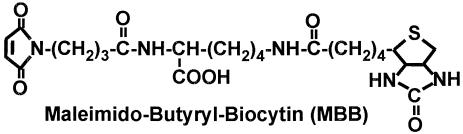Abstract
Retinitis pigmentosa (RP) point mutations in both the intradiscal (ID) and transmembrane domains of rhodopsin cause partial or complete misfolding of rhodopsin, resulting in loss of 11-cis-retinal binding. Previous work has shown that misfolding is caused by the formation of a disulfide bond in the ID domain different from the native Cys-110–Cys-187 disulfide bond in native rhodopsin. Here we report on direct identification of the abnormal disulfide bond in misfolded RP mutants in the transmembrane domain by mass spectrometric analysis. This disulfide bond is between Cys-185 and Cys-187, the same as previously identified in misfolded RP mutations in the ID domain. The strategy described here should be generally applicable to identification of disulfide bonds in other integral membrane proteins.
Keywords: G protein-coupled receptors, signal transduction, transmembrane domain, N-ethylmaleimide, biotin-avidin affinity chromatography
A large number of naturally occurring point mutations in rhodopsin are now known. The majority of such mutations are associated with retinitis pigmentosa (RP), and whereas they are found in all of the three domains—cytoplasmic, transmembrane (TM), and intradiscal (ID)—those in the latter two domains account for the bulk of them (1–5). Mutations in the ID domain were found to cause partial or complete misfolding in the ID domain, misfolding being defined by the loss of ability to bind 11-cis-retinal (6–10). Studies of naturally occurring RP mutations and of designed mutations in the ID domain showed that misfolding was caused by the formation of a disulfide bond different from that in native rhodopsin. More recent work has conclusively shown that the abnormal disulfide bond in misfolded RP mutants in the ID domain is between Cys-185 and Cys-187. On the one hand, the evidence came from studies of a rhodopsin mutant (in which Cys-110 was replaced by alanine) and of the RP mutants, C110F‡ and C110Y (11); on the other hand, it came from identification of the Cys-185–Cys-187 disulfide bond formed after reconstitution of rhodopsin from two complementary fragments (12). Further, studies of RP mutations in the TM domain of rhodopsin showed that they also cause misfolding by formation of an abnormal ID disulfide bond (13, 14). Identification of the abnormal disulfide bond in the latter case acquires special significance because, if this disulfide bond is the same as that shown above for misfolded RP mutations in the ID domain, between Cys-185 and Cys-187, then this result would show that defects in the packing of the helices in the TM domain are able to cause misfolding in the ID domain. Therefore, packing of the helices in the TM domain and folding to a tertiary structure in the ID domain must be coupled. Here we report the direct identification by mass spectrometry of the abnormal disulfide bond in misfolded RP mutants in the TM domain (15). From the RP mutants in the TM domain studied (14), we selected four (G89D, L125R, A164V, and H211P; Fig. 1) in the present study. Misfolding in all of the four mutants is now shown to be caused by a Cys-185–Cys-187 disulfide bond. We also confirm by the method now developed that the disulfide bond in wild-type (WT) rhodopsin, indirectly identified previously (16–18), is indeed between Cys-110 and Cys-187.
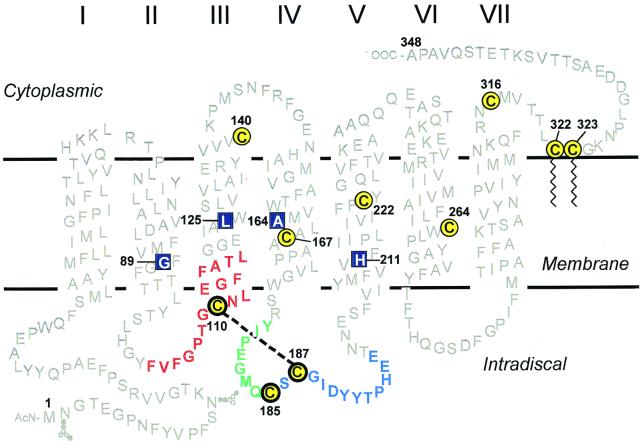
Figure 1.
