Abstract
Salmonella bacteria cause millions of infections and thousands of deaths every year. This pathogen has an unusually broad host range including humans, animals, and even plants. During infection, Salmonella expresses a variety of virulence factors and effectors that are delivered into the host cell triggering cellular responses through protein–protein interactions (PPIs) with host cell proteins which make the pathogen’s invasion and replication possible. To speed up proteomic efforts in elucidating Salmonella–host interactomes, we carried out a survey of the currently published Salmonella–host PPI. Such a list can serve as the gold standard for computational models aimed at predicting Salmonella–host interactomes through integration of large-scale biological data sources. Manual literature and database search of >2200 journal articles and >100 databases resulted in a gold standard list of currently 62 PPI, including primarily interactions of Salmonella proteins with human and mouse proteins. Only six of these interactions were directly retrievable from PPI databases and 16 were highlighted in databases featuring literature extracts. Thus, the literature survey resulted in the most complete interactome available to date for Salmonella. Pathway analysis using Ingenuity and Broad Gene Set Enrichment Analysis (GSEA) software revealed among general pathways such as MAPK signaling in particular those related to cell death as well as cell morphology, turnover, and interactions, in addition to response to not only Salmonella but also other pathogenic – viral and bacterial – infections. The list of interactions is available at http://www.shiprec.org/indicationslist.htm
Keywords: Interactome, Pathway analysis, Protein–protein interaction, Salmonella, Subnetwork analysis
1 Introduction
Salmonella are Gram-negative bacterial pathogens that are capable of infecting a wide range of animals including humans and farm animals such as cows, chicken, and pigs as well as pets such as reptiles, and even plants. Salmonella is a facultative endopathogen and the causative agent of various human diseases, reaching from enteritis to typhoid fever. According to the world health organization, Salmonellosis is the most frequent food-borne disease with around 1.5 billion infections world-wide yearly (http://www.who.int/mediacentre/factsheets/fs139/en/). Disease in mammals usually occurs by oral ingestion of contaminated food or water. Systemic infection of animals and humans depends on the ability of the bacteria to survive the harsh conditions of the gastric tract before entering intestinal epithelial and subsequently other host cells. After entering the small intestine, Salmonella traverses the intestinal mucous layer and can invade nonphagocytic enterocytes of the intestinal epithelium by bacterial-mediated endocytosis. Once the epithelial barrier has been breached, Salmonella can enter intestinal macrophages, sensing the phagosomal environment and activating various virulence mechanisms in order to survive in the microbicidal environment of the host cells.
Salmonella replicates within host cells in a membrane-bounded compartment, the Salmonella-containing vacuoles (SCVs). Intravacuolar bacterial replication depends on tightly controlled interactions with host cell vesicular compartments [1]. Salmonella type III secretion effector proteins subvert trafficking events and alter vacuole positioning by acting on host cell actin filaments, microtubule motors, and components of the Golgi complex. Salmonella replicates in SCVs in both nonphagocytic epithelial cells and macrophages. Once positioned, maturation is stalled and bacterial replication is initiated.
Salmonella encodes two distinct type III secretion systems (TTSSs) on chromosomal Salmonella pathogenicity islands 1 and 2 (SPI1 and SPI2). These two secretion systems are very well characterized compared with other known Salmonella secretion systems which contribute to virulence as well. Although the TTSS-1 inserts into the host cell membrane and translocates Salmonella effectors into the host cell, the TTSS-2 translocates effectors across the SCV membrane into the host cytosol. The majority of TTSS-1 translocated effectors promote actin cytoskeletal modification and rearrangements to force bacterial internalization [2]. Other TTSS-1 and -2 translocated effectors trigger various host pathways and act on location and maturation of the SCV, Salmonella-induced filaments (Sifs) formation as well as Salmonella replication, escape from the SCV, systemic spread and function to manipulate the host innate and adaptive immune response [3, 4].
Understanding the precise mechanisms for the communication between Salmonella and its hosts requires taking a system-wide view and determining the network of interactions between the Salmonella proteins and the host proteins. The use of system-wide approaches to study infectious diseases, and thus the protein–protein interaction (PPI) networks mediating the communication between pathogen and host, is expected to yield new approaches to design treatment strategies. Identification of the interactions allows inference of common proteins targeted by pathogens in host signal transduction and metabolic pathways [5–8]. Alternatively, alternate paths circumventing the pathogen disrupted paths in signal transduction pathways, can be identified through pathway analysis [9]. The information on Salmonella interactions can then be exploited for drug discovery. However, establishing Salmonella–host interactomes is in its infancy.
The identification of global networks of PPI has been accelerated by the development of new high-throughput technologies such as two-hybrid assays [10] and affinity purifications followed by mass spectrometry [11]. Thus, a vast amount of PPI data has been collected for a number of different organisms, but not yet for Salmonella proteins, neither within Salmonella, nor across its different hosts. Because of Salmonella’s broad host range, it is a particularly interesting question to what extent the different interactomes between Salmonella and its hosts overlap or differ, requiring essentially several interactomes to be determined. It is unlikely that multiple interactomes will be fully discovered through experimental proteomic efforts alone. Instead, integration with available data and transfer learning from one host to another host organism through computational methods will be particularly suitable in this case to speed up the studies of the multiple interactomes to be determined. Indeed, numerous in silico methods have been developed to predict PPI, both for intraspecies as well as interspecies interactome scenarios [12]. The most successful methods integrate multiple biological databases through machine-learning approaches such as supervised classification [13]. The PPI prediction task is cast as a binary classification problem, where the two classes are “interact” and “does not interact.” In order to develop the models to differentiate between the two classes, a so-called “gold standard” set is required in which pairs of proteins are labeled as interacting pairs based on experimental evidence. It is the goal of this review to survey the literature as well as PPI-related databases to develop such a gold standard. This review focuses on the known host–protein interaction partners for Salmonella TTSS-1 and -2 translocated effectors as well as their functions. We will discuss the impacts of the interactions at different stages of Salmonella infections. Finally, we will discuss the use of this knowledge for interactome modeling and drug development through pathway analysis.
2 Gathering the current Salmonella–host interactome: Literature search is essential
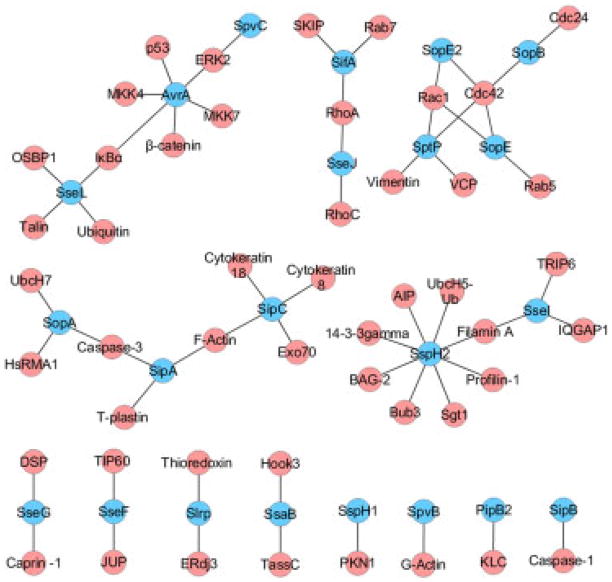
Thirty six pairwise interactions between Salmonella and host proteins in total have been reviewed previously by Haraga et al. 2008 [14], McGhie et al. 2009 [15], and Heffron et al. 2011 [16]. To expand and update the current Salmonella–host interactome, here, we describe the screening of more than 2200 journal articles, followed by survey of the retrieved papers, and over 100 databases (Supporting Information Table 1). This resulted in a data set of 62 known Salmonella–host PPI, of which the previously reviewed 36 pairs are a subset (Table 1, Fig. 1). The new, expanded data set now includes 22 Salmonella effectors and 57 direct and three indirect interactions (SipC-Cyto-keratin-8, SipA-T-plastin, SsaB-Hook3). The resulting interactome of 62 pairs represents the most complete Salmonella–host interactome available to date and its functional relevance is described in subsequent sections. The details of the manual curation process and its conclusions are provided below.
Table 1.
