Summary
TcdB, an intracellular bacterial toxin that inactivates small GTPases, is a major Clostridium difficile virulence factor. Recent studies have found that TcdB produced by emerging/hypervirulent strains of C. difficile is more potent than TcdB from historical strains, and in the current work, studies were performed to investigate the underlying mechanisms for this change in TcdB toxicity. Using a series of biochemical analyses we found that TcdB from a hypervirulent strain (TcdBHV) was more efficient at autoprocessing than TcdB from a historical strain (TcdBHIST). TcdBHV and TcdBHIST were activated by similar concentrations of IP6; however, the overall efficiency of processing was 20% higher for TcdBHV. Using an activity based fluorescent probe (AWP19) an intermediate, activated but uncleaved, form of TcdBHIST was identified, while only a processed form of TcdBHV could be detected under the same conditions. Using a much higher concentration (200 µM) of the probe revealed an activated uncleaved form of TcdBHV, indicating a preferential and more efficient engagement of intramolecular substrate than TcdBHIST. Futhermore, a peptide-based inhibitor (Ac-GSL-AOMK), was found to block the cytotoxicity of TcdBHIST at a lower concentration than required to inhibit TcdBHV. These findings suggest that TcdBHV may cause increased cytotoxicity due to more efficient autoprocessing.
Keywords: Clostridium difficile, Toxin B, NAP1, cysteine protease
INTRODUCTION
Clostridium difficile-associated disease (CDAD) is a serious health care problem for hospitalized patients and elderly patients in long-term nursing facilities (McFarland et al., 1989; Simor et al., 1993; Bartlett, 1992; Redelings et al., 2007; Gerding, 2010). The disease has also recently emerged in healthy individuals within the general population (Klein et al., 2006; Hirschhorn et al., 1994; Wilcox et al., 2008; CDC, 2008). In a common scenario, CDAD occurs when patients undergoing antibiotic treatments are infected with spores of C. difficile residing within the hospital (McFarland et al., 1989). It is thought that spores enter the new host by ingestion, survive the stomach’s acidic environment, and then germination is triggered by bile salts (Sorg and Sonenshein, 2008; Wilson et al., 1985; Wilson, 1983), although many details of this infection need further investigation. After germination, C. difficile colonizes the large intestines and releases toxins that cause localized inflammation and then systemic damage (Sullivan et al., 1982; Taylor et al., 1981; Abrams et al., 1980; Hamm et al., 2006; Libby et al., 1982). Thus, the activity of C. difficile toxins influences the outcome of CDAD.
CDAD frequency, severity, and mortality have increased over the past decade. A recent report by Karas et al. found a striking difference in the mortality rate prior to and following the year 2000 (Karas et al., 2010). Before 2000 the mortality rate of CDAD patients was estimated to be 3.64% and after 2000 the mortality rate increased to over 8.0% (Redelings et al., 2007). The increases in disease severity, frequency, and mortality are directly correlated with emergence of a hypervirulent strain of C. difficile termed North American Pulsovar 1 (NAP1), Restriction Endonuclease Assay Type BI, and Ribotype 027 C. difficile strain (referred to as C. difficile NAP1 herein) (McDonald et al., 2005; Muto et al., 2005). C. difficile NAP1 may exhibit several characteristics that could explain the strain’s role in increasing the morbidity and mortality of this disease. C. difficile NAP1 is fluoroquinolone resistant, and some groups have proposed these strains sporulate more efficiently than historical strains, and possibly express higher levels of the two major C. difficile toxins (TcdA and TcdB) (Akerlund et al., 2008; Bourgault et al., 2006; Drudy et al., 2006; Drudy and Kyne, 2007; Warny et al., 2005; Merrigan et al., 2010). Additionally, recent reports by us and others have revealed that TcdB expressed by C. difficile NAP1 (TcdBHV) is more cytotoxic than TcdB from a historical strain of C. difficile (TcdBHIST) (Stabler et al., 2009; Lanis et al., 2010). Many of the properties originally attributed to the increased virulence of the NAP1 strains are controversial, as recent reports suggest neither sporulation characteristics nor toxin levels correlate with hypervirulent strain type or clade (Carter et al., 2011; Burns et al., 2011). Thus, increased toxicity of TcdBHV is a reasonable explanation, at least in part, for the heightened virulence of hypervirulent strains of C. difficile.
TcdB (~270 kD) is an intracellular bacterial toxin that glucosylates small GTPases from the Rho family of proteins (Just et al., 1995). TcdB is thought to engage a yet undefined cell surface receptor and enter the cell via receptor-mediated endocytosis; acidification of the endosome and formation of ion-conducting channels occur during cell entry by TcdB (Florin and Thelestam, 1986; Florin and Thelestam, 1983; Qa'Dan et al., 2000; Barth et al., 2001; Papatheodorou et al., 2010; von Eichel-Streiber et al., 1992). Upon exposure to the cytoplasm TcdB binds inositol hexakisphosphate (IP6), which activates the toxin’s intramolecular cysteine protease domain (CPD) (Reineke et al., 2007; Egerer et al., 2007; Egerer et al., 2009). Following autoprocessing by the CPD, the glucosyltransferase domain is released into the cytoplasm where it inactivates target substrates (Reineke et al., 2007; Rupnik et al., 2005; Pfeifer et al., 2003). In a previous study we reported that TcdBHV entered cells rapidly and proposed that TcdBHV may be translocated into the cytosol at an earlier stage of endosomal trafficking than TcdBHIST (Lanis et al., 2010). Accelerated cell entry by TcdBHV was congruent with our observation that TcdBHV undergoes a hydrophobic transition at a higher pH than TcdBHIST (Lanis et al., 2010). Such a pH-induced structural change has previously been shown to be a prelude to membrane insertion by TcdB (Qa'Dan et al., 2000; Barth et al., 2001). Whether autoprocessing of TcdBHV might also be activated more quickly or efficiently by IP6 is not known, but such a phenotype would support a model in which TcdBHV can enter cells more rapidly than TcdBHIST.
In the current study we were interested in knowing if the previously observed TcdBHV phenotypes were related to highly efficient autoprocessing. The findings from this study support this notion and provide insight into a mechanism underlying the heightened toxicity of TcdB produced by a hypervirulent strain of C. difficile.
RESULTS
Comparison of TcdBHV and TcdBHIST Autoprocessing
We have reported previously that TcdBHV is able to undergo pH-induced hydrophobic transitions at a higher pH than TcdBHIST (Lanis et al., 2010). This allows TcdBHV to translocate into the cytoplasm at an earlier stage of endocytosis, as we confirmed using a chase experiment involving lysosomotropic inhibitors (Lanis et al., 2010). Yet, a conundrum of this observation is that earlier translocation into the cytoplasm should only increase the rate of intoxication, not the extent of cytotoxicity of TcdBHV. This led us to investigate other activities related to cell entry that might be enhanced or more efficient in TcdBHV.