A secondary structure model of rhodopsin showing the cytoplasmic, TM, and the ID domains. The 10 cysteines are shown in yellow with black circles; the three ID cysteines are highlighted by bolder circles. Amino acid sequences adjoining the three cysteines of interest (for identification of the disulfide bonds) are color coded for each cysteine: red for Cys-110, green for Cys-185, and blue for Cys-187. Native rhodopsin contains a disulfide bond between Cys-110 and Cys-187 (indicated by dashed line). The RP mutants studied here are located in the TM domain (shown in blue boxes): G89D (helix II), L125R (helix III), A164V (helix IV), and H211P (helix V).
The aim of the strategy was to isolate and sequence peptides adjoining the two cysteines participating in the disulfide linkage. Because of the hydrophobic nature of the opsin, the technique aimed at selective isolation of peptide segments adjoining the cysteines involved in the disulfide bonds (color coded in Fig. 1). In step one, all of the free cysteines in the protein were derivatized with N-ethylmaleimide (NEM). This step required denaturing conditions (0.5% SDS). Further, an acidic pH (pH 6) was used to avoid any disulfide-exchange reaction (19). Because of the low rate of reaction at pH 6, a high concentration of NEM was used for this derivatization. The derivatized protein was purified by binding to anti-rhodopsin 1d4-Sepharose. In step two, the protein, while bound to the Sepharose beads, was treated with DTT to reduce the disulfide bond. The sulfhydryl groups now formed were derivatized by treatment of the protein-Sepharose suspension with maleimido-butyryl-biocytin (MBB; Fig. 2). In WT rhodopsin and analogously, folded retinal-binding rhodopsin mutants (Fig. 3A), the free sulfhydryl groups formed from reduction of the Cys-110 and Cys-187 disulfide bond would react with the maleimido group in MBB (Cys-185 would have been derivatized with NEM in step one, along with all of the other sulfhydryl groups). In the misfolded proteins, the abnormal disulfide bond could be either between Cys-110 and Cys-185, or between Cys-185 and Cys-187. Positions for the attachment of MBB expected for a Cys-110–Cys-185 disulfide bond are shown in Fig. 3BI, and positions for the attachment of MBB expected for a Cys-185–Cys-187 disulfide bond are shown in Fig. 3BII. After derivatization with MBB, excess of the latter was removed from the derivatized proteins while they were bound to anti-rhodopsin 1d4-Sepharose. Subsequently, the proteins were eluted with the epitope nonapeptide. Step three involved digestion of the isolated derivatized proteins with proteinase K. The resulting MBB-carrying peptide fragments were purified by their selective binding to avidin-Sepharose (step four). Finally, after elution with excess biotin, the MBB peptides were subjected to MALDI-TOF mass spectrometry (step five).§
Figure 2.
Structure of maleimido-butyryl-biocytin (MBB).
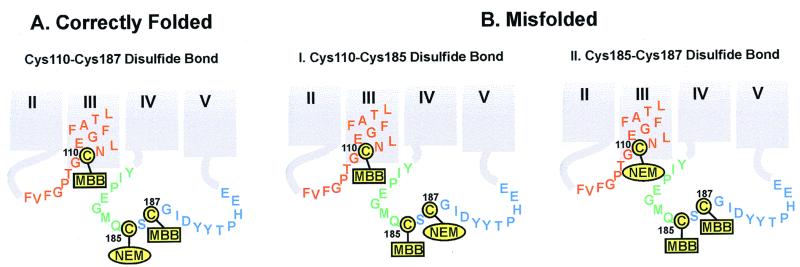
Figure 3.
Derivatizations as expected for ID cysteines in correctly folded (A) and misfolded (B) rhodopsins. The disulfide linkage in correctly folded A rhodopsin is between Cys-110 and Cys-187, as identified by mass spectrometry (Fig. 4). Thus, steps 1 and 2 of the strategy in the identification of disulfide bonds (see above) produced rhodopsin NEM-derivatized at Cys-185, whereas Cys-110 and Cys-187 were derivatized with MBB. In misfolded rhodopsin B, the disulfide bond could either be between Cys-110 and Cys-185 (I) or between Cys-185 and Cys-187 (II). Experimentally, the disulfide bond was found between Cys-185 and Cys-187 (Fig. 4). Thus, Cys-110 was NEM-derivatized, whereas Cys-185 and Cys-187 were MBB-derivatized (II). The color code for the amino acid residues adjacent to each of the cysteines is the same as in Fig. 1. The derivatized products were then subjected to steps 3–5 (above), yielding the MBB-derivatized peptides that were analyzed by MALDI-TOF (Fig. 4).
