Abstract
Recombination during meiosis shapes the complement of alleles segregating in the progeny of hybrids, and has important consequences for phenotypic variation. We examined allele frequencies, as well as crossover (XO) locations and frequencies in over 7000 plants from 17 F2 populations derived from crosses between 18 Arabidopsis thaliana accessions. We observed segregation distortion between parental alleles in over half of our populations. The potential causes of distortion include variation in seed dormancy and lethal epistatic interactions. Such a high occurrence of distortion was only detected here because of the large sample size of each population and the number of populations characterized. Most plants carry only one or two XOs per chromosome pair, and therefore inherit very large, non-recombined genomic fragments from each parent. Recombination frequencies vary between populations but consistently increase adjacent to the centromeres. Importantly, recombination rates do not correlate with whole-genome sequence differences between parental accessions, suggesting that sequence diversity within A. thaliana does not normally reach levels that are high enough to exert a major influence on the formation of XOs. A global knowledge of the patterns of recombination in F2 populations is crucial to better understand the segregation of phenotypic traits in hybrids, in the laboratory or in the wild.
Keywords: Arabidopsis thaliana, crossover, segregation distortion, SNP
Introduction
The genetic information of individuals is transmitted to the next generation after sexual reproduction. In the case of an F1 plant, each homologous chromosome is derived from a different parental strain. The two genomes will be recombined during meiosis, when double-stranded breaks form between homologs, to be later repaired as crossovers (XOs) or non-XOs. Meiotic recombination is thus at the core of trait segregation and linkage. Understanding early patterns of segregation and recombination following hybridization in natural or artificial populations is important for predicting phenotypic outcomes in descendents, and the degree to which linkage will lead to trait correlations.
Studies of XO distribution in Arabidopsis thaliana have been limited to selfed plants from crosses to the common laboratory strain Columbia (Col-0) and have not been generalized to other intraspecific crosses (Copenhaver et al., 2002; Drouaud et al., 2007; Kim et al., 2007). One aspect that is likely to affect all crosses is the small number of XOs per chromosome that take place during each meiotic division, with the consequence that the genomes of F2 plants are composed of mosaics of large genomic blocks from each grandparent.
A comprehensive study of recombination patterns in distinct F2 populations provides information for simulations of mapping populations. Indeed, deep-sequencing techniques now make it possible to identify the causal mutation from a bulked DNA sample (Schneeberger et al., 2009); in A. thaliana, this technique has so far only been applied to simple, recessive mutations, but it may have a much wider reach and become effective in mapping dominant mutations or complex genetic traits (Ehrenreich et al., 2010). Because estimation of population sizes needed to accurately map causal mutations/polymorphisms either by traditional genetic mapping or deep-sequencing depends on recombination frequencies in the mapping population, it is important to understand the recombinational landscape of large segregating samples to make informed decisions on experimental design.
Here, we present a detailed analysis of meiotic recombination in over 7000 F2 plants from 17 populations derived from crosses between 18 distinct A. thaliana accessions that we exploited previously to describe the genetic architecture of flowering time variation (Salomé et al., 2011). Importantly, we show that recombination frequencies do not correlate with genetic diversity between accessions. We also address how the recombinational landscape of F2 populations is affected by segregation distortion likely owing to segregation of genetic incompatibilities and how this in turn can correlate with phenotypic variation.
Materials and methods
Plant material and plant genotyping
Our laboratory has previously determined sequence polymorphisms in 20 Arabidopsis genomes using ultra-high-density microarrays (Clark et al., 2007). All 20 accessions were crossed in a full diallel; we chose 17 crosses according to a simple round robin design, such that most founding accessions are represented in two independent F2 populations. The list of founding accessions for each population is presented in Table 1.
Table 1. Estimated recombination rates (cM/Mb).
