wiencke JK, Bondy ML. Associations between polymorphisms in DNA repair genes and glioblastoma. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1118–26. doi: 10.1158/1055-9965.EPI-08-1078. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X, Zhang X, Zhang L, Guo Y, Hao B, Tan W, He F, Lin D. Adenosine diphosphate ribosyl transferase and x-ray repair cross-complementing 1 polymorphisms in gastric cardia cancer. Gastroenterology. 2006;131(2):420–7. doi: 10.1053/j.gastro.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Nosho K, Yamamoto H, Mikami M, Taniguchi H, Takahashi T, Adachi Y, Imamura A, Imai K, Shinomura Y. Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur J Cancer. 2006;42(14):2374–81. doi: 10.1016/j.ejca.2006.01.061. [DOI] [PubMed] [Google Scholar]
- Nozaki T, Fujihara H, Watanabe M, Tsutsumi M, Nakamoto K, Kusuoka O, Kamada N, Suzuki H, Nakagama H, Sugimura T. Parp-1 deficiency implicated in colon and liver tumorigenesis induced by azoxymethane. Cancer Sci. 2003;94(6):497–500. doi: 10.1111/j.1349-7006.2003.tb01472.x. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pero RW, Roush GC, Markowitz MM, Miller DG. Oxidative stress, DNA repair, and cancer susceptibility. Cancer Detect Prev. 1990;14(5):555–61. [PubMed] [Google Scholar]
- Prasad SC, Thraves PJ, Bhatia KG, Smulson ME, Dritschilo A. Enhanced poly(adenosine diphosphate ribose) polymerase activity and gene expression in Ewing’s sarcoma cells. Cancer Res. 1990;50(1):38–43. [PubMed] [Google Scholar]
- Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, Jakobsdottir M, Helgadottir H, Thorlacius S, Aben KK. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41(2):221–7. doi: 10.1038/ng.296. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaee-Behbahani N, Schmezer P, Ramroth H, Burkle A, Bartsch H, Dietz A, Becher H. Reduced poly(ADP-ribosyl)ation in lymphocytes of laryngeal cancer patients: results of a case-control study. Int J Cancer. 2002;98(5):780–4. doi: 10.1002/ijc.10234. [DOI] [PubMed] [Google Scholar]
- Rajaraman P, Hutchinson A, Wichner S, Black PM, Fine HA, Loeffler JS, Selker RG, Shapiro WR, Rothman N, Linet MS. DNA repair gene polymorphisms and risk of adult meningioma, glioma, and acoustic neuroma. Neuro Oncol. 2010;12(1):37–48. doi: 10.1093/neuonc/nop012. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10(4):293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J Biol Chem. 1998;273(23):14461–7. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- Samper E, Goytisolo FA, Menissier-de Murcia J, Gonzalez-Suarez E, Cigudosa JC, de Murcia G, Blasco MA. Normal telomere length and chromosomal end capping in poly(ADP-ribose) polymerase-deficient mice and primary cells despite increased chromosomal instability. J Cell Biol. 2001;154(1):49–60. doi: 10.1083/jcb.200103049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, Wang SS, Holford TR, Leaderer B, Yeager M, Welch R. Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Hum Genet. 2006;119(6):659–68. doi: 10.1007/s00439-006-0177-2. others. [DOI] [PubMed] [Google Scholar]
- Shiokawa M, Masutani M, Fujihara H, Ueki K, Nishikawa R, Sugimura T, Kubo H, Nakagama H. Genetic alteration of poly(ADP-ribose) polymerase-1 in human germ cell tumors. Jpn J Clin Oncol. 2005;35(2):97–102. doi: 10.1093/jjco/hyi028. [DOI] [PubMed] [Google Scholar]
- Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN, Liu-Mares W, Hu JJ. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29(11):2132–8. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staibano S, Pepe S, Lo Muzio L, Somma P, Mascolo M, Argenziano G, Scalvenzi M, Salvatore G, Fabbrocini G, Molea G. Poly(adenosine diphosphate-ribose) polymerase 1 expression in malignant melanomas from photoexposed areas of the head and neck region. Hum Pathol. 2005;36(7):724–31. doi: 10.1016/j.humpath.2005.04.017. others. [DOI] [PubMed] [Google Scholar]
- Stern MC, Butler LM, Corral R, Joshi AD, Yuan JM, Koh WP, Yu MC. Polyunsaturated fatty acids, DNA repair single nucleotide polymorphisms and colorectal cancer in the Singapore Chinese Health Study. J Nutrigenet Nutrigenomics. 2009;2(6):273–9. doi: 10.1159/000308467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MC, Conti DV, Siegmund KD, Corral R, Yuan JM, Koh WP, Yu MC. DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese Health Study. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2363–72. doi: 10.1158/1055-9965.EPI-07-0268. [DOI] [PubMed] [Google Scholar]
- Tong WM, Cortes U, Wang ZQ. Poly(ADP-ribose) polymerase: a guardian angel protecting the genome and suppressing tumorigenesis. Biochim Biophys Acta. 2001;1552(1):27–37. doi: 10.1016/s0304-419x(01)00035-x. [DOI] [PubMed] [Google Scholar]
- Tsutsumi M, Masutani M, Nozaki T, Kusuoka O, Tsujiuchi T, Nakagama H, Suzuki H, Konishi Y, Sugimura T. Increased susceptibility of poly(ADP-ribose) polymerase-1 knockout mice to nitrosamine carcinogenicity. Carcinogenesis. 2001;22(1):1–3. doi: 10.1093/carcin/22.1.1. [DOI] [PubMed] [Google Scholar]
- Wang M, Qin C, Zhu J, Yuan L, Fu G, Zhang Z, Yin C. Genetic variants of XRCC1, APE1, and ADPRT genes and risk of bladder cancer. DNA Cell Biol. 2010;29(6):303–11. doi: 10.1089/dna.2009.0969. [DOI] [PubMed] [Google Scholar]
- Wang XG, Wang ZQ, Tong WM, Shen Y. PARP1 Val762Ala polymorphism reduces enzymatic activity. Biochem Biophys Res Commun. 2007;354(1):122–6. doi: 10.1016/j.bbrc.2006.12.162. [DOI] [PubMed] [Google Scholar]
- Wiesmuller L, Ford JM, Schiestl RH. DNA Damage, Repair, and Diseases. J Biomed Biotechnol. 2002;2(2):45. doi: 10.1155/S1110724302001985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, Zhang Q, Millikan RE, Lerner S, Dinney CP. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78(3):464–79. doi: 10.1086/500848. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosunkaya E, Kucukyuruk B, Onaran I, Gurel CB, Uzan M, Kanigur-Sultuybek G. Glioma risk associates with polymorphisms of DNA repair genes, XRCC1 and PARP1. Br J Neurosurg. 2010;24(5):561–5. doi: 10.3109/02688697.2010.489655. [DOI] [PubMed] [Google Scholar]
- Zhai X, Liu J, Hu Z, Wang S, Qing J, Wang X, Jin G, Gao J, Shen H. Polymorphisms of ADPRT Val762Ala and XRCC1 Arg399Glu and risk of breast cancer in Chinese women: a case control analysis. Oncol Rep. 2006;15(1):247–52. [PubMed] [Google Scholar]
- Zhang Q, Li Y, Li X, Zhou W, Shi B, Chen H, Yuan W. PARP-1 Val762Ala polymorphism, CagA+ H. pylori infection and risk for gastric cancer in Han Chinese population. Mol Biol Rep. 2009;36(6):1461–7. doi: 10.1007/s11033-008-9336-y. [DOI] [PubMed] [Google Scholar]
- Zhang X, Miao X, Liang G, Hao B, Wang Y, Tan W, Li Y, Guo Y, He F, Wei Q. Polymorphisms in DNA base excision repair genes ADPRT and XRCC1 and risk of lung cancer. Cancer Res. 2005;65(3):722–6. others. [PubMed] [Google Scholar]
- Zhang Y, Newcomb PA, Egan KM, Titus-Ernstoff L, Chanock S, Welch R, Brinton LA, Lissowska J, Bardin-Mikolajczak A, Peplonska B. Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15(2):353–8. doi: 10.1158/1055-9965.EPI-05-0653. others. [DOI] [PubMed] [Google Scholar]