Salmonella –host PPI data set
| Salmonella protein | Description | Host protein | Human gene symbol | Host organism | Experiment | Reference | Database listed |
|---|---|---|---|---|---|---|---|
| AvrA | Avirulence Protein A | p53 | TP53 | Eukaryotic cells | Immunoprecipitation | [109] | |
| ERK2 | MAPK1 | NA | Yeast 2-hybrid assay | [70] | |||
| MKK7a)b) | MAP2K7 | NA | Yeast 2-hybrid assay | [70] | |||
| MKK4a)b) | MAP2K4 | Mammalian | Acetylation | [69] | |||
| IκBα (ubiquitinated)a)b) | NFKBIA | Human | Deubiquitination | [71] | |||
| β-Catenin (ubiquitinated)a)b) | CTNNB1 | Human | Deubiquitination | [71] | |||
|
| |||||||
| SipA or SspA | Salmonella Invasion Protein A | F-actina)b)c) | ACTA1d) | Human, E. coli, rabbit, mouse | Pull-down assay, cosedimentation, immunofluorescence | [23, 25, 110] | UniProt |
| T-plastina)b)c)e) | PLS3 | Human | Yeast 2-hybrid screen, pull-down assay | [111] | |||
| Caspase-3 | CASP3 | Human | Caspase-3 cleavage assay, immunoblot | [67] | |||
|
| |||||||
| SipB | Salmonella Invasion Protein B | Caspase-1c) | CASP1 | Mouse, E. coli | Western blotting, pull-down assays, immunoblotting | [85, 112] | IntAct, UniProt |
|
| |||||||
| SipC or SspC | Salmonella Invasion Protein C | F-Actina)b)c) | ACTA1d) | Human | Pull-down assay, coimmunoprecipitation | [24] | UniProt |
| Cytokeratin 8c)e) | KRT8 | ||||||
| Cytokeratin 18c) | KRT18 | Human | Yeast-2 hybrid assay | [26, 37] | |||
| Exo70 | EXOC7 | Human | Yeast-2 hybrid assay, pull-down assay | [27] | |||
|
| |||||||
| SopA | Salmonella Outer Protein A | UbcH7 (UBE2L3; UBCE7) | UBE2L3 | Human | Pull-down assay, fluorescence polarization | [113] | dip |
| Caspase-3 | CASP3 | Mouse | Caspase-3 cleavage assay | [67] | |||
| HsRMA1a)b) | RNF5 | Human | Yeast 2-hybrid assay, pull-down assay | [61] | |||
|
| |||||||
| SopB or SigD | Salmonella Outer Protein B | Cdc24 | CDC24 | Saccharomyces cerevisiae | Pull-down assay | [114] | |
| Cdc42b) | CDC42 | Human | Pull-down assay, immunoprecipitation, colocalization | [114, 115] | |||
|
| |||||||
| SopE | Salmonella Outer Protein E | Cdc42a)b)c) | CDC42 | Human | Hela cDNA library screening, bacteriophage plaque-binding assay, Surface plasmon resonance analysis, X-ray crystallography | [19–21] | Intact, PIG, BIND, UniProt |
| Rac1a)b)c) | RAC1 | Human | Hela cDNA library screening, bacteriophage plaque-binding assay, Surface plasmon resonance analysis | [19, 20] | |||
| Rab5c) | RAB5A | Mouse | Pull-down assay, ELISA, competition ELISA | [30, 31, 116] | |||
|
| |||||||
| SopE2 | Salmonella Outer Protein E2 | Cdc42a)b)c) | CDC42 | NA | Surface plasmon resonance analysis, HSQC-NMR spectroscopy | [20, 22] | |
| Rac1c) | RAC1 | NA | Surface plasmon resonance analysis | [20] | |||
|
| |||||||
| SptP | Salmonella Protein Tyrosine Phosphatase | Rac1a)b)c) | RAC1 | Human | Pull-down assay, Gel-filtration, X-ray crystallography | [28, 117] | Intact, PIG, UniProt |
| VCPb) | VCP | Pig, human | Pull-down assay, immunoprecipitation | [59] | |||
| Vimentina)b) | VIM | Human | Yeast-2 hybrid, pull-down assay | [74] | |||
| Cdc42a)b) | CDC42 | Human, S. cerevisiae | Activation assay | [28, 29] | |||
|
| |||||||
| PipB2 | NA | Kinesin Light Chain (KLC)a)b)c) | KLC1 | Mouse, human | Yeast 2-hybrid assay, pull-down assay, immunoprecipitation | [40] | UniProt |
|
| |||||||
| SifA | Sifs A | SKIP (PLEKHM2; KIAA0842)a)b)c) | PLEKHM2 | Human | Yeast 2-hybrid assay, pull-down assay, immunoprecipitation, Copurification, isothermal titration calorimetry, colocalization studies, X-ray crystallography | [42, 43, 47] | Dip, BIND, PIG, UniProt |
| Rab7a)c) | RAB7A | NA | Pull-down assay | [32] | |||
| RhoA | RHOA | NA | Surface plasmon resonance analysis | [46] | |||
|
| |||||||
| SpiC or SsaB | Salmonella Pathogenecity Island Protein C | Hook3a)b)c)e) | HOOK3 | Mouse | Pull-down assay, coimmunoprecipitation (yeast-2-hybrid and pull down with recombinant Hook3 was not successful) | [35] | UniProt |
| TassCa)b) | NIPSNAP3A | Human, mouse | Yeast-2-hybrid, co-purification (in vitro and in vivo) | [34] | UniProt | ||
|
| |||||||
| SpvB | Salmonella Plasmid Virulence Protein B | G-Actina)b)c) | ACTA1d) | Mouse | Copurification, mass spectrometry | [57, 58] | |
|
| |||||||
| SseF | Secretion System Effector F | TIP60 acetyltransferase | KAT5 | Human | Yeast 2-hybrid screen, (pull-down assay performed but the results are not shown) | [60] | |
| Junction Plakoglobin | JUP | Human | SILAC (stable isotope labeling of amino acids in cell culture) | [38] | |||
|
| |||||||
| SseG | Secretion System Effector G | Desmoplakin | DSP | Human | SILAC | [38] | |
| Caprin-1 | CAPRIN1 | Human | SILAC | [38] | |||
|
| |||||||
| SseI or SrfH | Secretion System Effector I | TRIP6a)b)c) | TRIP6 | Human, mouse | Yeast 2-hybrid assay, fluorescence microscopy, localization studies, immunoprecipitation | [62, 63] could not reproduce interaction may be due to using a different Salmonella strain | UniProt |
| Filamin Aa)b)c) | FLNA | Mouse | Yeast 2-hybrid assay | [56] | |||
| IQGAP1b) | IQGAP1 | Mouse | Copurification, immunoprecipitation, colocalization studies | [63] | UniProt | ||
|
| |||||||
| SseJ | Secretion System Effector J | RhoA | RHOA | Human | Yeast-2-hybrid assay, immunoprecipitation, size-exclusion chromatography, Gel filtration analyses | [33, 47] | UniProt |
| RhoC | RHOC | Human | Yeast 2-hybrid assay SILAC | [38, 47] | |||
|
| |||||||
| SseL | Secretion System Effector L | Ubiquitin A, B and C (mono- and polyubiquitinated proteins)c) | UBA52 UBB UBC | Human | Yeast 2-hybrid assay, coimmunoprecipitation | [88] | |
| IκBαa)b) | NFKBIA | Human, mouse | Deubiquitination | [72] | |||
| Oxysterol-binding protein 1 | OSBP | Human | SILAC, coimmunoprecipitation | [38] | |||
| Talin-1 | TLN1 | Human | SILAC | [38] | |||
|
| |||||||
| SspH2 | Salmonella- Secreted Protein H2 | Filamin Aa)b)c) | FLNA | Mouse | Yeast 2-hybrid assay, colocalization | [56] | |
| Profilin-1a)b)c) | PFN1 | Mouse, human | Yeast 2-hybrid assay, pull-down assay | [56] | |||
| UbcH5-Ub conjugate | UBE2D1 | Human | Heteronuclear 2-D-NMR spectroscopy | [118] | |||
| Sgt1 | SUGT1 | Human | SILAC, coimmunoprecipitation | [38] | |||
| AIP | AIP | Human | SILAC | [38] | |||
| Bub3 | BUB3 | Human | SILAC, coimmunoprecipitation | [38] | |||
| 14-3-3γ | YWHAG | Human | SILAC | [38] | |||
| BAG regulator 2 | BAG2 | Human | SILAC | [38] | |||
|
| |||||||
| Slrp | Salmonella Leucine-Rich Repeat Protein | Thioredoxin 1a) | TXN | Human, mammalian | Yeast 2-hybrid screen, copurification, coimmunoprecipitation | [87] | UniProt |
| ERdj3 | DNAJB11 | Human | Yeast 2-hybrid screen, pull-down assay, coimmunoprecipitation, colocalization | [83] | |||
|
| |||||||
| SspH1 | Salmonella- secreted protein H1 | PKN1a)b)c) | PKN1 | Human | Yeast 2-hybrid screen, coimmunoprecipitation | [73] | UniProt |
|
| |||||||
| SpvC | MAPK1 (PRKM1; ERK2; PRKM2) | MAPK1 | Human | Crystallography | [76] | DIP | |
The Salmonella effectors are translocated by TTSS-1: SipA, SipB, SipC, SipD, SopA, SopB, SopD, SopE, SopE2 TTSS-1 and -2: AvrA, GtgE, PagJ, SlrP, SpvC, SpvD, SptP, SseK1, SspH1, SteA, SteB TTSS-2: CigR, GogB, GtgA, PipB, PipB2, SifA, SifB, SopD2, SpiC, SpvB, SseB, SseC, SseD, SseF, SseG, SseI, SseJ, SseK2, SseK3, SseL, SspH2, SteC, SteD. Salmonella effectors where no protein-binding partner is known and that are consequently not listed here are given in bold face.
Reviewed in [15].
Reviewed by Heffron et al. 2011 [16].
Reviewed in [14].
For the network analysis, ACTA2, ACTB, ACTC1, ACTG1, and ACTG2 were also included in addition to ACTA1.
Indirect. SipA and T-plastin complex is mediated by F-actin; this complex cannot be formed in the absence of actin; Cytokeratin-18 bind to Cytokeratin-8; SpiC–Hook3 interaction may be indirect as speculated by [35].
Figure 1.
Current Salmonella–host interactome. The image was created using cytoscape software (www.cytoscape.org). Blue, Salmonella proteins; red, host proteins.
The literature search initially utilized PubMed’s Mesh database to search abstracts using multiple keywords (e.g. Salmonella effector+bind/degrade/cleave). However, a parallel basic search in PubMed revealed that the Mesh database was missing numerous relevant publications. We therefore entered individual Salmonella effector protein gene names and abbreviation (e.g. SopE) into the search field of PubMed and manually inspected the resulting publications. Each article was deemed relevant based on its title and abstract; the full-text article was retrieved and was searched for the occurrence of specific keywords such as “bind,” “interacts,” “directly interacts,” “cleaves,” and the effector’s name/abbreviation. Relevant articles were stored using the PMID reference code. This was repeated for all Salmonella effector proteins. In total, more than 2200 articles were screened. All papers that passed this first stage were then scrutinized for evidence supporting a direct interaction by thoroughly examining results and methodology. For papers passing this stage, the following data were extracted from the article to be incorporated into the overall data set: experimental method(s), strain of Salmonella and host organism studied. If multiple journal articles confirmed the same result, these findings were also extracted for incorporation into the data set.
The database search began with a Google search for “protein–protein interaction databases” which retrieved the “Jena Protein-Protein Interaction Website (PPI): Databases,” listing 112 databases and data collection tools for searching PPI (http://ppi.fli-leibniz.de/jcb_ppi_databases.html [last update of website: January 5, 2011]). Cross-comparison with other relevant hits from the Google search indicated that this listing is comprehensive and we tested each of the databases listed. The results of this database search are summarized in Supporting Information Table 1. In addition, “PSICQUIC VIEW” was used to search several databases including intact, dip, bind, string, and apid simultaneously (http://www.ebi.ac.uk/Tools/webservices/psicquic/view/main.xhtml). The UniProt database (http://www.uniprot.org/) was also used extensively since it not only includes links to IntAct’s “binary interactions” fields but also provides functional information and links. The database most directly related to pathogen–host interactions was the PIG database (http://molvis.vbi.vt.edu/pig/) but is restricted to Human interspecies interactions, and retrieves the same Salmonella–host interactions as when using PSICQUIC.