| Population | Grandparents | Chr-1 | Chr-2 | Chr-3 | Chr-4 | Chr-5 | Whole genome |
|---|---|---|---|---|---|---|---|
| P2 | Lov-5 × Sha | 4.2 | 3.8 | 3.3 | 4.5 | 4.0 | 3.9 |
| P3 | Bur-0 × Bay-0 | 3.3 | 3.1 | 3.6 | 3.8 | 4.4 | 3.7 |
| P6 | Van-0 × Bor-4 | 3.4 | 3.4 | 3.7 | 4.0 | 3.6 | 3.6 |
| P7 | NFA-8 × Van-0 | 3.7 | 3.8 | 3.5 | 3.9 | 3.6 | 3.7 |
| P8 | Est-1 × RRS7 | 3.1 | 3.3 | 3.5 | 3.7 | 3.1 | 3.3 |
| P9 | Tsu-1 × RRS10 | 2.8 | 3.7 | 3.1 | 3.9 | 3.4 | 3.4 |
| P10 | Bur-0 × Cvi-0 | 3.2 | 3.4 | 3.3 | 3.6 | 3.3 | 3.3 |
| P12 | Est-1 × Br-0 | 3.1 | 3.6 | 3.6 | 3.7 | 3.3 | 3.5 |
| P15 | Br-0 × C24 | 3.2 | 3.4 | 3.3 | 3.9 | 3.4 | 3.5 |
| P17 | Cvi-0 × RRS7 | 3.6 | 4.2 | 3.8 | 3.8 | 3.5 | 3.8 |
| P19 | Bay-0 × Lov-5 | 4.0 | 4.0 | 3.7 | 3.7 | 3.8 | 3.8 |
| P20 | Bor-4 × NFA-8 | 4.2 | 4.0 | 4.5 | 4.5 | 4.4 | 4.3 |
| P35 | Tamm-2 × Col-0 | 3.0 | 3.8 | 3.2 | 3.1 | 3.3 | 3.3 |
| P66 | Fei-0 × Col-0 | 3.5 | 3.2 | 3.2 | 3.8 | 3.8 | 3.5 |
| P129 | C24 × RRS10 | 2.6 | 3.4 | 3.5 | 3.5 | 3.0 | 3.2 |
| P145 | Sha × Fei-0 | 3.3 | 3.4 | 3.7 | 3.8 | 3.5 | 3.6 |
| P169 | Ts-1 × Tsu-1 | 3.0 | 3.4 | 3.1 | 3.8 | 3.2 | 3.3 |
| Min. | 2.6 | 3.1 | 3.1 | 3.1 | 3.0 | 3.2 | |
| Max. | 4.2 | 4.2 | 4.5 | 4.5 | 4.4 | 4.3 | |
| Mean | 3.4 | 3.6 | 3.5 | 3.8 | 3.6 | 3.6 |
A total of 7045 plants, derived from these 17 F2 populations, was genotyped using the MassArray technology (Jurinke et al., 2001) by Sequenom (San Diego, CA, USA). The genotype information has been published and is available from the Genetics website as supporting information (http://www.genetics.org/content/suppl/2011/03/15/genetics.111.126607.DC1/FileS1.zip) (Salomé et al., 2011). To genotype all F2 plants, we selected a single set of single-nucleotide polymorphism (SNP) markers chosen to be maximally informative in as many of the F2 populations as possible. We first classified SNPs according to the number of crosses in which they were predicted to be informative. Giving priority to highly informative SNP markers, we selected groups of four SNPs for each Mb of the A. thaliana genome, requiring that each parental pair be distinguished by at least two of these four SNPs. We then filled in additional SNPs, aiming a maximal inter-marker distance of less than 1 Mb for each F2 population, predicted to correspond to about 5 c (Lister and Dean, 1993). Raw genotype data were converted into the format A, B, H, indicating homozygosity for grandparent A or B, or heterozygosity, respectively, assessed for potential genotyping errors (see Supplementary Figure 1 for details), and corrected when appropriate.
XO landscape analysis
All XOs were identified for all five chromosomes of each F2 population. The presence of XO events in our genotype files was scored based on a text search for expected XO ‘words': a single XO would translate into the possible words A → H, B → H, H → A, or H → B. Double XO events between successive SNPs would appear as words A → B or B → A. XO positions were approximated as the mid-point of the interval between the markers flanking the recombination site. Note that, with the exception of double XOs that occur on the same chromosome, our plant genotypes are not phased (see Supplementary Figure 3 for details).
To measure recombination rates as a function of their proximity to centromeres, we segmented each chromosome into centromeric, centromere-adjacent and away-from centromere regions; these regions are indicated in Figure 3 (centromeres in gray, centromere-adjacent regions in light blue). Recombination rates between centromere-adjacent and away-from centromere regions were compared by Student's t-test (with Bonferroni correction for multiple testing).