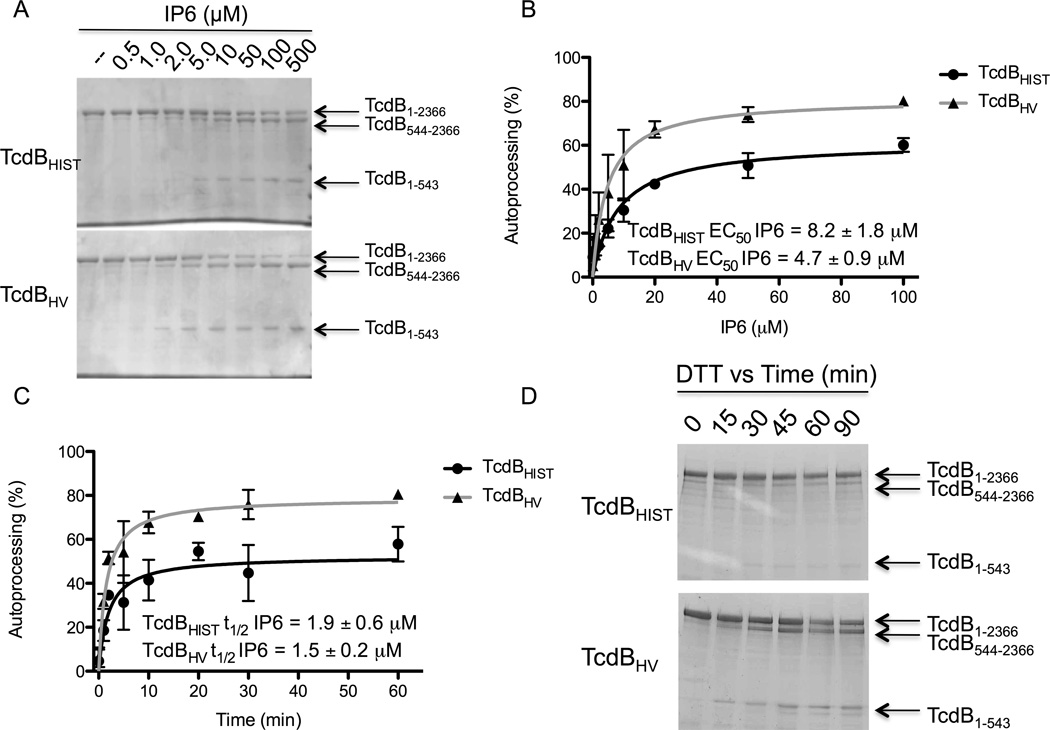
Autoproteolytic activity could be a critical step in cell entry, and as such, differences in this activity between TcdBHV compared to TcdBHIST might contribute to variations in cytotoxicity. We first performed an experiment to determine if TcdBHV and TcdBHIST differed in their sensitivity to activation by IP6. The two forms of TcdB were incubated with IP6, ranging in concentration from 500 nM to 500 µM. The toxins were allowed to incubate with the various concentrations of IP6 for 1 h and the reaction was resolved by SDS-PAGE. The results from this comparison are shown in Fig. 1A. Autoprocessing of TcdBHV and TcdBHIST was activated by similar concentrations of IP6, with an EC50 of 8.2 ± 1.9 µM and 4.7 ± 0.9 µM of IP6 respectively (Fig. 1B). These concentrations are comparable to the reported binding constant of IP6 at 2 µM (Egerer et al., 2009), and since intracellular concentrations of IP6 typically exceed 50 µM, the minor differences in sensitivity to IP6 most likely do not account for the variation in toxicity between TcdBHV and TcdBHIST (Irvine et al., 2001).
Fig. 1. In vitro processing of TcdBHIST and TcdBHV in response to inositol hexakisphosphate (IP6).
(A) Coomassie stained SDS-PAGE of TcdBHIST (top) or TcdBHV (bottom) that was treated with 500 nM to 500 µM of IP6. Full length TcdB (1-2366) and processed TcdB (544-2366 and 1-543) are indicated by the arrows. (B) Activation of autoprocessing by IP6. The percent autoprocessing of TcdBHIST (black) and TcdBHV (gray) was determined by comparing the relative amounts of TcdB1-543 and TcdB544-2366 to full-length toxin using densitometry. The activation constant (EC50), is defined as the concentration of IP6 at which half-maximal activity occurs. Error bars represent the S.D. of 4 independent experiments and toxin preparations. (C) Comparison of the rate of TcdB processing in response to IP6. TcdBHIST or TcdBHV were incubated with 100 µM of IP6 for the time points indicated and the reactions resolved by SDS-PAGE. The percent autoprocessing of TcdBHIST (black) and TcdBHV (gray) was determined by comparing the relative amounts of TcdB1-543 and TcdB544-2366 to full-length toxin using densitometry. The time to half-max (t1/2), is defined as the time at which half-maximal activity occurs. Error bars represent the S.D. of 4 independent experiments and toxin preparations. (D) Coomassie stained SDS-PAGE of TcdBHIST (top) or TcdBHV (bottom) that was treated with 5 mM DTT for up to 90 min. Full length TcdB (1-2366) and processed TcdB (544-2366 and 1-543) are indicated by the arrows.
Although we did not detect a significant difference in minimal concentrations of IP6 needed to activate TcdBHV compared to TcdBHIST, we did note that a higher percentage of total TcdBHV protein was processed under these conditions. To examine this phenotype further, in the next experiment the two forms of TcdB were incubated with 100 µM IP6 (a condition of excess IP6) and the extent of processing was determined at time-points between 1 min and 60 min by using densitometry to quantify the relative amounts of processed and unprocessed toxin. As shown in Fig. 1C, TcdBHV underwent almost 80% cleavage within the first hour of incubation. In contrast, maximal processing of TcdBHIST is under 60%. This difference in percent autoprocessing does not seem to be due to the rate of activation, as both toxins reached the half-maximal level of proteolysis by 2 min (Fig. 1C). Finally, we wanted to confirm that these were universal differences in CPD activity between TcdBHIST and TcdBHV and not a function of experimental conditions. Dithiothreitol (DTT) has also been shown to activate processing of TcdB, and in Fig. 1D the in vitro cleavage reaction has been repeated on TcdBHIST and TcdBHV using 5 mM DTT in place of IP6. While DTT is not as efficient an activator of proteolysis as IP6, the data indicate that DTT-mediated activation was substantially more efficient for TcdBHV in comparison to TcdBHIST. We examined this difference over a time-course of 90 min, and while TcdBHV was activated by DTT within 15 min and reached maximum activation within approximately 1 h, the activation of TcdBHIST by DTT during this time-course was just slightly above the level of detection (Fig. 1D). These data indicate that TcdBHV is more efficient than TcdBHIST in autoproteolytic processing, and that these differences are independent of rate and interaction with IP6.