Materials and Methods
Materials.
The 11-cis retinal was a gift from R. Crouch (University of South Carolina, Charleston, SC, and the National Eye Institute, National Institutes of Health, Bethesda). The detergent dodecyl maltoside (DM) was from Anatrace (Maumee, OH). All other reagents were from Sigma. Anti-rhodopsin monoclonal antibody rhodopsin-1d4 was purified from myeloma cell lines provided by R. S. Molday (University of British Columbia, Vancouver) and was coupled to cyanogen bromide-activated Sepharose 4B (Amersham Pharmacia) as described (20). The buffers used were buffer A [NaCl, 137 mM/KCl, 2.7 mM/KH2PO4, 1.8 mM/NaH2PO4, 10 mM (pH 7.2)], buffer B (buffer A plus 0.05% DM), buffer C [2 mM NaH2PO4/Na2HPO4 (pH 6.0)/0.05% DM], and buffer D (buffer C plus 150 mM NaCl). All buffers were purged with argon.
Methods.
Construction of the mutant opsin expression plasmids, transient expression in COS-1 cells, purification of the expressed proteins, and separation of folded and misfolded opsins, have been described elsewhere in detail (14).
Derivatization of Sulfhydryl Groups in Free Cysteines in Denatured Rhodopsin with NEM.
Separated, correctly folded rhodopsin mutants and the misfolded opsins (about 10 μg) in 300 μl of elution buffer C (containing epitope nonapeptide) or buffer D (containing epitope nonapeptide) were treated with NEM (100 mM) in the presence of 0.5% SDS for 3 h at room temperature. The extent of derivatization with NEM was determined by titration with 4′-4′dithiodipyridine (21, 22). The reaction mixtures were then concentrated to 100 μl by using Centricon 30 (Millipore), and the elution nonapeptide was removed by gel filtration by using Sephadex G50 medium (Pharmacia). The NEM-derivatized proteins were then bound to 1d4-Sepharose suspension by nutating for 2 h at 4°C.
Reduction of the Disulfide Bond in NEM-Derivatized Opsins or Mutant Rhodopsins with DTT and Derivatization of the Resulting Sulfhydryl Groups with MBB.
The NEM-derivatized protein [while bound to the rhodopsin-1d4-Sepharose column (step 1)] was washed with 10 ml of buffer C. Buffer C (1 ml), containing 0.1% SDS and 10 mM DTT, was added to the suspension of the Sepharose beads, and the mixture was incubated at 20°C for 60 min. The beads in a column were then washed with 5 ml of buffer C. Buffer B, containing 0.1% SDS and 0.1 μM MBB, was added to the suspension and nutated end-over-end for 2 h at 20°C. After washing the beads with buffer B (20 ml), the MBB-derivatized protein was eluted from the Sepharose beads with the epitope nonapeptide.
Digestion with Proteinase K.
The MBB-derivatized protein was digested at room temperature for 6 h (1 h for the mutants H211P and L125R) in 500 μl buffer B by using 1 μg Proteinase K (Sigma). The reaction was terminated by the addition of 0.1 mM PMSF.
Selective Binding of MBB-Containing Peptides to Avidin–Agarose and Subsequent Elution with Biotin.
The proteinase K digestion mixture was added to an avidin–agarose column equilibrated with buffer A and nutated end-over-end for 2 h at 20°C. The suspension was washed with water, and the MBB-derivatized peptides were eluted with a biotin solution (1 mg/ml) in water. The water was evaporated by using a speed vac, and the residue of MBB-derivatized peptides was dissolved in 5 μl of acetonitrile/water (1/1) containing 0.1% TFA.
MALDI-TOF of MBB-Labeled Peptides.