The exhaustive search of both publications and databases revealed an important conclusion related to the rate-limiting steps in developing pathogen–host interactomes: listing interactions in the databases and not integrating different databases is the current bottleneck in obtaining pathogen–-host interactomes, as demonstrated here for Salmonella. Only 16 of the 62 interactions gathered through the literature search can also be found in PPI databases. Furthermore, of these 16, only 6 are listed in the databases DIP, IntAct, PIG, and/or BIND and are thus retrievable in an automated fashion. In contrast, the other ten are merely found in the descriptions of the UniProt database (www.uniprot.org). This demonstrates the need for manual curation of thousands of articles to obtain a reliable interactome for Salmonella. This is significant because it is unlike the intraspecies interactions e.g. between all human proteins or all yeast proteins. Therefore, integration of information listed in different databases is essential because each database has a different coverage and there is sufficient diversity across databases. For example, the BIANA platform provides one such capability to retrieve interactions across many relevant databases through a single interface [17]. The only pathogen for which listing of interactions in databases is not the rate-limiting step is HIV-1, for which thousands of PPI are listed in several databases, most comprehensively in the HIV-1, Human Protein Interaction Database (http://www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions/index.html).
The gathering of reported PPI is an essential step in computer-assisted PPI prediction for training and for validation, representing the “gold standard” from which the algorithms learn to distinguish interacting from noninteracting protein pairs. Incorporating these literature-curated PPI in computational methods requires further processing, since the publications reporting the experimental work usually do not use standard identifiers and the genes derived are therefore often ambiguous. Many computational techniques (such as the network analysis described in Section 4) use information from various databases and to associate any properties from these to an interaction involving such genes will not be possible without an appropriate identifier. We therefore mapped the host proteins into gene symbols through the Entrez Gene mapping tool. Further problems arise while converting gene-level interactions to protein-level interactions since a single gene may have multiple gene products. We used the most popular protein symbol. Finally, not all of the interactions reported in the literature were with human proteins. Therefore, Table 1 lists the respective organism from which the host protein was derived. For simplicity, we mapped the protein names to the respective human gene symbols (column 4 in Table 1). The data set obtained based on the literature retrieved interactions and removing ambiguities in identifiers as much as possible using only human gene symbols and human protein symbols is available in excel format in Supporting Information 2 and in csv format at www.shiprec.org under the “Technologies” tab, direct link: http://www.shiprec.org/indicationslist.htm.
3 Specific impacts of Salmonella–host PPIs at different stages of Salmonella infection
Below, we propose a brief overview of the functional roles of the interactions in the gold standard.
3.1 Salmonella invasion of host cells
Upon contact with the host cell Salmonella inserts the SPI-1-encoded TTSS into the host cell membrane to translocate effectors into the cytosol of the targeted cell. Focusing on this secretion system, several effectors are necessary to promote Salmonella uptake by modifying the actin cytoskeleton, membrane ruffling, and bacterial engulfment. SipA, SopA, SopB, SopD, and SopE2 play a role in the process of Salmonella invasion of epithelial cells [18]. SopE and SopE2 are guanine nucleotide exchange factors (GEFs) which activate small Rho GTPases [19]. Both effectors bind and activate Cdc42 with SopE2 being a more efficient GEF for Cdc42 than SopE. Only SopE has been shown to efficiently activate Rac1 although a weak binding of SopE2 to Rac1 was observed [20–22]. The activation of these GTPases stimulates actin polymerization. Two other effectors, SipA and SipC, which directly bind F-actin are involved in Salmonella internalization [23, 24]. The C-terminal domain of SipC harbors the F-actin binding site, nucleates actin polymerization, and bundles F-actin. It has been shown that SipC is necessary for Salmonella invasion, whereas SipA enhances its nucleation and bundling efficiency [25]. It is supposed that SipA achieves localization of membrane ruffles thereby restricting them to the site of Salmonella–cell contact [23]. SipC binds to the filament protein Cytokeratin-18 as well as Exo70, a component of the exocyst complex [26, 27], resulting in activation of the Arp2/3 complex to stimulate actin polymerization and recruitment of exocytic vesicles to the sites of bacterial internalization possibly to supply membranes for macropinocytosis.
After internalization of Salmonella, SptP functions to reverse the process of membrane ruffling in order to establish normal cytoskeleton arrangement. The N-terminal domain of SptP possesses GAP activity and thereby inactivates Rac1 and Cdc42, resulting in the downregulation of signaling through those GTPases [28, 29].
3.2 The SCV, Sif formation, and Salmonella replication
After engulfment of bacteria within the SVC, it has been reported that the SCV acquires several endocytic markers. One of the early markers is the GTPase Rab5 which is recruited by SopE in the GTP-bound form and is required for the fusion of the SCV membrane with early endosomes [30, 31]. The SCV membrane is enriched with the GTPase Rab7 which participates in maturation of phagosomes and is involved in late endocytic transport. Rab7 interacts with RILP which can associate with the minus-end-directed dynein–dynactin motor complex, leading to the transport of the vesicle along microtubules toward the cell center. SifA may uncouple Rab7 from RILP which may facilitate the extension of tubules from the SCV [32]. Moreover, the SCV membrane is modified by SseJ activity leading to esterification of membrane cholesterol. This process is stimulated by the interaction of SseJ with the GTPase RhoA [33]. In order to prevent exposure to microbicidal compounds, Salmonella is able to prevent the fusion of SCVs with lysosomes. This may be achieved by the binding of SipC to TassC [34] and the inactivation of Hook3 by this Salmonella effector [35], followed by inhibiting the recruitment of the dynein motor complex (SifA–Rab7 interaction) [36]. The establishment of an intermediate filament network comprising Cytokeratin, Vimentin, and the adaptor protein 14-3-3 around the SCV is necessary to ensure localization of the SCV close to the nucleus [37]. SipC binds Cytokeratin-18 directly and Cytokeratin-8 indirectly [26] and SspH2 binds 14-3-3γ [38], but the precise functions of these interactions remain unclear.
Upon localization of the SCV close to the nucleus and the Golgi in epithelial cells [39], Salmonella replicates and Sifs are formed. Although there is evidence that Salmonella interacts with the Golgi in macrophages as well, an association of the SCV with the Golgi could not be observed in macrophages [39]. Sifs are membranous tubular structures extending from the SCV along microtubules to the cell periphery. The Sifs are thought to play a role in nutrient acquisition, movement of bacteria from cell to cell and may provide Salmonella with more space for replication. Upon extension of tubules, vesicles are observed at the cell periphery. Several effectors are thought to contribute to tubulation of the SCV membrane, the extension of the Sifs, and location of endocytic compartments to the cell periphery. Among those effectors, SifA and PipB2 take on a central role. PipB2 binds KLC, a subunit of the plus end-directed kinesin-1 microtubule motor complex that drives transport of cargo to the cell periphery, and thereby recruits kinesin-1 to the SCV membrane [40]. SifA, which is anchored to the SCV membrane by its C-terminal prenylated CaaX motif [41], binds SKIP [42, 43]. SKIP itself interacts with KLC [42]. Recent data suggest that SKIP does not downregulate the recruitment of kinesin-1 to the SCV as believed previously [42] but instead activates kinesin-1 and leads to its expulsion from vesicles [44]. SifA antagonizes Rab9 for binding SKIP and the fact that SifA-SKIP binding is tighter than Rab9-SKIP binding thus may enable SifA to recruit SKIP to the SCV membrane [45]. The interaction of the N-terminal domain of SifA with the C-terminal PH-domain of SKIP requires residues W197 and E201 of SifA’s WxxxE motif and SifA may thus mimic the mammalian GTPases [45]. The ability of SifA to bind Rab7 contributes to the formation of tubules.
Another activity of SifA is to interact with the GDPbound form of RhoA. Although this interaction does not trigger nucleotide exchange [46], SifA seems to promote RhoA-family GTPase signaling pathways on the phagosome membrane [47]. Moreover, the interaction of SseJ, which localizes to the SCV membrane [48] with GTP-bound RhoA and RhoC, is speculated to induce tubulation of the SCV membrane [38, 47]. Furthermore, it has been shown that SNX3 (sorting nexin 3) is essential for the formation of tubules and that SopB-mediated recruitment of Rab5 to the SCV is required for SNX3 tubulation. SNX3 tubules are formed within 30–60 min postinfection and go along with the recruitment of LAMP1 and Rab7 which is impaired when SXN3 is depleted [49]. It is very likely that so far unknown factors contribute to endosomal tubulation as well.
In epithelial cells, SseG and SseF play a role in keeping the SCV in the Golgi region [39, 50]. However, it is unclear how the interaction of SseG and SseF with Desmoplakin, Caprin-1, TIP60, and junction plakoglobin relate to this function. Rather, the SseF–TIP60 interaction appears to play a role in Salmonella replication in macrophages (see below). Moreover, TIP60 is generally known to be a multifunctional enzyme involved in diverse processes, including cell cycle, apoptosis, signaling, and DNA repair [51]. Caprin-1 has been shown to have a role in cell proliferation as the suppression of Caprin-1 expression resulted in the prolongation of the cell cycle [52]. Thus, it is unlikely that the interaction of SseG with Caprin-1 can be connected with SseG’s role in SCV positioning. Finally, SeeG interacts with Desmoplakin and SseF with junction plakoglobin [38]. Both host proteins are part of the desmosome, with a major role in cell–cell adhesion [53]. Thus, SseG and SseF may influence processes associated with the desmosome and are likely to have multiple functions in Salmonella pathogenicity.
An actin network is built around the SCV [54] referred to as vacuole-associated actin polymerizations (VAPs). SteC has Raf-like activity and was shown to be essential for the establishment of VAP and F-actin remodeling [55]. A number of effectors seem to help in inhibiting the formation of VAP. Both SseI and SspH2 bind Filamin A through their N-terminus and thereby preventing the crosslinking of F-actin by Filamin dimers [56]. The C-terminal domain of SspH2 interacts with Profilin-1 and thus prevents the interaction of Profilin-1 with G-actin in order to inhibit polymerization [56]. The C-terminal ADP-ribosyltransferase domain of SpvB modifies G-actin at residue Arg177 through which actin polymerization is inhibited, most likely by a steric mechanism. Activity of SpvB leads to reduced VAP formation as well as disruption of the cytoskeleton and apoptosis [57, 58].
Besides reversing the cytoskeleton to its normal shape after Salmonella entry, SptP dephosphorylates vasolin-containing protein (VCP), an AAA ATPase family protein, by its C-terminal tyrosine phosphatase activity. Dephosphorylation of VCP enhances replication of bacteria in the SCV [59]. Another effector that contributes to Salmonella replication is SseF which binds to TIP60 and thereby enhances the histone acetyltransferase activity of TIP60. Knockdown of TIP60 expression in macrophages resulted in a reduced replication of Salmonella [60]. Interaction of SopA with HsRMA1 results in ubiquitination of SopA [61]. Monoubiquitinated SopA either directly or in cooperation with an unknown host factor facilitates Salmonella escape from SCVs into the cytoplasm of host cells [61]. Polyubiquitinated SopA is degraded by the proteasome.