Physical, genetic maps and genetic distance calculations
The physical locations of SNP markers are based on the TAIR7 annotation of the Arabidopsis accession Col-0. Genetic distances for all populations were estimated in R/qtl (Broman et al., 2003) using the Haldane map function. Segregation distortion was determined by examining the frequencies of each parental allele with the geno.table function of R/qtl, which includes a P-value for χ2-tests of Mendelian segregation. Interaction between distorted loci was assessed by plotting recombination frequencies using the plot.rf function. Distortion was confirmed by a χ2-test between expected and observed allele frequencies.
Marey maps allow a direct visual comparison of the genetic and physical maps for many populations (Chakravarti, 1991). They also highlight local variation in recombination rates. In a Marey map, genetic map length in cM (obtained from R/qtl, see above) is plotted as a function or the physical position of a SNP marker. Variation in recombination rates along each chromosome of our 17 F2 populations was tested by the bootstrap method, using the boot package in R with 10 000 imputations. We tested whether the coefficient of variance associated with the distribution of recombination rates of each chromosome differed from 0 (which one would expect with no variation in recombination rate), using all recombination rate values for each chromosome as input for a bootstrap resampling.
Genetic distances between pairs of accessions were determined by using MEGA 4.0, using the Maximum Likelihood model (Tamura et al., 2007) with a set of 139 intermediate-frequency SNPs, which has also been used to genotype thousands of accessions (Platt et al., 2010) (Supplementary Figure 2a), or with our set of 402 optimized SNP markers (Supplementary Figure 2b).
SNP frequency
The presence of a SNP between two founding accessions was extracted from available re-sequencing data, available at the POLYMORPH website (http://polymorph-clark20.weigelworld.org/). The number of SNPs was then calculated for 1-Mb windows and a slide of 200 kb for each population, to retain the same window size as for recombination frequencies used throughout.
XO interference
From a data set containing all XO pairs mapping to the same chromosome (see Supplementary Figure 3 for details on selection of these informative pairs), inter-XO distances were calculated according to the physical (in bp) and genetic (in cM) maps. The distribution of inter-XO distances was compared to a random distribution: S(k)=(2*(4−k)+1)*N/16, where k is the rank of the class (between 1 and 4), and N is the number of double XOs (Drouaud et al., 2007). Inter-XO distance distributions were also compared to a gamma distribution with shape=(mean/s.d.)2 and scale=(s.d.)2/mean (mean being the mean inter-XO distance and s.d. the associated standard deviation). All gamma distributions were generated in R using the function dgamma (chromosome length, shape, rate).
Results
A common SNP set for genotyping all populations
We generated 17 large F2 populations derived from 18 A. thaliana accessions, which capture most of the genetic diversity of the species (Clark et al., 2007). These populations were also used to describe the genetic architecture of flowering time variation in A. thaliana (Salomé et al., 2011). A set of 402 SNPs was designed to genotype all plants from all populations; the power of these SNPs to discriminate between founding accessions is illustrated by the pairwise genetic distance calculated using these markers as input for Maximum Likelihood in MEGA 4.0 (Supplementary Figure 2a; Tamura et al., 2007). Pairwise genetic distance between all possible combinations of the 18 parental accessions showed a bimodal distribution, the upper mode (genetic distance above 0.6) consisting of the 17 pairs used to generate our populations. Genetic distance for all remaining possible pairs of accessions varied between 0.3 and 0.6, and was the same as that seen with a set of 139 SNPs chosen to be polymorphic across a worldwide sample of accessions (Supplementary Figure 2b; Warthmann et al., 2007; Platt et al., 2010).
7045 F2 plants were genotyped at 402 SNP markers (Salomé et al., 2011). After removal of SNPs that failed during genotyping, we retained 370 markers, which translated into an average of 237 (range 215–257) informative SNPs for each population. Supplementary Table S1 provides a summary of SNPs for each chromosome and population. The resulting physical maps provided very good coverage of all chromosomes. Supplementary Figure 2c shows the map positions of all 402 SNPs and Supplementary Figure 2d shows the distribution of informative SNPs for one exemplary population. The majority of SNPs provided useful information for 10–14 populations (Supplementary Figure 2e). About 95% of inter-SNP intervals were smaller than 1 Mb (Supplementary Figure 2f). The few intervals larger than 1 Mb mostly overlapped with centromere positions, which are characterized by lower SNP density (Clark et al., 2007).