Probing the Activation State of TcdBHV and TcdBHIST using an Activity-Based Fluorescent Probe
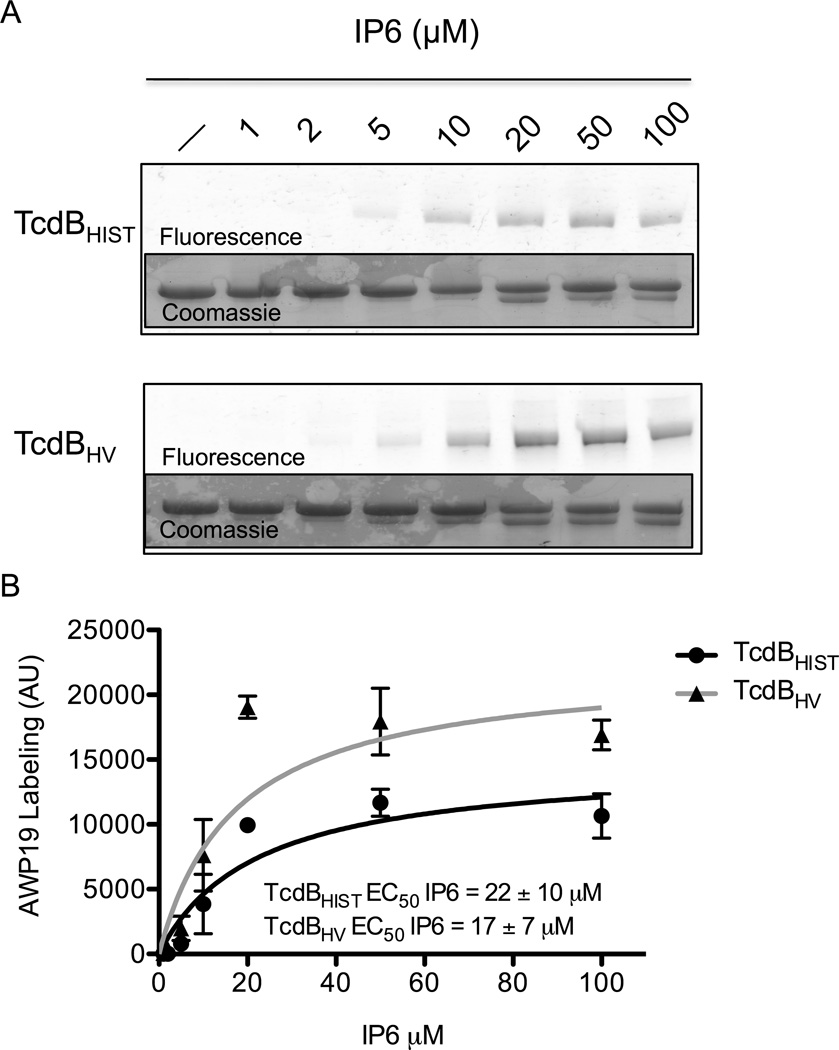
Previous work by Puri et al. described a fluorescent small molecule designed to interact with TcdB when the CPD domain has been activated (Puri et al., 2010). This probe is fluorescently labeled SL-AOMK that was designed from a peptide-based Ac-GSL-AOMK protease inhibitor and can interact with the protease domain by mimicking the natural substrate and binding covalently to the catalytic cysteine. The probe (AWP19) depends on availability of the active site to bind; therefore, AWP19 is a useful tool to compare the IP6 induced conformational changes and subsequent activation of the CPD between TcdBHIST and TcdBHV (Puri et al., 2010). Further work by Shen et al. demonstrated that this probe (AWP19) could be used to precisely measure the kinetics and substrate binding characteristics of the TcdB-CPD active site (Shen et al., 2011). Using a FITC conjugated version of AWP19, we examined the differences in IP6-induced changes to the active site between TcdBHIST and TcdBHV. In this experiment, both forms of TcdB were incubated with a range of IP6 concentrations and, following a 1 h incubation, AWP19 was added in 10-fold molar excess to the proteins. The reactions were then resolved by SDS-PAGE and the extent of labeling at each concentration of IP6 was determined by scanning for FITC fluorescence. As shown in Fig. 2A, both TcdBHIST and TcdBHV labeled with AWP19 could be detected following incubation with 5 µM IP6, although the level of labeled protein at this concentration was substantially less than that observed at higher concentrations of IP6. Densitometry of gels from 4 independent experiments using different toxin preparations indicates that the total labeling of TcdBHV is much higher than TcdBHIST, however, equal amounts of IP6 are required for half-maximal activity of both toxins (Fig 2B). The predominant labeling of TcdB by AWP19 is observed in the TcdB544-2366 fragment, indicating that the probe is only able to detect processed toxin under these conditions. Therefore, the difference in AWP19 labeling between the toxin variants can be attributed to the difference in maximal proteolysis between TcdBHIST and TcdBHV, consistent with the findings in Fig 1A.
Fig. 2. Comparison of cysteine protease activation with the activity-based probe AWP19.
(A) Representative fluorescent gel image of the IP6 induced labeling of TcdBHIST (top) or TcdBHV (bottom) by AWP19. The in vitro cleavage assay was allowed to come to completion with the indicated concentration of IP6, then the gel was imaged for FITC fluorescence. Inset: Coomassie stained gel verifying equal loading. (B) Densitometry indicating the average fluorescence intensity (arbitrary units) of TcdBHIST (black) and TcdBHV (gray) at the indicated IP6 concentrations. The activation constant (EC50), is defined as the concentration of IP6 at which half-maximal activity occurs. Error bars represent the S.D. of 4 independent experiments.
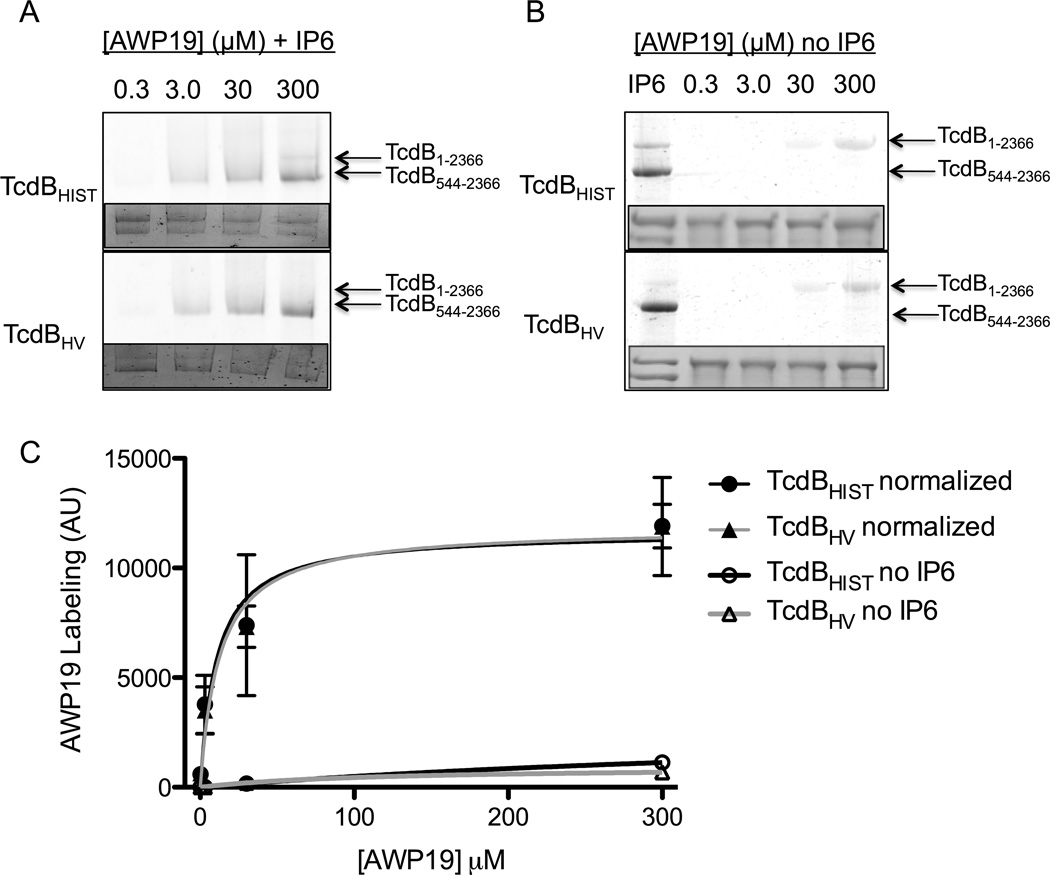
To continue utilizing the probe as an indicator of differential CPD activity, our next experiments concentrated on validating whether the probe interacts with equal affinity to both TcdBHIST and TcdBHV. To this end, 0.3 µM of toxin was incubated with or without 100 µM IP6, then AWP19 was added from 0.3 µM up to 300 µM. The samples were analyzed by SDS-PAGE for FITC fluorescence and the percent processing by coomassie stain. As in previous experiments, TcdBHV exhibits greater fluorescence than TcdBHIST (Fig 3A). However, once the fluorescence is normalized by the percent of processing it is clear that the extent of labeling is nearly identical between the different strains of TcdB (Fig. 3B). Another concern was whether the probe could differentially bind to the CPD without activation by IP6. While a prolonged 24 hr incubation of either TcdBHIST or TcdBHV with AWP19 does lead to some minimal labeling (Fig. 3B), the level of detection is much lower than with activated CPD as determined by densitometry (Fig. 3C). These experiments validate that there is no differential affinity of AWP19 to either TcdBHIST or TcdBHV, and the probe is a valuable tool for studying subtle differences in CPD activity between toxins.