3.3 Systemic infection
Salmonella can spread within its host leaving the intestine moving via the bloodstream or the lymphatic vessels to other organs like the spleen and the liver. The only effector that is known to contribute to this process is SseI although it has been shown that SseI alone is not the only Salmonella protein responsible for systemic infection. SseI-deficient Salmonella can still but to a lesser extent spread through the bloodstream and mutation of the TTSS-2 completely abolished Salmonella ability to spread systemically. Interaction of SseI with the LIM-domain of TRIP6 enhances dissemination of CD18+Salmonella-containing cells most likely by blocking TRIP6 in inhibiting cell motility through interaction with the Rac pathway [62]. IQGAP1 is another protein that is bound by SseI. This interaction facilitates the maintenance of chronic systemic infection by inhibiting migration of dendritic cells to the sites of infection thus keeping the host from clearing Salmonella from systemic sites of infection [63].
3.4 Activation and regulation of immune response
Salmonella manipulates and evades the host innate and adaptive immune response by a diverse set of mechanisms involving several Salmonella effectors and affecting inflammatory response, antigen (AG) presentation and T cells. Although at least six effectors that are exclusively translocated via TTSS-1 trigger inflammatory responses, effectors that require TTSS-2 or TTSS-1 and -2 for their translocation function to dampen the innate and adaptive immune response. It may be assumed that Salmonella first attracts polymorphonuclear neutrophils (PMNs) which the bacteria invade and then by manipulation of the host immune system escapes host defense and uses host cells for systemic spread in order to manifest a chronic or long-term infection [64].
Activation of the inflammatory response occurs through several mechanisms. The GEF activity of SopE and SopE2 causes exchange of GDP by GTP in Cdc42 and thereby activates this small GTPase [19, 65]. This leads to the activation of MAPK pathways, which in turn activate transcription factors resulting in the production of cytokines like IL-8 [19, 66]. Consequently, PMNs are attracted to the site of infection. Other Salmonella effectors that induce PMN migration and activate inflammatory response are SipA and SopA. SipA is cleaved into its two functional domains by host caspase-3. The C-terminal domain of SipA directly binds F-actin (see above) and the N-terminal domain is responsible for inducing PMN migration across the intestinal epithelium by triggering MAPK pathways leading to cytokine secretion [67]. SopA, a HECT-like E3 ubiquitin ligase, has been shown to induce PMN migration. This function has been attributed to its ubiquitin ligase activity [68]. Moreover, SopA contains two functional caspase-3 recognition motifs which are in close proximity to each other [67]. Mutation of both motifs renders SopA insensitive to cleavage by caspase-3 and leads to a reduced induction of PMN migration [67]. Beside the functional caspase-3 cleavage sites of SipA and SopA, other Salmonella effectors that comprise putative caspase-3 recognition motifs are AvrA, SopB, SifA, SopD, SptP, and SpvB [67]. Thus, Salmonella may exploit the host to cleave effector proteins into functional domains necessary for their activity.
Downregulation and inhibition of immune response is accomplished by Salmonella targeting several host proteins to inhibit MAPK signaling and/or activation of NF-kB using the effectors AvrA, SseL, SspH1, SptP, SseI, and SpvC. AvrA causes inhibition of JNK and NF-kB signaling through acetylation of MKK4 and MKK7 [69, 70] and inhibits NF-kB activation by deubiquitination of IkBa [71]. In cells, IkBa binds NF-kB and thereby inhibits this transcription factor. Ubiquitination of IkBa results in its degradation by the host proteasome and an increase of unbound active NF-kB [71]. Therefore, deubiquitination of IkBa by AvrA leads to increased levels of this NF-kB inhibitor protein. The function of SseL in interacting with IkBa is assumed to be the same as for AvrA [72]. Another host protein that has been recently identified to be targeted by SseL is OSBP1 [38]. The LLR-domain of the E3 ubiquitin ligase SspH1 interacts with PKN1. It is proposed that due to this interaction NF-kB is inhibited [73]. As described above, SptP resembles GAP activity which leads to the inactivation of Cdc42 and Rac1. Thus, SptP downregulates MAPK signaling. Moreover, the tyrosine phosphatase activity of SptP is involved in reversing MAPK activation [74]. Both the GAP and the tyrosine phosphatase activity of SptP lead to inhibition of Raf-induced ERK activation [75]. SseI blocks NF-kB signaling through a novel mechanism involving at least Nod1 and Nod2 [16]. SpvC interacts with a phosphorylated MAPK1-derived peptide that contains the TXY motif whose phosphorylation at T and Y is necessary for kinase activity [76, 77]. Due to its phosphothreonine lysase activity, SpvC inactivates the MAP kinases ERK and p38 thereby inhibiting MAPK signaling downstream of p38 and MAPK1 [78]. Moreover, a Salmonella strain overexpressing SpvC has been shown to reduce cytokine release from infected cells [77].
Interference with AG presentation by dendritic cells through MHC-I and MHC-2 molecules is proposed to be caused by the TTSS-2 translocated effectors SifA, SspH2, SlrP, PipB2, SopD2 and to a lesser extent by SseF and SseG. This activity allows Salmonella to suppress AG-dependent T-cell proliferation [79–81]. AG presentation to T cells is reduced by either direct or indirect ubiquitination of MHC-II by Salmonella effectors [82]. The only Salmonella–host interaction that is known so far and could contribute to the inhibition of AG-presentation is between SlrP and ERdj3. This interaction may cause the accumulation of unfolded proteins in the ER and thereby disturb AG presentation by MHC-I [83, 84].
3.5 Induction and inhibition of apoptosis
Salmonella both dampens and activates apoptosis of host cells. Induction of cell death may allow bacteria to evade host cells in order to infect new cells and the killing of cells may aim at reducing the host immune response. On the other hand, the prolongation of cell survival may provide Salmonella with a safe environment for replication, and permit systemic spread and manipulation of host defense. The following mechanisms are at play. SipB induces apoptosis of macrophages through its interaction with caspase-1. Interaction results in caspase-1 activation which induces apoptosis. Moreover, activation of caspase-1 leads to maturation of IL-1β, a caspase-1 substrate known to be an endogenous pyrogen, and thereby may contribute to the inflammatory response [85]. SipB has also been shown to induce apoptosis in a caspase-1 independent way which involves caspase-2, -3, -6, -8 and mitochondrial cytochrome C [86]. Another effector, SlrP, is thought to promote cell death by interacting with Thioredoxin 1 in the cytosol and the chaperone ERdj3 in the ER. SlrP-mediated ubiquitination of Thioredoxin 1 could cause a reduced redox activity of the target protein and its degradation [87]. Interaction with ERdj3 may lead to the accumulation of unfolded proteins in the ER [83]. The ADP-ribosyltransferase activity of SpvB may additionally contribute to cell death through disruption of cytoskeletal structures (see above) [58]. SseL plays a role in Salmonella-induced cytotoxicity in macrophages [88]. On the other hand, AvrA targeting MKK4 and MKK7 inhibits JNK signaling and thus slows down apoptosis [69, 70]. The proposed SpvC-mediated inhibition of p38 may have the same effect (see above).
4 Pathway and subnetwork analysis of host proteins
Next, we performed pathway and network analyses with the human genes implicated directly or by inference from other hosts (such as mouse) in interactions with Salmonella proteins using two complementary approaches.
In the first approach, pathway enrichment was calculated based on multiple pathway data sources using software GSEA (http://www.broadinstitute.org/gsea/index.jsp) on July 21, 2011 (reference: http://www.ncbi.nlm.nih.gov/pubmed/16199517). The software integrates several pathway sources to perform gene set enrichment analysis, which provides comprehensive insight on the enriched pathways in our gene set. These data sources include KEGG (http://www.genome.jp/kegg/), BioCarta (http://www.biocarta.-com), Reactome (http://www.reactome.org/), Signal transduction knowledge environment (ST, http://stke.sciencemag.org/) and Signaling gateway (SIG, http://www.grt.kyushu-u.ac.jp/spad/menu.html). The pathways with P-value less than 1×10−4 are summarized in Table 2. As expected from the large number of interactions involving actin and actin regulatory proteins, the pathways “Genes related to regulation of the actin cytoskeleton” and “Regulation of actin cytoskeleton” are among the most significantly targeted pathway. The analysis further revealed interference of Salmonella especially with pathways related to cell shape, cell growth, and interactions with other cells. This includes, for example, the pathways, Adherens junction, Neurotrophin signaling pathway, Focal adhesion, Genes involved in Sema4D-induced cell migration and growth-cone collapse, Genes involved in Apoptotic cleavage of cell adhesion proteins, Agrin in Postsynaptic Differentiation, Genes involved in Axon guidance, Integrin Signaling Pathway, Calpain and friends in Cell spread and mCalpain and friends in Cell motility. Numerous pathways are involved in immune response to pathogens, such as Pathogenic Escherichia coli infection, Genes involved in TRAF6-Mediated Induction of the antiviral cytokine IFN-αη cascade, Genes involved in Toll-Like Receptor 3 (TLR3) Cascade, How does Salmonella hijack a cell, Epithelial cell signaling in Helicobacter pylori infection, Genes involved in Toll Receptor Cascades, Toll-like receptor signaling pathway, and HIV-I Nef: negative effector of Fas and TNF. The occurrence of different signaling pathways related to pathogenic processes of various species supports the notion that there are conserved mechanisms of communication, even for pathogens as diverse as viruses and bacteria. The remaining over-represented pathways are frequently generic, such as general signaling mechanisms, such as Pathways in cancer, Ras Signaling Pathway, Genes involved in G α (12/13) signaling events and MAPK signaling pathway. Particularly interesting is the retrieval of Phospholipids as signaling intermediaries because numerous lipidation events are known to modulate Salmonella interactions with hosts [89] and phosphoinositide signaling plays an important role in Salmonella invasion [90].
Table 2.