Frequent segregation distortion
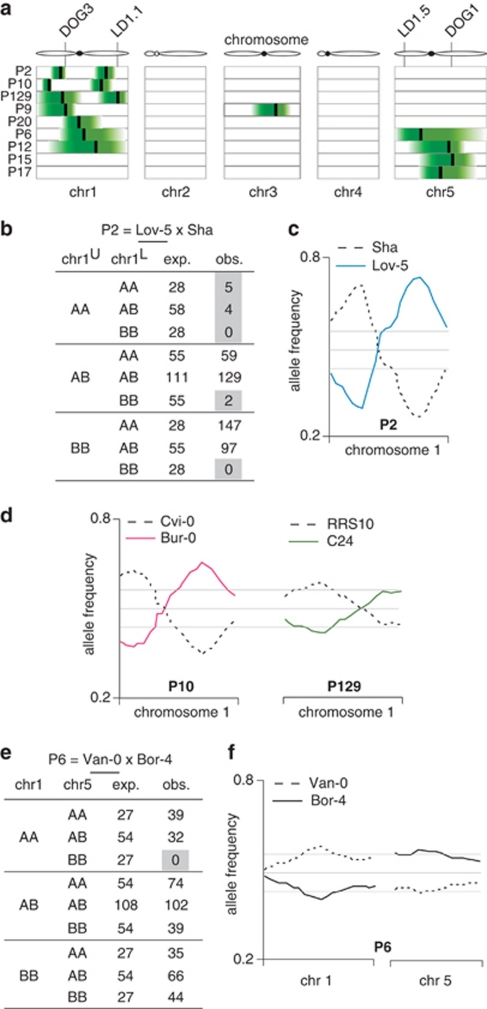
Several cases of segregation distortion have been reported in A. thaliana recombinant inbred lines (RILs), whereby one or two (interacting) alleles in certain genomic regions are under-represented; the causal genes have been identified in at least two cases (Loudet et al., 2002; O'Neill et al., 2008; Balasubramanian et al., 2009; Bikard et al., 2009; Vlad et al., 2010). Allele frequencies across all 17 F2 populations conformed globally to the expected 1:2:1 Mendelian segregation pattern (Supplementary Table S1). However, in 9 out of 17 populations, allele frequencies departed from expectations in at least one genomic region, based on χ2-tests for a series of contiguous SNP markers. Significantly distorted regions are shown in Figure 1a. The occurrence of segregation distortion in an F2 population was not related to sequence divergence between the founding accessions: mean genetic distance between accessions showing some distortion was identical to that of accessions not showing any distortion (mean=0.48, t-test P-value=0.7). The most striking example was on chromosome 1 of the P2 (Lov-5 × Sha) population (Figures 1b and c), where two genomic regions were almost devoid of homozygous combinations of parental alleles (Lov-5 for the region on the upper arm, Sha for the lower arm). The observed allele frequencies are consistent with a lethal effect associated with each distorted allele, without interaction between alleles (Figure 1b). Lov-5 and Sha were the grandparents for two additional populations (P19, Bay-0 × Lov-5 and P145, Sha × Fei-0); neither showed segregation distortion, suggesting a specific effect in the Lov-5 × Sha cross caused by the Lov-5 and Sha alleles at the distorted loci. Distortion was also found for chromosome 1 in two additional F2 populations, P10 (Bur-0 × Cvi-0) and P129 (C24 × RRS10) (Figure 1d). The overlap in confidence intervals among population pairs suggests that the same causal loci are responsible in different populations (P2 and P129 for the upper arm locus, P2 and P10 for the lower arm locus).
Figure 1.
Common genomic regions involved in segregation distortion across F2 populations. (a) Heat map of the genomic location of alleles causing segregation distortion. The physical locations of distorted regions are indicated as a vertical bar, at the SNP with the strongest difference in allele frequency. The green areas indicate increase of allele frequencies that deviate by more than 5% from expected frequencies. (b) Expected and observed allele frequencies for chromosome 1 in the P2=Lov-5 × Sha population. A=Lov-5; B=Sha. Deviation from expected frequencies is highly significant (P<0.001). (c) Allele frequencies for the P2 population along chromosome 1. (d) Two other populations, P10=Bur-0 × Cvi-0 and p129=C24 × RRS10, show a bimodal distortion on chromosome 1. (e) Expected and observed allele frequencies for chromosomes 1 and 5 in the P6=Van-0 × Bor-4 population. A=Bor-4; B=Van-0. Deviation from expected frequencies is highly significant (P<0.001). Note that expected values were rounded to the nearest integers, and therefore add up to 432 for a total number of 431 observed plant genotypes. (f) Allele frequencies for the P6 population along chromosomes 1 and 5.