One microliter of the sample (1–10 pM) was analyzed by mass spectrometry with a Voyager instrument (PerSeptive Biosystems, Framingham, MA). The sample was transferred to the sample plate and allowed to dry by evaporation. The sample was then overlayed with 0.5 μl of the matrix compound (α-cyano-4-hydroxy cinnamic acid) at a concentration of 10 mg/ml. The program, ms product (http://prospector.ucsf.edu/ucsfhtml3.4/msprod.htm), was used to calculate the predicted monoisotopic and average m/z to identify the peptides containing MBB-labeled cysteines. Experiments for WT and all of the mutant rhodopsins were repeated at least three times.
Results
WT Rhodopsin Contains a Cys-110–Cys-187 Disulfide Bond.
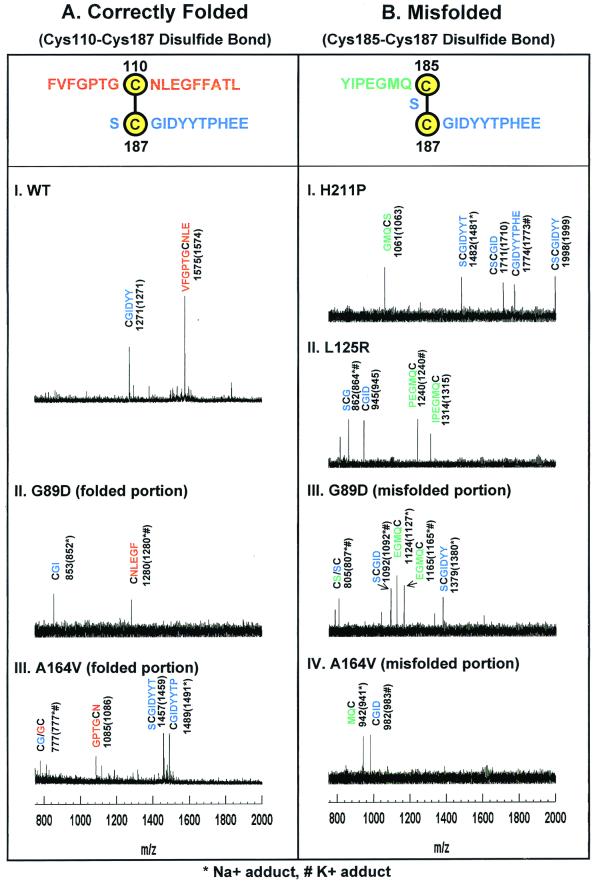
The strategy (Fig. 2) was first applied to WT rhodopsin. As seen in Fig. 4AI, the major experimentally observed m/z of 1,575 corresponded to the peptide VFGPTGCNLE + MBB (expected average, m/z 1,574). This result showed Cys-110 as one of the cysteines participating in the disulfide bond. The second major ion, detected at m/z 1,271, corresponded to CGIDYY + MBB (expected, m/z 1,271). This result confirmed Cys-187 as the second cysteine involved in the disulfide bond with Cys-110.
Figure 4.
MALDI-TOF Analysis of peptide fragments generated from the derivatized correctly folded (A) and misfolded (B) rhodopsins. The adjoining peptide sequences expected for the disulfide bonds are highlighted at the top in A and B. Rhodopsins were derivatized at cysteines as in the strategy outlined above. Because step 4 of the strategy selects only MBB-labeled peptides, all peptide assignments shown here include the mass for the MBB adducts of cysteines. The y axis shows intensity in arbitrary units. The x axis shows m/z in the range of 750–2,000. (A) Mass spectrometric results for WT rhodopsin and correctly folded portions of the A164V and G89D mutants. The observed signals in the mass spectra correspond to MBB-labeled Cys-110 peptides (red) and Cys-187 peptides (blue). (B) Mass spectrometric results for the misfolded portion of A164V and G89D (III and IV) and of completely misfolded H211P and L125R (I and II). Only peptides corresponding to a Cys-185–Cys-187 disulfide bond were observed. MBB-labeled Cys-185 containing peptides are shown in green, and MBB-labeled Cys-187 peptides are shown in blue.
Correctly Folded Portions of Mutants G89D (TM Helix II) and A164V (TM Helix IV) Contain a Cys-110–Cys-187 Disulfide Bond.