Pathways overrepresented in the 54 proteins based on multiple data sources
| Description | P-value | Data source |
|---|---|---|
| Adherens junction | 5.52×10−8 | KEGG |
| Role of MAL in Rho-Mediated Activation of SRF | 1.88×10−7 | Biocarta |
| Fas Signaling Pathway | 1.91×10−7 | ST |
| Neurotrophin signaling pathway | 3.10×10−6 | KEGG |
| Pathogenic E. coli infection | 3.88 × 10−6 | KEGG |
| Genes related to regulation of the actin cytoskeleton | 4.80×10−6 | SIG |
| Colorectal cancer | 5.20×10−6 | KEGG |
| Calpain and friends in Cell spread | 7.32×10−6 | BIOCARTA |
| Focal adhesion | 1.26×10−5 | REACTOME |
| Erk and PI-3 Kinase Are Necessary for Collagen Binding in Corneal Epithelia | 2.46×10−5 | Biocarta |
| Genes involved in Sema4Dinduced cell migration and growth-cone collapse | 2.46×10−5 | REACTOME |
| Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway | 2.91×10−5 | ST |
| Influence of Ras and Rho proteins on G1 to S Transition | 3.43×10−5 | Biocarta |
| Amyotrophic lateral sclerosis (ALS) | 3.84×10−5 | KEGG |
| Genes involved in TRAF6-Mediated Induction of the antiviral cytokine IFN-αη cascade | 3.84×10−5 | REACTOME |
| Genes involved in Sema4D in semaphoring signaling | 5.35×10−5 | REACTOME |
| Genes involved in Apoptotic cleavage of cell adhesion proteins | 5.91×10−5 | REACTOME |
| Genes involved in Toll-Like Receptor 3 (TLR3) Cascade | 6.49×10−5 | REACTOME |
| NOD-like receptor signaling pathway | 8.25×10−5 | KEGG |
| Genes related to CD40 signaling | 9.03×10−5 | SIG |
| How does salmonella hijack a cell | 1.01×10−4 | Biocarta |
| Genes involved in Semaphorin interactions | 1.11×10−4 | REACTOME |
| Agrin in Postsynaptic Differentiation | 1.28×10−4 | Biocarta |
| Epithelial cell signaling in H. pylori infection | 1.29×10−4 | KEGG |
| Regulation of actin cytoskeleton | 1.54×10−4 | KEGG |
| Genes involved in Axon guidance | 1.55×10−4 | REACTOME |
| Integrin Signaling Pathway | 1.58×10−4 | Biocarta |
| JNK MAPK Pathway | 1.58×10−4 | ST |
| Role of PI3K subunit p85 in regulation of Actin Organization and Cell Migration | 1.96×10−4 | Biocarta |
| Leukocyte transendothelial migration | 2.04×10−4 | KEGG |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 2.18×10−4 | KEGG |
| Integrin Signaling Pathway | 2.46×10−4 | ST |
| Genes involved in p75 NTR receptor-mediated signaling | 3.11×10−4 | REACTOME |
| Keratinocyte Differentiation | 3.36×10−4 | Biocarta |
| Genes involved in Toll Receptor Cascades | 3.89×10−4 | Biocarta |
| MAPKinase Signaling Pathway | 4.10×10−4 | Biocarta |
| Trefoil Factors Initiate Mucosal Healing | 4.52×10−4 | Biocarta |
| Apoptotic Signaling in Response to DNA Damage | 5.21×10−4 | Biocarta |
| Pathways in cancer | 5.47×10−4 | KEGG |
| Ras Signaling Pathway | 5.96×10−4 | Biocarta |
| Genes involved in G α (12/13) signaling events | 6.23×10−4 | REACTOME |
| MAPK signaling pathway | 6.48×10−4 | KEGG |
| Genes involved in Further platelet releasate | 6.78×10−4 | REACTOME |
| Genes involved in Smooth Muscle Contraction | 6.78×10−4 | REACTOME |
| mCalpain and friends in Cell motility | 7.66×10−4 | Biocarta |
| HIV-I Nef: Negative effector of Fas and TNF | 8.18×10−4 | Biocarta |
| Toll-like receptor signaling pathway | 8.51×10−4 | KEGG |
| Phospholipids as signaling intermediaries | 9.65×10−4 | Biocarta |
In a second approach, Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com/) was carried out. For a given canonical signaling pathway in IPA, Fisher’s exact test was performed to measure the probability (P-value) that the pathway is randomly selected. To control the error rate, the P-values were corrected by Benjamini–Hochberg method [91] and those pathways whose P-values were less than 1.0×10−5 (or score >5, here score=−log P) were defined as the significant pathways. The results are summarized in Supporting Information Table 3. A mapping of the human genes involved in the respective pathways, together with the Salmonella-human PPI targeting them, is provided in Supporting Information Table 4. This analysis supports the conclusion that current host targets of Salmonella effectors affect numerous ubiquitous signaling pathways through interactions with hub proteins involved in crosstalk among different pathways.
Table 3.
Subnetworks over-represented in the 54 proteins
| ID | Molecules in subnetwork | Scorea) | Number of focus moleculesb) | Top functions |
|---|---|---|---|---|
| 1 | ACTA1c), ACTA2, ACTB, ACTC1, ACTG1, ACTG2, Actin, Alpha actin, Alpha Actinin, Alpha catenin, atypical protein kinase C, Cadherin, CASP1, CASP3, ERK1/2, Fgf, G-Actin, Hsp27, Hsp90, Ige, JUP, KAT5, OSBP, PFN1, Phosphatidylinositol4,5 kinase, Pld, PP2A, Profilin, RAB9A, Ras homolog, Rho gdi, RHOC, RhoGap, Rock, Rsk | 29 | 14 | Cell Death, Cellular Assembly and Organization, Skeletal and Muscular System Development and Function |
| 2 | 14-3-3, Alcohol group acceptor phosphotransferase, BAG5, BCR, Caspase, CD3, Cdc2, CDC42, DSP, Ephb, EXOC7, F Actin, Fcer1, Filamin, FLNA, IQGAP, IQGAP1, KRT8, KRT18, Laminin, MAP2K4, MAP2K7, MAP2K4/7, MAP3K, Mek, NFkB (complex), Pak, Pkg, PKN1, Raf, Rap1, Sapk, TCR, TXN, VAV | 24 | 12 | Cellular Assembly and Organization, Gastrointestinal Disease, Genetic Disorder |
| 3 | 26s Proteasome, AIP, Akt, APC/APC2, Calmodulin, Cbp/p300, Ck2, CTNNB1, DNAJB11, FSH, Gpcr, hCG, Histone h3, Histone h4, Hsp70, Ifn gamma, IgG, Ikb, IL12 (complex), Insulin, Interferon alpha, Jnk, Lh, Pka, PLC, PLS3, RNA polymerase II, TP53, UBB, UBC, UBE2D1, UBE2L3, Ubiquitin, VCP, YWHAG | 20 | 11 | Cancer, Cellular Development, Cellular Growth and Proliferation |
| 4 | AMPK, Ap1, Calpain, Collagen type I, Collagen type IV, ERK, Estrogen Receptor, Fibrinogen, Focal adhesion kinase, G protein beta gamma, Gef, IFN Beta, IL1, Integrin, LDL, Mapk, MAPK1, NFKBIA, P38 MAPK, p85 (pik3r), Pdgf (complex), PDGF BB, PI3 K (complex), Pkc(s), RAB5A, RAB7A, Rac, RAC1, Ras, RHOA, STAT5a/b, Tgf beta, TLN1, TRIP6, Vegf | 14 | 8 | Cardiovascular System Development and Function, Cell-To-Cell Signaling and Interaction, Cell Signaling |
| 5 | AFAP1L2, AGT, AKIRIN2, ARL6, ARL4C, BUB3, C11orf1, CAPRIN1, CES2, COX7A2, GIPC2, HNF4A, HNRNPA0, HOOK3, IL6, IRF6, LPP, MRPL44, NIPSNAP3A, POLR3G, PRCP, PTK2, PTK7, REG1A, SBNO2, SETDB1, SLC14A2, SLC16A6, SUGT1, TDO2, TXNDC9, UBA52, UTP3, XPNPEP2, ZFP64 | 10 | 6 | Increased Levels of CRPd), Cell Morphology, Cellular Development |
Scores were log-transformed by the P-values, which were calculated by Fisher’s exact test.
The number of focus molecules is the number of the genes with the bold font.
The genes with the bold front are these genes existed in our gene set.
CRP is abbreviated for c-reactive protein.
The IPA analysis was then used to investigate subnet-work enrichment in the resulting global network. To this end, the genes of interest were first overlaid onto a global molecular network developed based on the Ingenuity Pathways Knowledge Base and then subnetworks for these genes were extracted from the global network based on their connectivity using the algorithm developed by IPA (http://www.ncbi.nlm.nih.gov/pubmed/16136080). Similar to pathway analysis in IPA, for each subnetwork, IPA computed a score based on the P-value calculated by Fisher’s exact test and the P-value indicated the likelihood of the genes in the subnetwork were found by chance. Additionally, the top three biological functions according to the IPA score were assigned to each network. We identified the enriched subnetworks in our gene set if a given subnetwork has one score higher than 5. The results are summarized in Table 3. A mapping of the Salmonella proteins involved in interactions with the proteins in these subnetworks is provided in Supporting Information Table 5. Five subnetworks were identified. It is particularly interesting to examine the topranked functional terms associated with these networks: (i) Cell Death, Cellular Assembly and Organization, Skeletal and Muscular System Development and Function, (ii) Cellular Assembly and Organization, Gastrointestinal Disease, Genetic Disorder, (iii) Cancer, Cellular Development, Cellular Growth and Proliferation, (iv) Cardiovascular System Development and Function, Cell-To-Cell Signaling and Interaction, Cell Signaling, and (v) Increased Levels of CRP, Cell Morphology, Cellular Development.
Subnetwork 1 demonstrates the impact Salmonella has on “cellular assembly and organization” and “cell death.” Much of the actin skeleton modulations are related to this, in addition to direct induction or suppression of apoptosis, described in Section 3, above. This includes, in particular, the roles of SipA and SipC in actin polymerization and bundling, the proposed role of SseJ to induce SCV tubulation through interacting with RhoC, the function of SpvB to disrupt the cytoskeleton and promote apoptosis, the fact that the interaction of SspH2 with profilin inhibits actin polymerization and that the SipB–caspase-1 interaction induces apoptosis. The interactions between SseF and JUP and between SseL and OSBP may fit into this subnetwork by influencing cytoskeletal rearrangement and membrane dynamics. This is supported by the finding that JUP is part of the desmosome [53] and the manipulation of immune response is a proposed role for OSBP based on its involvement in MAPK signaling [92]. Subnetwork 2 represents the disruptive nature of the epithelial cells being infected with Salmonella. Specifically, there are several host proteins that have been shown or proposed to have a role in Salmonella invasion and internalization when interacting with the identified Salmonella protein-binding partner. These are Cytokeratin-8 and -18, Exo70, F-actin, and Cdc42 with the Salmonella proteins SipC, SopB, SopE, and SopE2. The interaction of SptP with Cdc42 also fits into this subnetwork because it results in the reversal of membrane ruffles after bacterial internalization. As the interaction of SseI and SspH2 with Filamin A is thought to prevent VAP formation, this can be categorized under the same top function, namely “cellular assembly and organization” as the interactions for subnetwork 2 listed above. The other focus host proteins within subnetwork 2 most likely fall into the function “gastrointestinal disease” based on their roles in MAPK signaling, inhibition of immune response, and cell death. The function of the interaction between SseG and DSP may be the same as for the interaction between SseF and JUP listed in subnetwork 1.