At least three F2 populations showed strong distortion in a genomic region on chromosome 5 that includes the DELAY OF GERMINATION-1 (DOG1) gene (Bentsink et al., 2006). This suggests that variation in seed dormancy, and thus germination date, may be the underlying cause for segregation distortion (Figure 1a). In our design, we thinned populations randomly to single seedlings per pot after release from vernalization and removed later-germinating seedlings in pots already occupied by established plants, which led to an inadvertent selection against late-germinating genotypes. The under-represented allele in the P17 population (Cvi-0 × RRS7) originated from the Cvi-0 accession, which is known to show stronger seed dormancy (Bentsink et al., 2006). Distortion around the DOG1 genomic region was also seen in the P12 population (Est-1 × Br-0), with under-representation of the Est-1 allele on chromosome 5, suggesting that the DOG1-Est-1 allele increases seed dormancy.
Finally, we found one additional case of distortion likely due to genetic incompatibility in the P6 population (Bor-4 and Van-0) (Figures 1e and f). A clear lethal epistatic interaction could be detected between two loci, on chromosomes 1 and 5: absence of the reciprocal homozygous combination for a region on chromosome 1 for Bor-4 and chromosome 5 for Van-0 suggests that this genotype combination is lethal. The observation that only homozygous combinations are missing indicates that the incompatibility is caused by recessive alleles. The two genomic regions identified in the P6 population do not overlap with previously reported examples of deleterious epistatic interactions (Loudet et al., 2002; O'Neill et al., 2008; Balasubramanian et al., 2009; Bikard et al., 2009; Vlad et al., 2010), and might constitute a new case of genetic incompatibility.
XO landscape in F2 populations
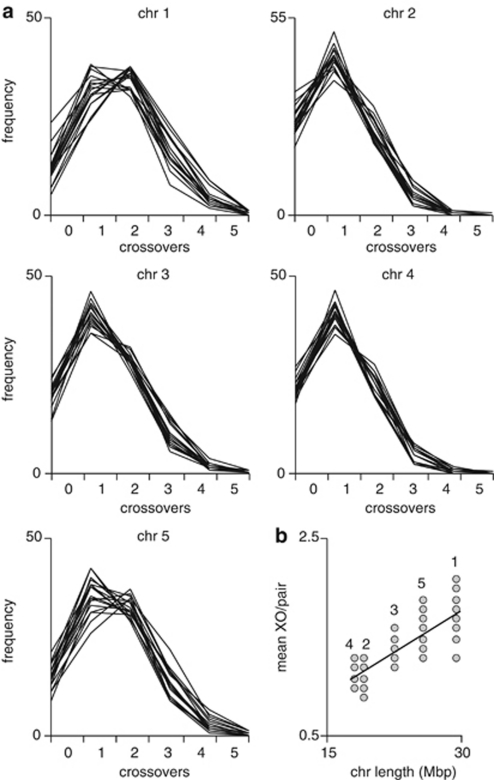
The mean inter-SNP distance of about 500 kb for all chromosomes allowed us to determine the precise location of all XO events (to within, on average, 250 kb). The number of XOs per chromosome pair varied from 0 to 6, with a mean of 1.4 per chromosome (range 0.9–2.1; Figure 2a and Supplementary Table S2). Mean XO number per chromosome pair was positively correlated with chromosome physical length (Figure 2b, R2=0.72, P<2.2e−16), indicating that longer chromosomes accumulate more XO events. Similarly, the incidence of multiple XO events per chromosome pair was highly positively correlated with chromosome physical length: in nearly half of F2 plants, only one XO was detected along chromosomes 2 and 4, with roughly 20% having no XO (Figure 2a). By contrast, two XOs were much more frequent along chromosomes 1 and 5, and the fraction of individuals without any XO on these chromosomes decreased to 10%. This correlation was highly significant (R2=0.33, P<2.2e−16 for all XO numbers; R2-values for XO=[0,1,2,3,4] are given in Supplementary Figure 4).
Figure 2.
XO frequencies across F2 populations. (a) Box-and-whisker plots of number of individuals in each of the 17 F2 populations, with the indicated number of XOs per chromosome. (b) Mean XO number is correlated with chromosome physical length. Mean XO numbers are taken from Supplementary Table S2 and plotted as a function of chromosome length in Mb. R2-value for linear regression is 0.72 (P<2.2 e−16). Chromosome number is indicated above the relevant data points.