Next, the four RP mutants in different TM helices (Fig. 1) were studied. Previously, the mutants L125R (TM helix III) and H211P (TM helix V) were found to cause complete misfolding, whereas G89D (TM helix II) and A164V (TM helix IV) caused partial misfolding (14). In the latter two cases, correctly folded and misfolded portions of the expressed proteins were separated as described elsewhere (10, 14). Mass spectrometric data obtained with the correctly folded portion of the mutant G89D are shown in Fig. 4AII. The two main ions seen, with experimental m/z 1,280 and m/z 853, corresponded to the peptides, CNLEGF + MBB (expected m/z 1,280 for one Na+ and one K+ adduct) and CGI + MBB (expected m/z 852 for one Na+ adduct). The finding of these peptides identified the presence of a Cys-110–Cys-187 disulfide bond in the correctly folded portion of mutant G89D. In the correctly folded portion of the mutant A164V (Fig. 4AIII), the peptide (m/z 1,085; GPTGCN + MBB) identified from the experimentally observed ion (expected m/z 1,086) showed Cys-110 involvement in the disulfide bond. The ions at m/z 1,457 and m/z 1,489, identified, respectively, SCGIDYYT + MBB (expected m/z 1,459) and CGIDYYTP + MBB (expected m/z 1,491 for Na+ adduct) demonstrating as the second cysteine. (The ion at m/z 777 could arise from labeling of either Cys-187 or Cys-110, corresponding to peptide CG + MBB or GC + MBB, respectively.) Thus, the correctly folded mutant, A164V, like the WT protein, contains a Cys-110-Cys-187 disulfide bond.
Misfolded Rhodopsin RP Mutants Contain a Cys-185–Cys-187 Disulfide Bond.
Misfolded mutants H211P and L125R.
Mass spectrometric studies of the completely misfolded proteins from H211P and L125R are shown in Fig. 4B, I and II, respectively. In the spectrum of H211P, one ion was observed at m/z 1,061, and was assigned to the Cys-185 containing peptide GMQCS + MBB (expected m/z 1,063). Two other signals observed in the same mutant, at m/z 1,482 and m/z 1,774, were identified as the Cys-187-containing peptides, SCGIDYYT and CGIDYYTPHE (shown in blue in Fig. 4B). Further, the signals observed, m/z 1,711 and m/z 1,998, corresponded to peptides CSCGID and CSCGIDYY, both Cys-185 and Cys-187 derivatized with MBB. In the spectrum of L125R, the ions seen with experimental m/z 862 and m/z 945 corresponded to the Cys-187-containing peptides, SCG (expected m/z 864 for the Na+ and K+ adduct) and CGID (expected m/z 945), respectively. The ions at m/z 1,240 and m/z 1,314 were assigned to the Cys-185-containing peptides, PEGMQC + MBB (expected m/z 1,240 with K+ adduct) and IPEGMQC + MBB (expected m/z 1,315), respectively. Thus, the completely misfolded mutants H211P and L125R both contain a Cys-185–Cys-187 disulfide bond.
Misfolded portions of the proteins from G89D and A164V.
Mass spectrometric data on the misfolded portions of G89D and A164V are shown in III and IV, respectively, of Fig. 4B. The ions observed at m/z 1,124 and at m/z 1,165 in the spectrum of the misfolded portion of G89D were both assigned to the Cys-185-containing peptide EGMQC + MBB with expected m/z 1,127 for the Na+ adduct and m/z 1,165 for the Na+/K+ adduct. The signals seen at m/z 1,092 and m/z 1,379 corresponded to the Cys-187-containing peptides SCGID + MBB and SCGIDYY + MBB. The signal at m/z 805 could be caused by either the Cys-185-containing peptide, CS + MBB, or the Cys-187-containing peptide, SC + MBB.
In the spectrum of the misfolded portion of mutant A164V, two signals were seen at m/z 942 and m/z 982. These corresponded to the peptides MQC + MBB (expected m/z 941 for a Na+ adduct) and CGID (expected m/z 983 for a K+ adduct). These two peptides identified a Cys-185–Cys-187 disulfide bond in the misfolded portion of mutant A164V. Thus, the misfolded portions of the proteins from both G89D and A164V contain a Cys-185–Cys-187 disulfide bond.