The general aspects of cell growth that also come out in the numerous generic pathways targeted, including those involved in cancer, are represented by subnetwork 3. Subnetworks 4 and 5 are related to the cell morphology and interactions aspect that is also observed by the very large number of related pathways being targeted. Presumably, much of Salmonella survival is related to the turnover of the infected cell and how loosely it is embedded in its native tissue. Cells that shed frequently and are degraded rapidly will automatically remove Salmonella-infected cells, limiting the establishment of an infection. On the other hand, it may be that rapid degradation of Salmonella-containing cells may conversely also contribute to a higher rate of infection. Bacteria inside cells are spread within the host and their release from the cells enables Salmonella to infect new cells.
5 Significance of the data set
This review provides the most complete Salmonella–host interactome to date. It stratified several previously reviewed interactions as indirect and added numerous new interactions, so that the current set of interactions includes 62 pairs. These pairs were derived from careful analysis of the primary literature and we consider the labels therefore of high quality. Although the primary literature and experimental evidence given for every PPI listed here has been carefully examined, the level of confidence, that a reported interaction is true, varies depending on the nature of the performed experiments as well as the amount of evidence given. To assist the reader in making their own judgments, the experiments done supporting an interaction are also listed in Table 1. It is important to note, however, that there is inherent bias in the choice of experiments and proteins investigated. Thus, identification of novel PPI focuses often on the proteins that are already known or thought to be interesting key molecules in pathogenicity, both on the Salmonella and on the host side. For example, most studies to date have focused on Salmonella proteins translocated via the bacterial TTSS-1 and TTSS-2 and many host proteins are those that are known key signaling molecules. A systematic high-throughput investigation of Salmonella–host PPI has not yet been carried to our knowledge. Moreover, any experimental approach depends on the nature of the protein itself. Thus, most data available for Salmonella are for proteins which are comparatively easy to handle, e.g. soluble proteins, as opposed to membrane proteins.
Despite this bias, the data set of known Salmonella–host PPI presented here is the most complete set of interactions available and makes it suitable to assist computational modeling. It can be used as a training set in a machine-learning approach and can serve to validate models predicting the pathogen–host interactome. In comparison to predicting intraspecies PPI, where much work has been done especially for the model organism yeast [93–95] and for human [96–99], the prediction of pathogen–host interactions is still in its infancy. Current study in interspecies PPI prediction includes domain profile-based approaches [5, 100], interolog- and homology-based approaches [6, 101, 102], and structural similarity-based approaches [103]. Supervised classification-based approaches have so far only been applied to the HIV–human interactome due to the availability of the data [12, 104]. It is significant to note that the homology-based models for bacteria use PPI databases such as DIP and iPFAM. The work presented here for Salmonella demonstrates that these machine readable databases lack many of the published known interactions (we only found 10% of the already published direct Salmonella–host interactions in PPI databases). This creates two problems. First, such approaches will suffer from false positives due to the large number of general domain overlap in proteins including those that do not interact. At the same time, they will likely miss critical interactions because bacteria and host have diverged greatly so that their genome’s similarity is generally low. The few cross-species pairs listed in databases cannot provide sufficient numbers to compensate for this loss in information due to sequence divergence. The only current computational approach to model Salmonella–host interactions uses such a homology-based approach [101]. Thus, our new data set opens the door to greatly improving the prospects for accurate prediction of the Salmonella–host interactome, by enabling application of machine-learning approaches to a larger data set and allowing the integration of multiple information sources, rather than basing predictions on homology alone.
Modeling the pathogen–host interactome can have a range of diverse objectives. It helps to acquire new insights into the communication between the two organisms, to get a better understanding of the infection process on a molecular level, to make comparison of different pathogen–host interactomes possible and last but not least modeling can push the rational development of drugs and vaccines to fight diseases. Computational machine-learning methods to model host–pathogen PPI and the host response in general will likely have great impact here. For example, modeling the host response to Brucella melitensis, Mycobacterium avium Paratuberculosis, and Salmonella enterica Typhimurium has provided strategies to develop vaccines on a rational basis [105]. Protein network analysis combined with tissue expression for human proteins and experimental validation of predicted interactions using GeneChip data was used to delineate the interactions between the gastric cancer causing pathogen H. pylori and the human [106]. In order to identify common strategies of pathogens and discover conserved host target proteins during infection, experimentally identified or computationally predicted interactomes have been compared [105, 107, 108]. The validity of this strategy for future Salmonella studies can also be found from the retrieval of a number of pathways related to other pathogens, including viruses. Thus, the present analysis presents an important milestone in the mapping of Salmonella–host interactions to the functional consequences of these interactions in human signal transduction pathways.
Acknowledgments
This work was funded by the Federal Ministry of Education and Research (BMBF), partner of the ERASysBio+initiative supported under the EU ERA-NET Plus scheme in FP7.
Abbreviations
- AG
antigen
- GEF
guanine nucleotide exchange factor
- PMN
polymorphonuclear neutrophil
- PPI
protein–protein interaction
- SCV
Salmonella-containing vacuole
- Sifs
Salmonella induced filaments
- SPI
Salmonella pathogenicity island
- TTSS
type III secretion system
- VAP
vacuole-associated actin polymerization
- VCP
vasolin-containing protein
Footnotes
The authors have declared no conflict of interest.
References
- 1.Salcedo SP, Holden DW. Bacterial interactions with the eukaryotic secretory pathway. Curr Opin Microbiol. 2005;8:92–98. doi: 10.1016/j.mib.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Ly KT, Casanova JE. Mechanisms of Salmonella entry into host cells. Cell Microbiol. 2007;9:2103–2111. doi: 10.1111/j.1462-5822.2007.00992.x. [DOI] [PubMed] [Google Scholar]
- 3.Galan JE. Salmonella interactions with host cells: Type III secretion at work. Annu Rev Cell Dev Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 4.Ramsden AE, Holden DW, Mota LJ. Membrane dynamics and spatial distribution of Salmonella-containing vacuoles. Trends Microbiol. 2007;15:516–524. doi: 10.1016/j.tim.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Dyer MD, Murali TM, Sobral BW. Computational prediction of host-pathogen protein-protein interactions. Bioinformatics. 2007;23:i159–i166. doi: 10.1093/bioinformatics/btm208. [DOI] [PubMed] [Google Scholar]
- 6.Davis FP, Barkan DT, Eswar N, McKerrow JH, et al. Host pathogen protein interactions predicted by comparative modeling. Protein Sci: A Publication of the Protein Society. 2007;16:2585–2596. doi: 10.1110/ps.073228407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuart LM, Boulais J, Charriere GM, Hennessy EJ, et al. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 8.Uetz P, Dong YA, Zeretzke C, Atzler C, et al. Herpes-viral protein networks and their interaction with the human proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 9.Balakrishnan S, Tastan O, Carbonell J, Klein-Seetharaman J. Alternative paths in HIV-1 targeted human signal transduction pathways. Biomed Chromatogr Genomics. 2009;10:S30. doi: 10.1186/1471-2164-10-S3-S30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parrish JR, Gulyas KD, Finley RL., Jr Yeast two-hybrid contributions to interactome mapping. Curr Opin Biotechnol. 2006;17:387–393. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Krogan NJ, Cagney G, Yu H, Zhong G, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 12.Tastan O, Qi Y, Carbonell JG, Klein-Seetharaman J. Prediction of interactions between HIV-1 and human proteins by information integration. Pac Symp Biocomput. 2009;14:516–527. [PMC free article] [PubMed] [Google Scholar]
- 13.Qi Y, Bar-Joseph Z, Klein-Seetharaman J. Evaluation of different biological data and computational classification methods for use in protein interaction prediction. Proteins Struct Funct Bioinform. 2006;63:490–500. doi: 10.1002/prot.20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 15.McGhie EJ, Brawn LC, Hume PJ, Humphreys D, et al. Salmonella takes control: effector-driven manipulation of the host. Curr Opinion Microbiol. 2009;12:117–124. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heffron F, Nieman G, Yoon H, Kidwai A, et al. Salmonella-secreted Virulence Factors. In: Porwollik S, editor. Salmonella: From Genome to Function. Caister Academic Press; Norfolk: 2011. pp. 187–223. [Google Scholar]
- 17.Garcia-Garcia J, Guney E, Aragues R, Planas-Iglesias J, et al. Biana: a software framework for compiling biological interactions and analyzing networks. Biomed Chromatogr Bioinform. 2010;11:56. doi: 10.1186/1471-2105-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raffatellu M, Wilson RP, Chessa D, Andrews-Polymenis H, et al. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect Immun. 2005;73:146–154. doi: 10.1128/IAI.73.1.146-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardt WD, Chen LM, Schuebel KE, Bustelo XR, et al. Styphimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 20.Friebel A, Ilchmann H, Aepfelbacher M, Ehrbar K, et al. SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell. J Biol Chem. 2001;276:34035–34040. doi: 10.1074/jbc.M100609200. [DOI] [PubMed] [Google Scholar]