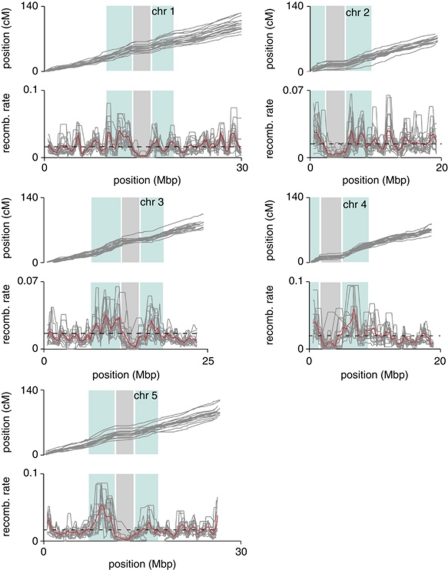
XO frequencies varied along each chromosome (bootstrap P<0.001) (Figure 3). For all populations, XO events were virtually absent from a single region that closely matched the expected position of centromeres (The Arabidopsis Genome Initiative, 2000; Clark et al., 2007). In several instances, we observed a suppression of recombination over several consecutive SNP markers in pairs of F2 populations that shared one founding accession. This pattern is suggestive of an inversion in the common grandparent. Such patterns were found in an approximately 1- to 2-Mb region on the lower arm of chromosome 3 in Sha; a region of about 2–3 Mb on the upper arm of chromosome 5 in RRS7; and finally, a small region of about 200 kb on the lower arm of chromosome 1 in Bay-0. That we could detect both parental alleles across the predicted inversions demonstrated that the absence of recombination does not result from a deletion. A possible inversion in Sha on the upper arm of chromosome 3 had been inferred before from the absence of recombination in the Bay-0 × Sha and Col-0 × Sha RILs (Loudet et al., 2002). The small inversion in Bay-0 on chromosome 1 was not detected previously, probably owing to insufficient marker density; alternatively this could be a new event that occurred in the maintenance of our Bay-0 stock and may not be reflected in the original RIL population. We did not observe any example of translocations, as demonstrated by the perfect co-linearity of all SNP markers across all F2 populations.
Figure 3.
Variation in recombination frequencies across populations and chromosomes in F2 populations. Upper panels: Marey maps (Chakravarti, 1991) for all F2 populations. Lower panels: Variation in recombination rates (as number of XOs per plant) along the chromosome and across F2 populations. Dashed line: Mean genome-wide recombination rate. Solid gray lines: Recombination rate of individual two populations. Solid red line: Mean local recombination rate. In both sets of panels, centromeres are shaded in gray, whereas peri-centromeric regions are shaded in light blue.
Outside the centromeres, XO frequencies varied extensively (Figure 3). While SNP marker density was insufficient for identification of local hotspots, regions adjacent to centromeres tended to have elevated mean recombination rates, except chromosome 2 (Figure 3 and Supplementary Figure 5). XO frequency adjacent to centromeres in individual F2 populations also followed this trend of increased recombination: 9 populations for chromosome 1; 2 for chromosome 2; 16 for chromosome 3; 15 for chromosome 4; and 9 for chromosome 5 with P<0.05 (Student's t-test with Bonferroni correction for multiple testing).