Fig. 3. AWP19 probe binding affinity.
(A) Representative fluorescent gel image of the IP6 induced labeling of TcdBHIST (top) or TcdBHV (bottom) in response to AWP19 concentration. The in vitro cleavage assay was allowed to come to completion in the presence of 100 µM IP6, then the gel was imaged for FITC fluorescence. (B) Representative fluorescent gel image of the labeling of TcdBHIST (top) or TcdBHV (bottom) in response to AWP19 alone. The probe indicated concentration of probe was incubated with 0.3 µM of TcdB overnight, then the gel was imaged for FITC fluorescence. The lane marked IP6 indicateds the control in which IP6 was included. (C) Densitometry indicating the average fluorescence intensity (arbitrary units) of TcdBHIST (black) and TcdBHV (gray) at the indicated AWP19 concentrations which has either been normalized to the percent processing or incubated without IP6. Error bars represent the S.D. of triplicate samples.
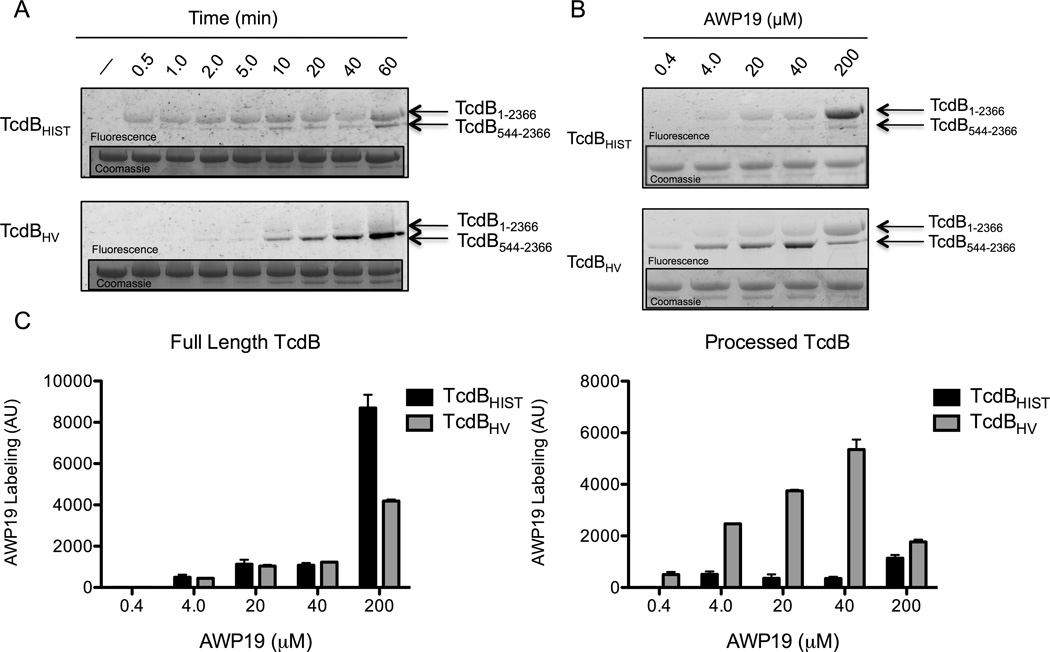
We next used the AWP19 probe to examine activation of TcdB over a specific time-course. TcdBHIST or TcdBHV were incubated with IP6 and AWP19 simultaneously, and the level of activated protein was again determined by examining the extent of fluorescent protein resolved by SDS-PAGE. Interestingly, when IP6 and AWP19 were added to proteins at the same time, a full-length (unprocessed) form of TcdBHIST was detected. In contrast only the processed form of TcdBHV was detected in this assay and at a much later time-point (Fig. 4A).
Fig. 4. AWP19 labeling of TcdB in response to rate of IP6 activation and AWP19 probe concentration.
(A) Representative fluorescent gel image of the AWP19 labeling at the indicated time points after activation of 0.5 µM of TcdBHIST (top) or TcdBHV (bottom) with 25 µM IP6. Full length TcdB (1-2366) and processed TcdB (544-2366) are indicated by the arrows. Inset: Coomassie stained gel verifying equal loading. (B) Representative fluorescent gel image of AWP19 labeling in response to increasing concentrations of the fluorescent probe. 0.4 µM of TcdBHIST (top) or TcdBHV (bottom) were incubated simultaneously with 25 µM of IP6 and the indicated concentration of AWP19 for 1 h. Full lengths TcdB (1-2366) and processed TcdB (544-2366) are indicated by the arrows. Inset: Coomassie stained gel verifying equal loading. (C) Densitometry indicating the average fluorescence intensity (arbitrary units) of TcdBHIST (black) and TcdBHV (gray) at the indicated AWP19 concentrations. Error bars represent the S.E.M. of triplicate samples.
We envisioned two possible explanations for detecting activated, but unprocessed, TcdBHIST. First, the kinetics of autoproteolysis may occur in a manner slow enough to capture an intermediate form of TcdBHIST, but occur much faster in TcdBHV. Alternatively, the binding affinity for the intramolecular substrate could be stronger in TcdBHV than in TcdBHIST, which would preclude competitive binding of AWP19 in TcdBHV. Only after the substrate has been cleaved could TcdBHV then bind the probe. We reasoned that if the latter was true, we should be able to detect the intermediate form of TcdBHV by adding higher concentrations of AWP19 to the reaction, thereby shifting the reaction to favor binding of the probe rather than the intramolecular domain. Therefore, we next assayed AWP19 labeling of TcdB under the conditions of an increasing ratio of probe to toxin. When TcdBHIST and TcdBHV were incubated with a 50-fold excess (20 µM) of AWP19 the full-length (unprocessed) form of TcdBHV could be detected (Fig. 4B). Further increases in probe concentration resulted in greater percentages of labeled full-length TcdBHV and a 500-fold excess (200 µM) shifted the predominant AWP19 labeling to the unprocessed form of TcdBHV (Fig. 4B–C). Additionally, the addition of excess probe can inhibit the processing of TcdBHV, supporting the idea that the probe is outcompeting the intramolecular substrate for position in the active site (Fig. 4B inset). In contrast, AWP19 labeling of the processed form of TcdBHIST was much less evident under these conditions, consistent with the data in Fig. 4A, and the uncleaved form of the toxin was predominantly labeled under all probe concentrations (Fig. 4B–C).