- 21.Buchwald G, Friebel A, Galan JE, Hardt WD, et al. Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 2002;21:3286–3295. doi: 10.1093/emboj/cdf329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams C, Galyov EE, Bagby S. solution structure, backbone dynamics, and interaction with Cdc42 of Salmonella guanine nucleotide exchange factor SopE2. Biochemistry. 2004;43:11998–12008. doi: 10.1021/bi0490744. [DOI] [PubMed] [Google Scholar]
- 23.Zhou D, Mooseker MS, Galan JE. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- 24.Myeni SK, Zhou D. The C terminus of SipC binds and bundles F-actin to promote Salmonella invasion. J Biol Chem. 2010;285:13357–13363. doi: 10.1074/jbc.M109.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGhie EJ, Hayward RD, Koronakis V. Cooperation between actin-binding proteins of invasive Salmonella: SipA potentiates SipC nucleation and bundling of actin. EMBO J. 2001;20:2131–2139. doi: 10.1093/emboj/20.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson SA, Omary MB, Jones BD. Identification of cytokeratins as accessory mediators of Salmonella entry into eukaryotic cells. Life Sci. 2002;70:1415–1426. doi: 10.1016/s0024-3205(01)01512-0. [DOI] [PubMed] [Google Scholar]
- 27.Nichols CD, Casanova JE. Salmonella-directed recruitment of new membrane to invasion foci via the host exocyst complex. Curr Biol: CB. 2010;20:1316–1320. doi: 10.1016/j.cub.2010.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y, Galan JE. A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Pachon JM, Martin H, North G, Rotger R, et al. A novel connection between the yeast Cdc42 GTPase and the Slt2-mediated cell integrity pathway identified through the effect of secreted Salmonella GTPase modulators. J Biol Chem. 2002;277:27094–27102. doi: 10.1074/jbc.M201527200. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee K, Parashuraman S, Raje M, Mukhopadhyay A. SopE acts as an Rab5-specific nucleotide exchange factor and recruits non-prenylated Rab5 on Salmonella-containing phagosomes to promote fusion with early endosomes. J Biol Chem. 2001;276:23607–23615. doi: 10.1074/jbc.M101034200. [DOI] [PubMed] [Google Scholar]
- 31.Madan R, Krishnamurthy G, Mukhopadhyay A. SopE-mediated recruitment of host Rab5 on phagosomes inhibits Salmonella transport to lysosomes. Methods Mol Biol. 2008;445:417–437. doi: 10.1007/978-1-59745-157-4_27. [DOI] [PubMed] [Google Scholar]
- 32.Harrison RE, Brumell JH, Khandani A, Bucci C, et al. Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol Biol Cell. 2004;15:3146–3154. doi: 10.1091/mbc.E04-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christen M, Coye LH, Hontz JS, LaRock DL, et al. Activation of a bacterial virulence protein by the GTPase RhoA. Sci Signal. 2009;2:ra71. doi: 10.1126/scisignal.2000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee AH, Zareei MP, Daefler S. Identification of a NIPSNAP homologue as host cell target for Salmonella virulence protein SpiC. Cell Microbiol. 2002;4:739–750. doi: 10.1046/j.1462-5822.2002.00225.x. [DOI] [PubMed] [Google Scholar]
- 35.Shotland Y, Kramer H, Groisman EA. The Salmonella SpiC protein targets the mammalian Hook3 protein function to alter cellular trafficking. Mol Microbiol. 2003;49:1565–1576. doi: 10.1046/j.1365-2958.2003.03668.x. [DOI] [PubMed] [Google Scholar]
- 36.Marsman M, Jordens I, Kuijl C, Janssen L, et al. Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol Biol Cell. 2004;15:2954–2964. doi: 10.1091/mbc.E03-08-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guignot J, Servin AL. Maintenance of the Salmonella-containing vacuole in the juxtanuclear area: a role for intermediate filaments. Microb Pathog. 2008;45:415–422. doi: 10.1016/j.micpath.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Auweter SD, Bhavsar AP, de Hoog CL, Li Y, et al. Quantitative mass spectrometry catalogues salmonella pathogenicity island-2 effectors and identifies their cognate host binding partners. J Biol Chem. 2011;286:24023–24035. doi: 10.1074/jbc.M111.224600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salcedo SP, Holden DW. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 2003;22:5003–5014. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henry T, Couillault C, Rockenfeller P, Boucrot E, et al. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc Natl Acad Sci USA. 2006;103:13497–13502. doi: 10.1073/pnas.0605443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinicke AT, Hutchinson JL, Magee AI, Mastroeni P, et al. A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J Biol Chem. 2005;280:14620–14627. doi: 10.1074/jbc.M500076200. [DOI] [PubMed] [Google Scholar]
- 42.Boucrot E, Henry T, Borg JP, Gorvel JP, et al. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 2005;308:1174–1178. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- 43.Diacovich L, Dumont A, Lafitte D, Soprano E, et al. Interaction between the SifA virulence factor and its host target SKIP is essential for Salmonella pathogenesis. J Biol Chem. 2009;284:33151–33160. doi: 10.1074/jbc.M109.034975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumont A, Boucrot E, Drevensek S, Daire V, et al. SKIP, the host target of the Salmonella virulence factor SifA, promotes kinesin-1-dependent vacuolar membrane exchanges. Traffic. 2010;11:899–911. doi: 10.1111/j.1600-0854.2010.01069.x. [DOI] [PubMed] [Google Scholar]
- 45.Jackson LK, Nawabi P, Hentea C, Roark EA, et al. The Salmonella virulence protein SifA is a G protein antagonist. Proc Natl Acad Sci USA. 2008;105:14141–14146. doi: 10.1073/pnas.0801872105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arbeloa A, Garnett J, Lillington J, Bulgin RR, et al. EspM2 is a RhoA guanine nucleotide exchange factor. Cell Microbiol. 2010;12:654–664. doi: 10.1111/j.1462-5822.2009.01423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohlson MB, Huang Z, Alto NM, Blanc MP, et al. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe. 2008;4:434–446. doi: 10.1016/j.chom.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freeman JA, Ohl ME, Miller SI. The Salmonella enterica serovar typhimurium translocated effectors SseJ and SifB are targeted to the Salmonella-containing vacuole. Infect Immun. 2003;71:418–427. doi: 10.1128/IAI.71.1.418-427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun V, Wong A, Landekic M, Hong WJ, et al. Sorting nexin 3 (SNX3) is a component of a tubular endosomal network induced by Salmonella and involved in maturation of the Salmonella-containing vacuole. Cell Microbiol. 2010;12:1352–1367. doi: 10.1111/j.1462-5822.2010.01476.x. [DOI] [PubMed] [Google Scholar]
- 50.Abrahams GL, Muller P, Hensel M. Functional dissection of SseF, a type III effector protein involved in positioning the salmonella-containing vacuole. Traffic. 2006;7:950–965. doi: 10.1111/j.1600-0854.2006.00454.x. [DOI] [PubMed] [Google Scholar]
- 51.Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 52.Wang B, David MD, Schrader JW. Absence of caprin-1 results in defects in cellular proliferation. J Immunol. 2005;175:4274–4282. doi: 10.4049/jimmunol.175.7.4274. [DOI] [PubMed] [Google Scholar]
- 53.Delva E, Tucker DK, Kowalczyk AP. The desmosome. Cold Spring Harb Perspect Biol. 2009;1:a002543. doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guiney DG, Lesnick M. Targeting of the actin cytoskeleton during infection by Salmonella strains. Clin Immunol. 2005;114:248–255. doi: 10.1016/j.clim.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 55.Poh J, Odendall C, Spanos A, Boyle C, et al. SteC is a Salmonella kinase required for SPI-2-dependent F-actin remodelling. Cell microbiol. 2008;10:20–30. doi: 10.1111/j.1462-5822.2007.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miao EA, Brittnacher M, Haraga A, Jeng RL, et al. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol Microbiol. 2003;48:401–415. doi: 10.1046/j.1365-2958.2003.t01-1-03456.x. [DOI] [PubMed] [Google Scholar]
- 57.Tezcan-Merdol D, Nyman T, Lindberg U, Haag F, et al. Actin is ADP-ribosylated by the Salmonella enterica virulence-associated protein SpvB. Mol Microbiol. 2001;39:606–619. doi: 10.1046/j.1365-2958.2001.02258.x. [DOI] [PubMed] [Google Scholar]
- 58.Margarit SM, Davidson W, Frego L, Stebbins CE. A steric antagonism of actin polymerization by a salmonella virulence protein. Structure. 2006;14:1219–1229. doi: 10.1016/j.str.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 59.Humphreys D, Hume PJ, Koronakis V. The Salmonella effector SptP dephosphorylates host AAA+ATPase VCP to promote development of its intracellular replicative niche. Cell Host Microbe. 2009;5:225–233. doi: 10.1016/j.chom.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Li D, Qu D, Zhou D. Involvement of TIP60 acetyltransferase in intracellular Salmonella replication. Biomed Chromatogr Microbiol. 2010;10:228. doi: 10.1186/1471-2180-10-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Higashide W, Dai S, Sherman DM, et al. Recognition and ubiquitination of Salmonella type III effector SopA by a ubiquitin E3 ligase, HsRMA1. J Biol Chem. 2005;280:38682–38688. doi: 10.1074/jbc.M506309200. [DOI] [PubMed] [Google Scholar]
- 62.Worley MJ, Nieman GS, Geddes K, Heffron F. Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc Natl Acad Sci USA. 2006;103:17915–17920. doi: 10.1073/pnas.0604054103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McLaughlin LM, Govoni GR, Gerke C, Gopinath S, et al. The Salmonella SPI2 effector SseI mediates long-term systemic infection by modulating host cell migration. PLoS Pathog. 2009;5:e1000671. doi: 10.1371/journal.ppat.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheminay C, Schoen M, Hensel M, Wandersee-Steinhauser A, et al. Migration of Salmonella typhimurium –harboring bone marrow-derived dendritic cells towards the chemokines CCL19 and CCL21. Microb Pathog. 2002;32:207–218. doi: 10.1006/mpat.2002.0497. [DOI] [PubMed] [Google Scholar]
- 65.Friebel A, Ilchmann H, Aepfelbacher M, Ehrbar K, et al. SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell. J Biol Chem. 2001;276:34035–34040. doi: 10.1074/jbc.M100609200. [DOI] [PubMed] [Google Scholar]
- 66.Huang FC, Werne A, Li Q, Galyov EE, et al. Cooperative interactions between flagellin and SopE2 in the epithelial interleukin-8 response to Salmonella enterica serovar typhimurium infection. Infect Immun. 2004;72:5052–5062. doi: 10.1128/IAI.72.9.5052-5062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srikanth CV, Wall DM, Maldonado-Contreras A, Shi HN, et al. Salmonella pathogenesis and processing of secreted effectors by caspase-3. Science. 2010;330:390–393. doi: 10.1126/science.1194598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Higashide WM, McCormick BA, Chen J, et al. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol. 2006;62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 69.Jones RM, Wu H, Wentworth C, Luo L, et al. Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe. 2008;3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 70.Du F, Galan JE. Selective inhibition of type III secretion activated signaling by the Salmonella effector AvrA. PLos Pathog. 2009;5:e1000595. doi: 10.1371/journal.ppat.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye Z, Petrof EO, Boone D, Claud EC, et al. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007;171:882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Negrate G, Faustin B, Welsh K, Loeffler M, et al. Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-kappaB, suppresses IkappaBalpha ubiquitination and modulates innate immune responses. J Immunol. 2008;180:5045–5056. doi: 10.4049/jimmunol.180.7.5045. [DOI] [PubMed] [Google Scholar]