XO interference
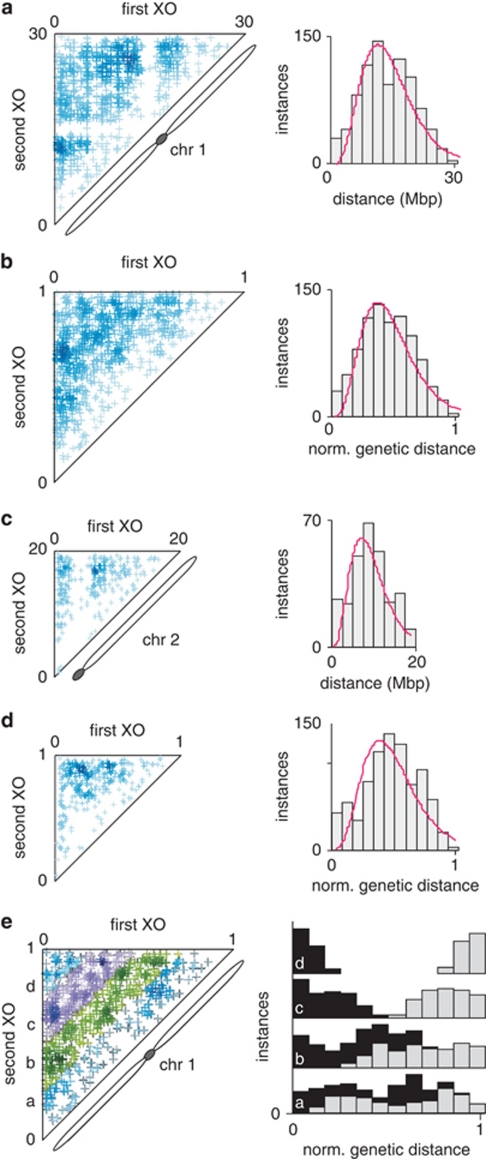
When multiple XOs occur along the same chromatid, they are not randomly distributed: a first XO event prevents others from occurring close by, owing to XO interference (Copenhaver et al., 2002; Lam et al., 2005). Quantifying XO interference in different cross combinations is important for estimating the extent to which genomes can recombine. To measure the strength of XO interference in our populations, we identified pairs of chromosomes in which one had undergone two XO events (Supplementary Figure 3). We then plotted the position of both XOs in a two-dimensional plot using the physical position (Figures 4a and c, and Supplementary Figures 6a, c and e) or the genetic map position of each XO (Figures 4b and d, and Supplementary Figures 6b, d and f). As expected, virtually no double XO events mapped to the centromeres when physical positions were used (Figure 4a for the pericentric chromosome 1; Figure 4c for the acrocentric chromosome 2; data for remaining chromosomes are provided in Supplementary Figure 6). The adoption of genetic map positions for all XO events alleviated the confounding effect of reduced recombination over the centromeres. Very few double XO events occurred in close proximity to each other, as illustrated by the scarcity of double XOs along the diagonal of the plots. Double XO events that mapped close to the diagonal were separated by less than one quarter of a chromosome length (Figure 4e for chromosome 1, using genetic map lengths), and were greatly under-represented when compared with a random distribution of inter-XO distances (Supplementary Figure 7; P<0.001, Pearson's χ2-test with Yates' continuity correction).
Figure 4.
Positive XO interference in F2 populations. Positions of first and second XOs for all double XO pairs, according to their physical (a, c) or genetic (b, d) positions along the chromosome. Darker blue indicates higher densities in double XOs. The panels on the left show the density of double XOs, whereas those on the right show the distribution of inter-XO distances separating the two XOs of a double XO pair. Magenta line: Gamma distribution of scale=(s.d.)2/mean and shape=(mean/s.d.)2. (e) Inter-XO distance dictates the relative positions of each XO when inter-XO distance is greater than ¼ chromosome length. Inter-XO distances were sorted in quartiles (left) and the positions of individual XOs were plotted for each quartile (right). Black bar: First XO. Gray bar: Second XO.
Our data clearly demonstrate a strong positive XO interference affecting all chromosomes (Figure 4 and Supplementary Figure 6). Mean inter-XO distances were approximately equal to half of a chromosome length, irrespective of the chromosome under investigation (Figure 4 and Supplementary Figure 6). This observation implies that inter-XO distances between adjacent XO events are controlled in part by the length of each chromosome, and is consistent with a gamma model for the locations of XOs on meiotic four-strand bundles (Foss and Stahl, 1995; Broman and Weber, 2000). A gamma distribution offered a better fit for our inter-XO distance data than a random distribution, especially for chromosomes 1 and 2 (Figure 4), without accounting for interference-independent XOs (Copenhaver et al., 2002).
The shape parameter of the gamma distribution provides a measure for the strength of interference (shape=1 being expected for no interference and shape >1 for positive interference). From the distributions shown in Figure 4 and Supplementary Figure 6, XO interference in our populations ranged from 4.2 to 4.9, with a mean of 4.5. Mapping functions used to derive genetic maps from recombination frequencies do not always account for interference: for example, the Haldane map function assumes no interference, but has been used extensively to estimate the genetic maps of Arabidopsis RILs (Simon et al., 2008). Our published genetic maps also made use of the Haldane function (Salomé et al., 2011). We therefore tested the effect of XO interference by re-analyzing our populations using the Kosambi (interference=2.6) and Carter–Falconer (interference=7.6) map functions. Total genetic map lengths shrank by 3.2% on average (range 2.8–3.8) when incorporating the moderate degree of interference of the Kosambi map function, and by 3.4% using Carter–Falconer (range 2.8–4.0). Supplementary Figure 8 illustrates the small effect of interference on the total genetic length of the P2 population: map shortening is spread over the whole genome, so that all inter-SNP distances in cM will be reduced by about 3% using the Kosambi or Carter–Falconer maps.