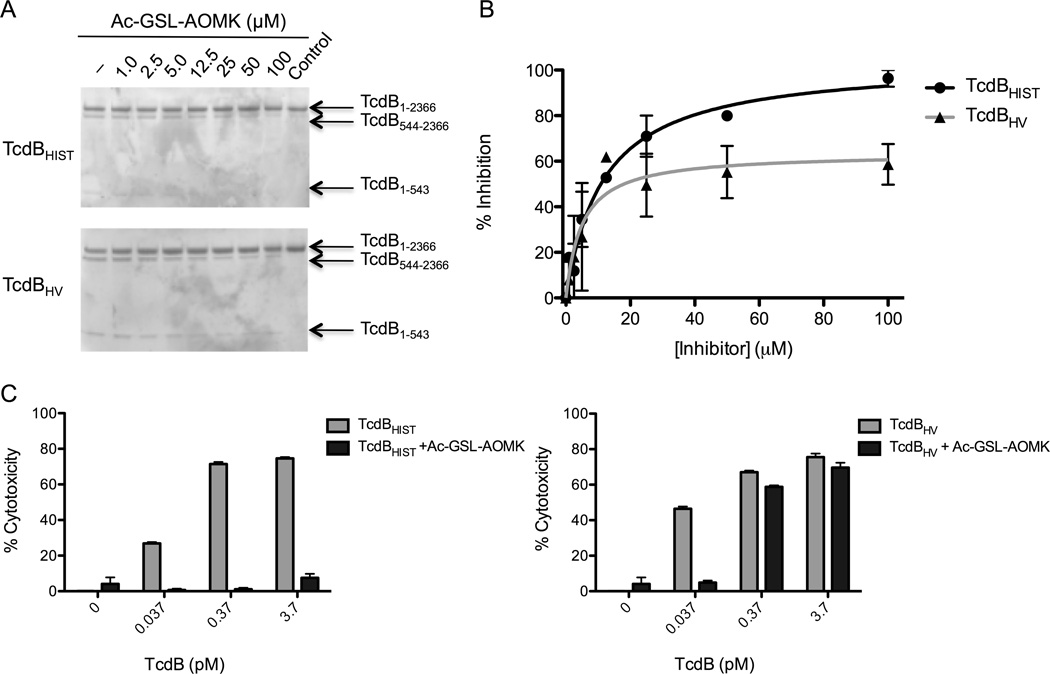
Inhibition of Toxin function with the CPD inhibitor Ac-GSL-AOMK
The data thus far indicated that the entire cleavage process occurs much more efficiently in TcdBHV, possibly due to an increased affinity for intramolecular substrate. To further investigate this process, we utilized a chemical inhibitor, previously described by Puri et al. (Puri et al., 2010), to inhibit the CPD. We reasoned that a greater processing efficiency and a potential increased affinity to substrate in TcdBHV might affect the binding and inhibitory capacity of the CPD inhibitor. The toxins were pre-incubated with up to 100 µM of Ac-GSL-AOMK and then IP6 was added to 25 µM. Consistent with the difference in AWP19 labeling, we also observed a noticeable difference in the inhibition of processing between TcdBHIST and TcdBHV. The difference in the concentration of inhibitor necessary to initiate blockage of IP6-induced processing of TcdBHIST or TcdBHV seems to be minimal, as evidence of inhibition was detectable in both toxins at concentrations around 12.5 µM Ac-GSL-AOMK (Fig. 5A). Densitometry of gels from 3 independent experiments revealed that the percent inhibition of TcdBHV reaches a maximum around 60% while nearly 100% of proteolysis is blocked in TcdBHIST (Fig. 5B). Just as AWP19 showed no effect in the absence of IP6, the inhibitor also had no effect on either TcdBHIST or TcdBHV without IP6 (data not shown), indicating that the inhibitor cannot bind in the absence of activation. Together, these experiments demonstrate that differences in proteolytic inhibition are not due to differential affinity or nonspecific binding of the inhibitor and support the interpretations from Fig. 4 in which TcdBHV has a structure that reduces such competitive binding of the inhibitor to the active site.
Fig. 5. Chemical inhibition of the TcdB CPD with Ac-GSL-AOMK.
(A) 2.5 µg of TcdBHIST (top) or TcdBHV (bottom) were pre-incubated with 1 µM to 100 µM of Ac-GSL-AOMK for 1 h. Then 25 µM IP6 was added and the samples were incubated for 1 h and separated by SDS-PAGE. Full length TcdB (1-2366) and processed TcdB (544-2366 and 1-543) are indicated by the arrows. (B) Quantification of the percentage of inhibition of TcdBHIST (black) and TcdBHV (gray) as determined by densitometry. (C) CHO cell cytotoxicity of TcdBHIST (left) or TcdBHV (right) that have been treated with 100 µM of the CPD inhibitor Ac-GSL-AOMK. Error bars represent the S.E.M. of triplicate samples.
Next, we compared the inhibition of toxin function in a cell culture model. For these experiments, TcdBHIST and TcdBHV were pre-incubated with 100 µM of the inhibitor in media and the mixture added to CHO cells. Consistent with the in vitro data, we found that Ac-GSL-AOMK provided protection to TcdBHIST treated cells but was not as functional against TcdBHV. Once toxin concentrations approach the TCD50 for TcdBHV, the inhibitor is able to reduce cytopathic effects presumably because a 60% inhibition is sufficient at these low toxin levels. In comparison, the inhibitor prevented cytotoxicity of cells treated with 100-fold higher concentrations of TcdBHIST (Fig. 5C).
Discussion
In the current study we investigated the autoprocessing of TcdB from a hypervirulent strain of C. difficile and compared this with a well-studied form of TcdB from a historical strain. These data indicate TcdBHV autoprocessing occurs at a higher efficiency than autoprocessing by TcdBHIST. Based on the earlier studies and the data presented in the current work, a common theme has emerged. TcdBHV is more efficient during processes of cellular intoxication than TcdBHIST This important difference in the efficiency with which TcdBHV functions may be a fundamental determinant of the increased cytotoxicity of this toxin variant.
Not all large clostridial toxins (LCTs) appear to exhibit the same biochemistry and efficiency of autoprocessing, which supports the idea that TcdBHIST and TcdBHV could differ in their intramolecular proteolytic cleavage. For example, TcdA requires a much higher concentration of IP6 for activation compared to TcdB despite maintaining similar binding of IP6 (Egerer et al., 2007; Pruitt et al., 2009). Unlike other LCTs Clostridium sordellii lethal toxin (TcsL) requires a low pH for efficient autoprocessing (Guttenberg et al., 2011). Interestingly, only full length TcsL required low pH for activation, while a recombinant fragment of just the glucosyltransferase and CPD domain did not (Guttenberg et al., 2011). These data suggest conformational differences encoded outside of the CPD influence the efficiency of autoprocessing, which is also a plausible explanation for the differences in the two forms of TcdB. In fact, this is the first study that utilizes native holotoxin to explore the function of the CPD in context of the full toxin molecule. Our data support the prediction that a structural difference impacts the variation in autoprocessing activity between TcdBHV and TcdBHIST.