Discussion
Methods for the identification of disulfide bonds in water-soluble proteins were established in the 1960s (for example, ref. 23). However, integral membrane proteins have so far proven intractable for corresponding chemical-structural studies. Indeed, it is for this reason that the wealth of information now available on primary structures of receptors, membrane transporters, and ion channels has been made possible only by the development of technology for the sequencing of the corresponding genes. In the present work, we have developed a strategy for direct identification of disulfide bonds in correctly folded rhodopsin and its mutants, and misfolded nonretinal binding opsins. The method should be applicable generally to corresponding studies of integral membrane proteins.
The strategy was first applied to identification of the native disulfide bond in rhodopsin. The conclusion that rhodopsin contains a Cys-110–Cys-187 disulfide bond had been arrived at, albeit indirectly, in a number of earlier studies (16–18). Direct identification of this disulfide bond as now accomplished is of central significance in the superfamily of G protein-coupled receptors (GPCRs). A disulfide bond at positions equivalent to Cys-110 and Cys-187 in rhodopsin is now known to be conserved in most of the GPCRs (ref. 24; see also, www.gcrdb.uthscsa.edu). This conservation implies the importance of this disulfide bond in a unitary mechanism for the activation of GPCRs (25). Other studies demonstrated misfolding in rhodopsin mutants prepared by designed mutagenesis (6–8), as well as in naturally occurring RP mutants (9, 10). Methods were developed for the separation and characterization of the misfolded opsin, and the conclusion was drawn that misfolding caused by RP mutations in the ID domain was caused by the formation of an abnormal disulfide bond (8). More recently, studies described the unequivocal identification of this disulfide bond as that between Cys-185 and Cys-187 (11, 12). In the present work, the abnormal disulfide bond present in four misfolded opsins resulting from RP mutations in TM helices II–V (Fig. 1) has been identified in every case as that between Cys-185 and Cys-187. The total results now firmly establish that the packing of the seven helices in the TM domain and folding in the ID domain are coupled (14).
Finally, it is noted that the Cys-185–Cys-187 disulfide bond between cysteines separated only by the single Ser-186 is unusual and, presumably, results from strain introduced in the ID tertiary structure from the defective packing of the TM helices in RP mutants. There is a precedent for the presence of a disulfide bond with identical sequence, Cys-Ser-Cys, in mengovirus coat protein (26, 27).
Acknowledgments
We are deeply indebted to Dr. Kevin Ridge (Center for Advanced Research in Biotechnology, Rockville, MD), who originally suggested the basic outline for the strategy developed in this article. We are grateful to Professor U. L. RajBhandary (Massachusetts Institute of Technology), Dr. Ivan Corriera (Whitehead Institute), and Professor Jane S. Richardson (Duke University) for valuable discussions. We acknowledge the enthusiastic assistance of Ms. Judy Carlin in the preparation of the manuscript. This work was supported by National Institutes of Health Grant GM28289 and National Eye Institute Grant EY11717 (to H.G.K.). J.K.-S. was the recipient of a Howard Hughes Predoctoral Fellowship, and J.H. was the recipient of a Howard Hughes Medical Institute Physician Postdoctoral Fellowship.
Abbreviations
- WT
wild type
- RP
retinitis pigmentosa
- TM
transmembrane
- ID
intradiscal
- PSB
protonated Schiff base
- NEM
N-ethylmaleimide
- MBB
maleimido-butyryl-biocytin
- MALDI-TOF
matrix-assisted laser desorption ionization–time of flight
Footnotes
See commentary on page 4819.
Amino acid substitutions in rhodopsin mutants are designated by using single letter code for the original amino acid, followed by sequence number, and the single letter code for the substituted amino acid.
This is paper 42 in the series “Stucture and Function in Rhodopsin.” Paper 41 is ref. 15.