- 73.Haraga A, Miller SI. A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell Microbiol. 2006;8:837–846. doi: 10.1111/j.1462-5822.2005.00670.x. [DOI] [PubMed] [Google Scholar]
- 74.Murli S, Watson RO, Galan JE. Role of tyrosine kinases and the tyrosine phosphatase SptP in the interaction of Salmonella with host cells. Cell Microbiol. 2001;3:795–810. doi: 10.1046/j.1462-5822.2001.00158.x. [DOI] [PubMed] [Google Scholar]
- 75.Lin SL, Le TX, Cowen DS. SptP, a Salmonella typhimurium type III-secreted protein, inhibits the mitogen-activated protein kinase pathway by inhibiting Raf activation. Cell Microbiol. 2003;5:267–275. doi: 10.1046/j.1462-5822.2003.t01-1-00274.x. [DOI] [PubMed] [Google Scholar]
- 76.Chen L, Wang H, Zhang J, Gu L, et al. Structural basis for the catalytic mechanism of phosphothreonine lyase. Nat Struct Mol Biol. 2008;15:101–102. doi: 10.1038/nsmb1329. [DOI] [PubMed] [Google Scholar]
- 77.Mazurkiewicz P, Thomas J, Thompson JA, Liu M, et al. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol Microbiol. 2008;67:1371–1383. doi: 10.1111/j.1365-2958.2008.06134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu Y, Li H, Long C, Hu L, et al. Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase. Mol Cell. 2007;28:899–913. doi: 10.1016/j.molcel.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 79.Cheminay C, Mohlenbrink A, Hensel M. Intracellular Salmonella inhibit antigen presentation by dendritic cells. J Immunol. 2005;174:2892–2899. doi: 10.4049/jimmunol.174.5.2892. [DOI] [PubMed] [Google Scholar]
- 80.Tobar JA, Gonzalez PA, Kalergis AM. Salmonella escape from antigen presentation can be overcome by targeting bacteria to Fc gamma receptors on dendritic cells. J Immunol. 2004;173:4058–4065. doi: 10.4049/jimmunol.173.6.4058. [DOI] [PubMed] [Google Scholar]
- 81.Halici S, Zenk SF, Jantsch J, Hensel M. Functional analysis of the Salmonella pathogenicity island 2-mediated inhibition of antigen presentation in dendritic cells. Infect Immun. 2008;76:4924–4933. doi: 10.1128/IAI.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sigel E. Dangers of ‘single-point pharmacology’ in receptor studies. GABAA receptor: complex picture confused. Trends Pharmacol Sci. 1990;11:103. doi: 10.1016/0165-6147(90)90192-b. [DOI] [PubMed] [Google Scholar]
- 83.Bernal-Bayard J, Cardenal-Munoz E, Ramos-Morales F. The Salmonella type III secretion effector, salmonella leucine-rich repeat protein (SlrP), targets the human chaperone ERdj3. J Biol Chem. 2010;285:16360–16368. doi: 10.1074/jbc.M110.100669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Granados DP, Tanguay PL, Hardy MP, Caron E, et al. ER stress affects processing of MHC class I-associated peptides. Biomed Chromatogr Immunol. 2009;10:10. doi: 10.1186/1471-2172-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hersh D, Monack DM, Smith MR, Ghori N, et al. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jesenberger V, Procyk KJ, Yuan J, Reipert S, et al. Salmonella-induced caspase-2 activation in macrophages: a novel mechanism in pathogen-mediated apoptosis. J Exp Med. 2000;192:1035–1046. doi: 10.1084/jem.192.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernal-Bayard J, Ramos-Morales F. Salmonella type III secretion effector SlrP is an E3 ubiquitin ligase for mammalian thioredoxin. J Biol Chem. 2009;284:27587–27595. doi: 10.1074/jbc.M109.010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rytkonen A, Poh J, Garmendia J, Boyle C, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meresse S. Is host lipidation of pathogen effector proteins a general virulence mechanism? Front Microbiol. 2011;2:73. doi: 10.3389/fmicb.2011.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brumell JH, Grinstein S. Role of lipid-mediated signal transduction in bacterial internalization. Cell Microbiol. 2003;5:287–297. doi: 10.1046/j.1462-5822.2003.00273.x. [DOI] [PubMed] [Google Scholar]
- 91.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
- 92.Wang PY, Weng J, Anderson RG. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 2005;307:1472–1476. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- 93.Jansen R, Yu H, Greenbaum D, Kluger Y, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 94.Wu X, Zhu L, Guo J, Zhang DY, et al. Prediction of yeast protein-protein interaction network: insights from the Gene Ontology and annotations. Nucleic Acids Res. 2006;34:2137–2150. doi: 10.1093/nar/gkl219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pitre S, Dehne F, Chan A, Cheetham J, et al. PIPE: A protein-protein interaction prediction engine based on the re-occurring short polypeptide sequences between known interacting protein pairs. Biomed Chromatogr Bioinform. 2006;7:365. doi: 10.1186/1471-2105-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scott MS, Barton GJ. Probabilistic prediction and ranking of human protein-protein interactions. Biomed Chromatogr Bioinform. 2007;8:239. doi: 10.1186/1471-2105-8-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McDowall MD, Scott MS, Barton GJ. PIPs: Human protein-protein interaction prediction database. Nucleic Acids Res. 2009;37:D651–D656. doi: 10.1093/nar/gkn870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qi Y, Dhiman HK, Bhola N, Budyak I, et al. Systematic prediction of human membrane receptor interactions. Proteomics. 2009;9:5243–5255. doi: 10.1002/pmic.200900259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pan XY, Zhang YN, Shen HB. Large-scale prediction of human protein-protein interactions from amino acid sequence based on latent topic features. J Proteome Res. 2010;9:4992–5001. doi: 10.1021/pr100618t. [DOI] [PubMed] [Google Scholar]
- 100.Evans P, Dampier W, Ungar L, Tozeren A. Prediction of HIV-1 virus-host protein interactions using virus and host sequence motifs. Biomed Chromatogr Med Genomics. 2009;2:27. doi: 10.1186/1755-8794-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krishnadev O, Srinivasan N. Prediction of protein-protein interactions between human host and a pathogen and its application to three pathogenic bacteria. Int J Biol Macromol. 2011;48:613–619. doi: 10.1016/j.ijbiomac.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 102.Tyagi N, Krishnadev O, Srinivasan N. Prediction of protein-protein interactions between Helicobacter pylori and a human host. Mol BioSyst. 2009;5:1630–1635. doi: 10.1039/b906543c. [DOI] [PubMed] [Google Scholar]
- 103.Doolittle JM, Gomez SM. Structural similarity-based predictions of protein interactions between HIV-1 and Homo sapiens. Virol J. 2010;7:82. doi: 10.1186/1743-422X-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nouretdinov I, Gammerman A, Qi Y, Klein-Seetharaman J. Determining confidence of predicted interactions between HIV-1 and human proteins using conformal method. Pac Symp Biocomput. 2012 [PMC free article] [PubMed] [Google Scholar]
- 105.Adams LG, Khare S, Lawhon SD, Rossetti CA, et al. Enhancing the role of veterinary vaccines reducing zoonotic diseases of humans: linking systems biology with vaccine development. Vaccine. 2011 doi: 10.1016/j.vaccine.2011.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim WK, Kim K, Lee E, Marcotte EM, et al. Identification of disease specific protein interactions between the gastric cancer causing pathogen, H-pylori, and human hosts using protein network modeling and gene chip analysis. Biochip J. 2007;1:179–187. [Google Scholar]
- 107.Dyer MD, Neff C, Dufford M, Rivera CG, et al. The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS One. 2010;5:e12089. doi: 10.1371/journal.pone.0012089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dyer MD, Murali TM, Sobral BW. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008;4:e32. doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu S, Ye Z, Liu X, Zhao Y, et al. Salmonella typhimurium infection increases p53 acetylation in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2010;298:G784–G794. doi: 10.1152/ajpgi.00526.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hayward RD, Koronakis V. Direct modulation of the host cell cytoskeleton by Salmonella actin-binding proteins. Trends Cell Biol. 2002;12:15–20. doi: 10.1016/s0962-8924(01)02183-3. [DOI] [PubMed] [Google Scholar]
- 111.Zhou D, Mooseker MS, Galan JE. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc Natl Acad Sci USA. 1999;96:10176–10181. doi: 10.1073/pnas.96.18.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim BH, Kim HG, Kim JS, Jang JI, et al. Analysis of functional domains present in the N-terminus of the SipB protein. Microbiology. 2007;153:2998–3008. doi: 10.1099/mic.0.2007/007872-0. [DOI] [PubMed] [Google Scholar]
- 113.Diao J, Zhang Y, Huibregtse JM, Zhou D, et al. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol. 2008;15:65–70. doi: 10.1038/nsmb1346. [DOI] [PubMed] [Google Scholar]
- 114.Rodriguez-Escudero I, Rotger R, Cid VJ, Molina M. Inhibition of Cdc42-dependent signalling in Saccharomyces cerevisiae by phosphatase-dead SigD/SopB from Salmonella typhimurium. Microbiology. 2006;152:3437–3452. doi: 10.1099/mic.0.29186-0. [DOI] [PubMed] [Google Scholar]
- 115.Rogers LD, Kristensen AR, Boyle EC, Robinson DP, et al. Identification of cognate host targets and specific ubiquitylation sites on the Salmonella SPI-1 effector SopB/SigD. J Proteomics. 2008;71:97–108. doi: 10.1016/j.jprot.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 116.Parashuraman S, Mukhopadhyay A. Assay and functional properties of SopE in the recruitment of Rab5 on Salmonella-containing phagosomes. Methods Enzymol. 2005;403:295–309. doi: 10.1016/S0076-6879(05)03025-9. [DOI] [PubMed] [Google Scholar]
- 117.Stebbins CE, Galan JE. Modulation of host signaling by a bacterial mimic: structure of the Salmonella effector SptP bound to Rac1. Mol Cell. 2000;6:1449–1460. doi: 10.1016/s1097-2765(00)00141-6. [DOI] [PubMed] [Google Scholar]
- 118.Levin I, Eakin C, Blanc MP, Klevit RE, et al. Identification of an unconventional E3 binding surface on the UbcH5~Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc Natl Acad Sci USA. 2010;107:2848–2853. doi: 10.1073/pnas.0914821107. [DOI] [PMC free article] [PubMed] [Google Scholar]