Results from the activation probe (AWP19) provide the basis for a model to explain differences in the efficiency of autoprocessing by TcdBHIST and TcdBHV. The first major difference revealed by the studies using AWP19 is that TcdBHIST transitions from activated state to autocleavage more slowly than TcdBHV. As shown in Fig. 4A, full-length unprocessed TcdBHIST was detected within less than a minute of addition of IP6, suggesting the protein was activated but had not yet engaged and cleaved intramolecular substrate. With extended IP6 incubation, TcdBHIST shifted to its processed form. In contrast, only trace levels of TcdBHV were detected in the unprocessed state following addition of IP6. Detection with AWP19 was substantially slower for TcdBHV, but in contrast to TcdBHIST the prominent species found was the processed form of the toxin. An interpretation of these data is that TcdBHV is more efficient at autoprocessing because the protein is in a conformation that highly favors intramolecular substrate. Thus, limited detection of unprocessed TcdBHV can be explained by the fact that endogenous substrate blocks binding by AWP19 and only after cleavage is complete can the probe access the catalytic region. In contrast, TcdBHIST is less efficient at autoprocessing due to limitations in its capacity to interact with intramolecular substrate. This results in AWP19 competing with substrate and labeling the activated form TcdBHIST.
The two strains of TcdB also demonstrate differential sensitivity to Ac-GSL-AOMK in the activation assay, and TcdBHV was much more resistant than TcdBHIST when the effects of the inhibitor were examined in a cellular intoxication assay. These results fit nicely with a model wherein subtle conformational differences account for the variation in autoprocessing. The CPD inhibitor, Ac-GSL-AOMK, is smaller in size than AWP19, which contains the bulkier FITC compound. Despite the smaller structure of Ac-GSL-AOMK access to the CPD is still restricted to activation by IP6 in both types of TcdB. How the inhibitor is able to effectively block TcdBHIST but not TcdBHV is related to the difference in binding of the AWP19 probe to the CPD. The probe has an equal affinity to both TcdBHIST and TcdBHV when the toxin has been pre-activated and normalized for the percent of processing. So, the difference in probe and inhibitor affinity to activated unprocessed toxin seems to be related to a difference in TcdBHV conformation that restricts binding to the CPD active site. The results of the inhibitor assays reveal another fundamental difference in the extent of toxic activity between TcdBHIST and TcdBHV.
Variation in structure and function between TcdBHIST and TcdBHV could also have additional consequences on cytotoxicity, such as altering the way host cells defend against these toxins. S-nitrosylation of C. difficile toxins in vivo was recently reported by Savidge et al. as a host mechanism to inhibit toxin function (Savidge et al., 2011). The authors found that IP6 and IP7 induced conformational changes to the toxin allowed for nitrosylation of the catalytic cysteine, leading to inhibition of toxin processing and a subsequent reduction in virulence (Savidge et al., 2011). Our studies show that TcdBHV is processed much more efficiently, and also provide evidence that access to the catalytic cysteine is much more restricted than in TcdBHIST. The way in which s-nitrothiols are able to interact with variants of TcdB inside the cell provide yet another explanation for the increased cytotoxicity of TcdBHV.
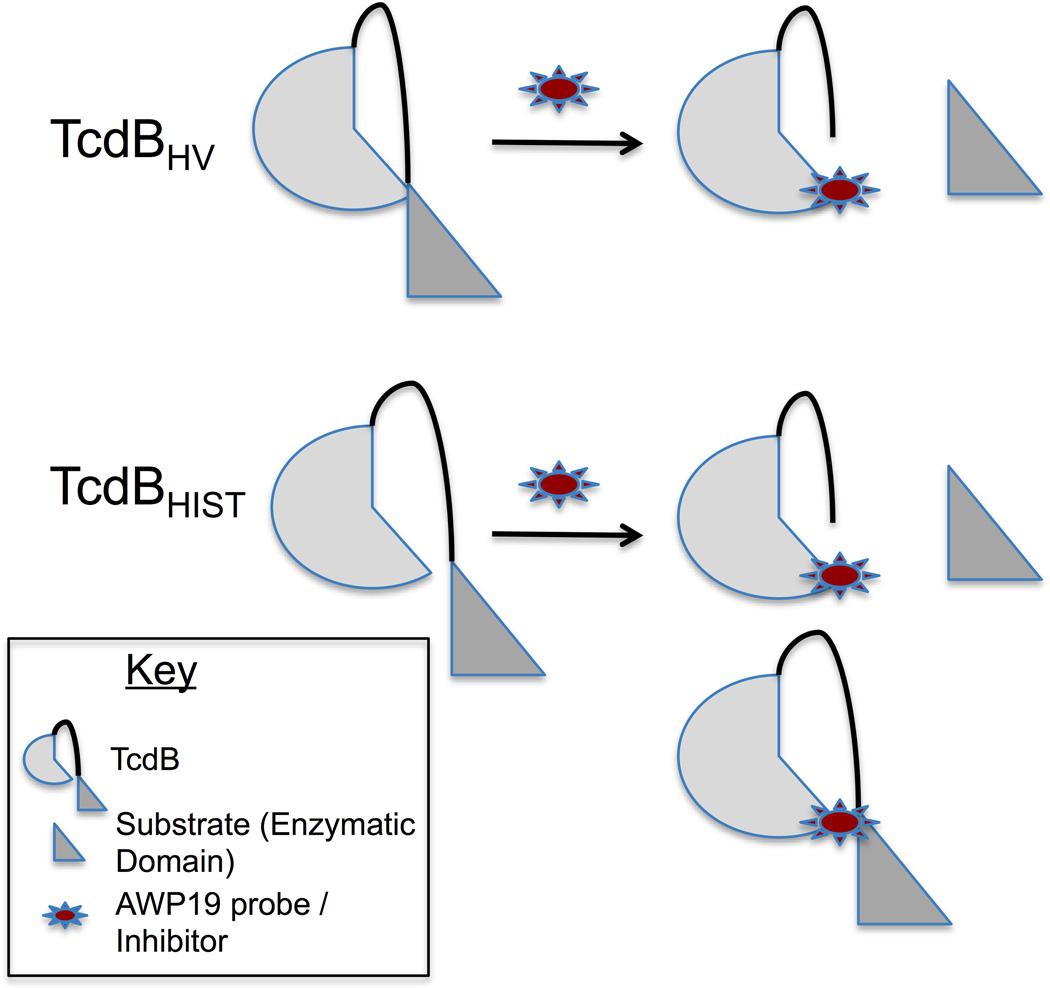
TcdBHIST and TcdBHV exhibit differences in their primary sequence, rates of cell entry, efficiency of autoprocessing, and cytotoxicity. The findings to date all point to the fact that TcdBHV is more cytotoxic by virtue of some fundamental differences in the structure of this protein. An appealing model is one in which TcdBHV is a more flexible molecule than TcdBHIST. Fig. 6 illustrates this model in which the increased flexibility of TcdBHV allows the toxin to more readily access intramolecular substrate. Therefore, this intramolecular interaction prevents binding to the probe or inhibitor until after the substrate has been cleaved. Conversely, TcdBHIST seems to maintain a structure that limits access of the CPD to the intramolecular substrate. In this way the interaction with the substrate is not sufficient to block binding by the probe and therefore the activated CPD can become labeled regardless of proteolysis (Fig. 6). This model supports the increased processing efficiency of TcdBHV and might also explain our previous observations on the extent of pH-induced conformational changes in TcdBHV (Lanis et al., 2010). Further structural studies will be needed to refine this model and determine how sequence changes in TcdBHV influence the overall folding and induced conformational changes of this protein.
Fig. 6. Intramolecular interactions of TcdBHIST and TcdBHV.
A working model demonstrating the fundamental differences in the CPD conformation and activity between TcdBHIST and TcdBHV. TcdBHV (top) undergoes an intramolecular interaction that precludes binding to the probe until after substrate is cleaved. The intramolecular reaction of TcdBHIST (bottom) is not sufficient to block binding by the probe, thus cleaved and uncleaved toxin can be labeled.