References
- 1.Berson E L. Invest Opthalmol Visual Sci. 1993;34:1659–1676. [PubMed] [Google Scholar]
- 2.Dryja T P, Berson E L. Invest Opthalmol Visual Sci. 1995;36:1197–1200. [PubMed] [Google Scholar]
- 3.Sung C H, Davenport C M, Hennessey J C, Maumenee I H, Jacobson S G, Heckenlively J R, Nowakowski R, Fishman G, Gouras P, Nathans J. Proc Natl Acad Sci USA. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macke J P, Davenport C M, Jacobson S G, Hennessey J C, Gonzalez-Fernandez F, Conway B P, Heckenlively J, Palmer R, Maumenee I H, Sieving P, et al. Am J Hum Genet. 1993;53:80–89. [PMC free article] [PubMed] [Google Scholar]
- 5.Inglehearn C F, Keen T J, Bashir R, Jay M, Fitzke F, Bird A C, Crombie A, Bhattacharya S S. Hum Mol Genet. 1992;1:41–45. doi: 10.1093/hmg/1.1.41. [DOI] [PubMed] [Google Scholar]
- 6.Doi T, Molday R S, Khorana H G. Proc Natl Acad Sci USA. 1990;87:4991–4995. doi: 10.1073/pnas.87.13.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anukanth A, Khorana H G. J Biol Chem. 1994;269:19738–19744. [PubMed] [Google Scholar]
- 8.Ridge K D, Lu Z, Liu X, Khorana H G. Biochemistry. 1995;34:3261–3267. doi: 10.1021/bi00010a016. [DOI] [PubMed] [Google Scholar]
- 9.Kaushal S, Khorana H G. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Garriga P, Khorana H G. Proc Natl Acad Sci USA. 1996;93:4554–4559. doi: 10.1073/pnas.93.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwa J, Reeves P J, Klein-Seetharaman J, Davidson F, Khorana H G. Proc Natl Acad Sci USA. 1999;96:1932–1935. doi: 10.1073/pnas.96.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kono M, Yu H, Oprian D D. Biochemistry. 1998;37:1302–1305. doi: 10.1021/bi9721445. [DOI] [PubMed] [Google Scholar]
- 13.Garriga P, Liu X, Khorana H G. Proc Natl Acad Sci USA. 1996;93:4560–4564. doi: 10.1073/pnas.93.10.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwa J, Garriga P, Liu X, Khorana H G. Proc Natl Acad Sci USA. 1997;94:10571–10576. doi: 10.1073/pnas.94.20.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cha K, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 2000;97:3016–3021. doi: 10.1073/pnas.97.7.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Saleeh S, Gore M, Akhtar M. Biochem J. 1987;246:131–137. doi: 10.1042/bj2460131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karnik S S, Sakmar T P, Chen H B, Khorana H G. Proc Natl Acad Sci USA. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karnik S S, Khorana H G. J Biol Chem. 1990;265:17520–17524. [PubMed] [Google Scholar]
- 19.Moffatt J G, Khorana H G. J Am Chem Soc. 1961;83:663–675. [Google Scholar]
- 20.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1988;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein-Seetharaman J, Hwa J, Cai K, Altenbach C, Hubbell W L, Khorana H G. Biochemistry. 1999;38:7938–7944. doi: 10.1021/bi990013t. [DOI] [PubMed] [Google Scholar]
- 22.Grassetti D R, Murray J F., Jr Arch Biochem Biophys. 1967;119:41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- 23.Brown J R, Hartley B S. Biochem J. 1966;101:214–228. doi: 10.1042/bj1010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldwin J M, Schertler G F, Unger V M. J Mol Biol. 1997;272:144–164. doi: 10.1006/jmbi.1997.1240. [DOI] [PubMed] [Google Scholar]
- 25.Khorana H G. J Biomol Struct Dyn. 2000;11:1–16. doi: 10.1080/07391102.2000.10506598. [DOI] [PubMed] [Google Scholar]
- 26.Krishnaswamy S, Rossmann M G. J Mol Biol. 1990;211:803–844. doi: 10.1016/0022-2836(90)90077-Y. [DOI] [PubMed] [Google Scholar]
- 27.Luo M, Vriend G, Kamer G, Rossmann M G. Acta Crystallogr B. 1989;45:85–92. doi: 10.1107/s0108768188010894. [DOI] [PubMed] [Google Scholar]