Experimental Procedures
Purification of Native TcdB
TcdBHIST and TcdBHV were isolated from C. difficile 10463 and C. difficile BI17 (provided by Dale Gerding) respectively, as previously described (Lanis et al., 2010; Krivan and Wilkins, 1987; Qa'Dan et al., 2000) The protein purity was assessed by SDS-PAGE, and the concentration determined by the Bradford method (Bio-Rad).
In vitro TcdB processing assays
The autoproteolysis assays were performed in 25 µl of 20 mM Tris-HCl pH 8.0, containing 2.5 µg of either TcdBHIST or TcdBHV and the indicated concentration of either Inositol hexakisphosphate (IP6), or dithiothreitol (DTT), to induce cleavage (all purchased from Sigma). Unless otherwise indicated, the samples were incubated at 37°C for 1 h, then boiled for 5 min in SDS sample buffer containing β-mercaptoethanol (BME) to halt the reaction. The samples were then separated by 8% SDS-PAGE and the toxin fragments visualized by coomassie blue stain.
Compound synthesis
FITC-AWP19 was synthesized by combining H2N-aminohexanoic-SL-AOMK (1 equiv.) with 5(6)-Carboxyfluorescein N-hydroxysuccinimide ester (Sigma) and N,N-Diisopropylethylamine (Sigma) (5 equiv.) in DMSO for one hour and then purifying directly by HPLC. The identity and purity of the compound was characterized by LCMS.
AWP19 probe labeling of TcdB
For most of the experiments, processing of 0.3 µM TcdBHIST or TcdBHV was first stimulated with the indicated concentration of IP6 (Sigma) in 24 µl of 20 mM Tris-HCl, pH 8.0. The reaction was allowed to complete for 1h at 37°C, then AWP19 was added to a final concentration of 5 µM, bringing the total volume to 25 µl. The AWP19 labeling reaction was then continued at 37°C for one additional hour unless otherwise noted. 10 µl of SDS sample buffer containing BME was then added to the samples, and the samples were heated for 5 min at 95°C. 35 µl of each sample was resolved by 8% SDS-PAGE. Fluorescence of bands labeled by the AWP19-FITC probe was detected using an Alpha Innotech FluorChem Q imager, and then the gel was stained with coomassie to guarantee equal loading. For rate of AWP19 labeling experiments, 25 µM IP6 and 0.4 µM to 200 µM of AWP19 were added to 0.4 µM TcdBHIST or TcdBHV in 20 mM Tris-HCl, pH 8.0, simultaneously and incubated at 37°C for the time points indicated. The samples were then analyzed as described above.
Inactivation of TcdB with Ac-GSL-AOMK
The in vitro inhibition assays were performed in 25 µl of 20 mM Tris-HCl pH 8.0, containing 2.5 µg of either TcdBHIST or TcdBHV and up to 100 µM Ac-GSL-AOMK. The samples were incubated at 37°C for 30 min then IP6 (Sigma) was added to a final concentration of 25 µM. The reactions were incubated for 1 additional h at 37°C then heated for 5 min at 95°C in SDS sample buffer containing β-mercaptoethanol (BME) to halt the reaction. The samples were then separated by 8% SDS-PAGE and the toxin fragments visualized by coomassie blue stain.
To assess the inactivation of TcdB in cell culture, CHO-K1 cells (ATCC) were seeded in 96 well plates at a density of 1–2 × 104 cells per well in F12-K media (ATCC) supplemented with 10% FBS. Prior to the assay, 0.037, 0.37, or 3.7 pM TcdBHIST and TcdBHV were preincubated with 100 µM of the inhibitor Ac-GSL-AOMK for 30 min in 100 µl of F12-K media. 100 µl of this mixture was then added to each well in triplicate, and the cells were incubated at 37°C in the presence of 6% CO2 for 24 hrs and cell viability was measured by CCK-8 (Dojindo).
Data Quantification and Non-linear Regression
Labeling and percent processing reactions were quantified using the program ImageJ (http://imagej.nih.gov/ij, NIH). The values were corrected for background and control and plotted against IP6 concentration, time, AWP19 concentration, or inhibitor concentration respectively. The graphs were curve-fit and the EC50 was determined using the Michaelis-Menten function on GraphPad Prism.
References
- Abrams GD, Allo M, Rifkin GD, Fekety R, Silva J., Jr Mucosal damage mediated by clostridial toxin in experimental clindamycin- associated colitis. Gut. 1980;21:493–499. doi: 10.1136/gut.21.6.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerlund T, Persson I, Unemo M, Noren T, Svenungsson B, Wullt M, Burman LG. Increased sporulation rate of epidemic Clostridium difficile type 027/NAP1. J Clin Microbiol. 2008;46:1530–1533. doi: 10.1128/JCM.01964-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H, Pfeifer G, Hofmann F, Maier E, Benz R, Aktories K. Low pH-induced formation of ion channels by Clostridium difficile toxin B in target cells. J Biol Chem. 2001;276:10670–10676. doi: 10.1074/jbc.M009445200. [DOI] [PubMed] [Google Scholar]
- Bartlett JG. Antibiotic-associated diarrhea. Clin. Infect. Dis. 1992;15:9. doi: 10.1093/clind/15.4.573. [DOI] [PubMed] [Google Scholar]
- Bourgault AM, Lamothe F, Loo VG, Poirier L the CDAD-CSI Study Group. In vitro susceptibility of Clostridium difficile clinical isolates from a multi-institutional outbreak in southern Quebec, Canada. Antimicrob Agents Chemother. 2006;50:3473–3475. doi: 10.1128/AAC.00479-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DA, Heeg D, Cartman ST, Minton NP. Reconsidering the sporulation characteristics of hypervirulent Clostridium difficile BI/NAP1/027. PLoS One. 2011;6:e24894. doi: 10.1371/journal.pone.0024894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter GP, Douce GR, Govind R, Howarth PM, Mackin KE, Spencer J, et al. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical Isolate of Clostridium difficile. PLoS Pathog. 2011;7:e1002317. doi: 10.1371/journal.ppat.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Centers for Disease Control and Prevention. Surveillance for community-associated Clostridium difficile--Connecticut, 2006. Morb Mortal Wkly Rep. 2008:4. [PubMed] [Google Scholar]
- Drudy D, Kyne L, O'Mahony R, Fanning S. gyrA mutations in fluoroquinolone-resistant Clostridium difficile PCR-027. Emerg Infect Dis. 2007;13:2. doi: 10.3201/eid1303.060771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drudy D, Quinn T, O'Mahony R, Kyne L, O'Gaora P, Fanning S. High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J Antimicrob Chemother. 2006;58:1264–1267. doi: 10.1093/jac/dkl398. [DOI] [PubMed] [Google Scholar]
- Egerer M, Giesemann T, Herrmann C, Aktories K. Auto-catalytic processing of Clostridium difficile toxin B - binding of inositol hexakisphosphate. J Biol Chem. 2009;284:7. doi: 10.1074/jbc.M806002200. [DOI] [PubMed] [Google Scholar]
- Egerer M, Giesemann T, Jank T, Satchell KJF, Aktories K. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J Biol Chem. 2007;282:25314–25321. doi: 10.1074/jbc.M703062200. [DOI] [PubMed] [Google Scholar]
- Florin I, Thelestam M. Lysosomal involvement in cellular intoxication with Clostridium difficile toxin B. Microb Pathog. 1986;1:373–385. doi: 10.1016/0882-4010(86)90069-0. [DOI] [PubMed] [Google Scholar]
- Florin TM., I Internalization of Clostridium difficile cytotoxin into cultured human lung fibroblasts. Biochim Biophys Acta. 1983;763:10. doi: 10.1016/0167-4889(83)90100-3. [DOI] [PubMed] [Google Scholar]
- Gerding D. Global epidemiology of Clostridium difficile infection in 2010. Infect Control Hosp Epidemiol. 2010;31:S32–S34. doi: 10.1086/655998. [DOI] [PubMed] [Google Scholar]
- Guttenberg G, Papatheodorou P, Genisyuerek S, Wei L, Jank T, Einsle O, Aktories K. Inositol hexakisphosphate-dependent processing of Clostridium sordellii lethal toxin and Clostridium novyi α-toxin. J Biol Chem. 2011;286:14779–14786. doi: 10.1074/jbc.M110.200691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm EE, Voth DE, Ballard JD. Identification of Clostridium difficile toxin B cardiotoxicity using a zebrafish embryo model of intoxication. Proc Natl Acad Sci U S A. 2006;103:14176–14181. doi: 10.1073/pnas.0604725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn LR, Trnka Y, Onderdonk A, Lee MLT, Platt R. Epidemiology of community-acquired Clostridium difficile-associated diarrhea. J Infect Dis. 1994;169:127–133. doi: 10.1093/infdis/169.1.127. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Schell MJ. Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol. 2001;2:12. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- Karas JA, Enoch DA, Aliyu SH. A review of mortality due to Clostridium difficile infection. J Infect. 2010;61:1–8. doi: 10.1016/j.jinf.2010.03.025. [DOI] [PubMed] [Google Scholar]
- Klein EJ, Boster DR, Stapp JR, Wells JG, Qin X, Clausen CR, et al. Diarrhea etiology in a children's hospital emergency department: A prospective cohort study. Clin Infect Dis. 2006;43:807–813. doi: 10.1086/507335. [DOI] [PubMed] [Google Scholar]
- Krivan HC, Wilkins TD. Purification of Clostridium difficile toxin A by affinity chromatography on immobilized thyroglobulin. Infect Immun. 1987;55:1873–1877. doi: 10.1128/iai.55.8.1873-1877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanis JM, Barua S, Ballard JD. Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog. 2010;6:e1001061. doi: 10.1371/journal.ppat.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby JM, Jortner BS, Wilkins TD. Effects of the two toxins of Clostridium difficile in antibiotic-associated cecitis in hamsters. Infect Immun. 1982;36:822–829. doi: 10.1128/iai.36.2.822-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald LC, Killgore GE, Thompson A, Owens RC, Jr, Kazakova SV, Sambol SP, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- McFarland LV, Mulligan ME, Kwok RYY, Stamm WE. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989;320:204–210. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, et al. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol. 2010;192:4904–4911. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto CA, Pokrywka M, Shutt K, Mendelsohn AB, Nouri K, Posey K, et al. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect Control Hosp Epidemiol. 2005;26:273–280. doi: 10.1086/502539. [DOI] [PubMed] [Google Scholar]
- Papatheodorou P, Zamboglou C, Genisyuerek S, Guttenberg G, Aktories K. Clostridial glucosylating toxins enter cells via clathrin- mediated endocytosis. PLoS One. 2010;5:e10673. doi: 10.1371/journal.pone.0010673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G, Schirmer J, Leemhuis J, Busch C, Meyer DK, Aktories K, Barth H. Cellular uptake of Clostridium difficile toxin B: translocation of the n-terminal catalytic domain into the cytosol of eukaryotic cells. J Biol Chem. 2003;278:44535–44541. doi: 10.1074/jbc.M307540200. [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Chagot B, Cover M, Chazin WJ, Spiller B, Lacy DB. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing in Clostridium difficile toxin A. J Biol Chem. 2009;284:21934–21940. doi: 10.1074/jbc.M109.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri AW, Lupardus PJ, Deu E, Albrow VE, Garcia KC, Bogyo M, Shen A. Rational design of inhibitors and activity-based probes targeting Clostridium difficile virulence factor TcdB. Chem Biol. 2010;17:1201–1211. doi: 10.1016/j.chembiol.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qa'Dan M, Spyres LM, Ballard JD. pH-induced conformational changes in Clostridium difficile toxin B. Infect Immun. 2000;68:2470–2474. doi: 10.1128/iai.68.5.2470-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelings MD, Sorvillo F, Mascola L. Increase in Clostridium difficile-related mortality rates, United States, 1999–2004. Emerg Infect Dis. 2007;13:3. doi: 10.3201/eid1309.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke J, Tenzer S, Rupnik M, Koschinski A, Hasselmayer O, Schrattenholz A, et al. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446:415–419. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- Rupnik M, Pabst S, Rupnik M, von Eichel-Streiber C, Urlaub H, Soling HD. Characterization of the cleavage site and function of resulting cleavage fragments after limited proteolysis of Clostridium difficile toxin B (TcdB) by host cells. Microbiology. 2005;151:199–208. doi: 10.1099/mic.0.27474-0. [DOI] [PubMed] [Google Scholar]
- Savidge TC, Urvil P, Oezguen N, Ali K, Choudhury A, Acharya V, et al. Host S-nitrosylation inhibits clostridial small molecule-activated glucosylating toxins. Nat Med. 2011;17:1136–1141. doi: 10.1038/nm.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Lupardus PJ, Gersch MM, Puri AW, Albrow VE, Garcia KC, Bogyo M. Defining an allosteric circuit in the cysteine protease domain of Clostridium difficile toxins. Nat Struct Mol Biol. 2011;18:364–371. doi: 10.1038/nsmb.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simor AE, Yake SL, Tsimidis K. Infection due to Clostridium difficile among elderly residents of a long-term-care facility. Clin Infect Dis. 1993;17:672–678. doi: 10.1093/clinids/17.4.672. [DOI] [PubMed] [Google Scholar]
- Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler R, He M, Dawson L, Martin M, Valiente E, Corton C, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NM, Pellett S, Wilkins TD. Purification and characterization of toxins A and B of Clostridium difficile. Infect Immun. 1982;35:1032–1040. doi: 10.1128/iai.35.3.1032-1040.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NS, Thorne GM, Bartlett JG. Comparison of two toxins produced by Clostridium difficile. Infect Immun. 1981;34:1036–1043. doi: 10.1128/iai.34.3.1036-1043.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eichel-Streiber C, Sauerborn M, Kuramitsu HK. Evidence for a modular structure of the homologous repetitive C-terminal carbohydrate-binding sites of Clostridium difficile toxins and Streptococcus mutans glucosyltransferases. J Bacteriol. 1992;174:6707–6710. doi: 10.1128/jb.174.20.6707-6710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- Wilcox MH, Mooney L, Bendall R, Settle CD, Fawley WN. A case control study of community-associated Clostridium difficile infection. J Antimicrob Chemother. 2008;62:388–396. doi: 10.1093/jac/dkn163. [DOI] [PubMed] [Google Scholar]
- Wilson KH. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol. 1983;18:1017–1019. doi: 10.1128/jcm.18.4.1017-1019.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KH, Sheagren JN, Freter R. Population dynamics of ingested Clostridium difficile in the gastrointestinal tract of the Syrian hamster. J Infect Dis. 1985;151:355–361. doi: 10.1093/infdis/151.2.355. [DOI] [PubMed] [Google Scholar]