Recombination rates and SNP density
We wished to test whether recombination frequencies in our 17 F2 populations were correlated with the genetic diversity of the founding accessions. We first calculated the recombination rate (in cM/Mb) for each chromosome and population, and plotted this information as a function of physical chromosome length in Mb. There was little variation in mean recombination rate between chromosomes (Supplementary Figure 9a), suggesting that the higher number of XOs detected on longer chromosomes (Figure 2b) can be solely explained by the longer physical length of these chromosomes. Individual populations showed variation in recombination rates from one chromosome to the next (Supplementary Figure 9b). There was little correlation between the recombination rates of the five chromosomes in a given population: within-population R2-values ranged from 0 to 0.92, with a mean of 0.4.
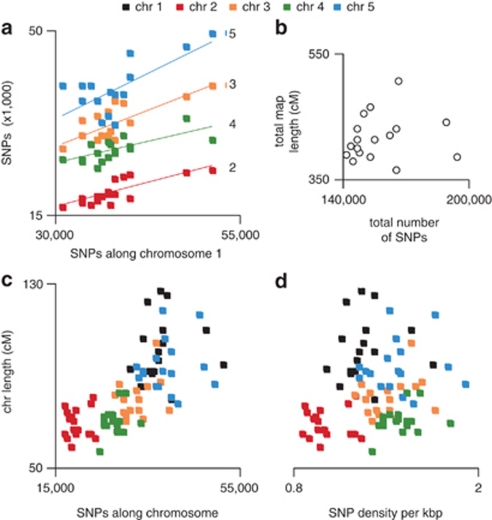
To explore correlations between genetic map lengths and polymorphism levels, we took into account all SNPs known from whole-genome re-sequencing efforts (Clark et al., 2007) (http://polymorph-clark20.weigelworld.org/). The number of SNPs that distinguished any two grandparents varied quite extensively across the 17 F2 populations, and reflected the physical length of each chromosome. For example, chromosome 1 (about 30 Mb) accounted for most differentiating SNPs, while chromosomes 2 and 4 (each about 18 Mb) had the least. Sequence differentiation was positively correlated between chromosomes (Figure 5a). The lower overall SNP count along chromosome 2 was striking, when considering that chromosomes 2 and 4 have about the same physical length. Our results therefore demonstrate that there is no significant correlation between recombination rates and sequence diversity (Figure 5a). Consistent with this, total map length (in cM) showed no correlation with total SNP counts (Figure 5b; R2=0.04, P=0.56).
Figure 5.
Effect of sequence diversity between parental accessions on genetic map lengths. (a) The number of SNPs along chromosomes 2–5 between each pair of founding accessions, plotted as a function of the number of SNPs along chromosome 1. R2-value for the multiple linear regression is 0.65 (P=3.5e−15). (b) Total genetic map length in cM as a function of the total number of SNPs between founding accessions. R2-value is 0.04 (P=0.56). (c) Chromosome length in cM as a function of the number of SNPs between founding accessions. R2-value for the multiple linear regression is 0.48 (P=1.15e−13). (d) Chromosome length in cM as a function of SNP density (per kb). R2-value for the multiple linear regression is 0.002 (P=0.01).
To confirm these results, we plotted chromosome length in cM as a function of either SNP count (Figure 5c) or SNP density (Figure 5d and Supplementary Tables S3 and S4) for each chromosome. The apparent strong correlation between chromosome length and SNP count completely disappeared when plotted against SNP density, indicating that chromosome physical length was driving the spurious pattern apparent in Figure 5c. A similar lack of correlation was observed between recombination rate and genome-wide SNP density (Supplementary Figure 10). We conclude that sequence diversity, at least as measured with SNPs, is not an important factor affecting recombination between the genomes of related accessions.
Discussion
There were two main goals of our study. First, we wished to thoroughly describe the locations and frequencies of the XOs that take place during meiosis of different F1 hybrid plants in A. thaliana, in order to further our understanding of the early genetic events that shape the segregation of parental alleles in F2 plants and the degree to which this varies among genotypes. Second, we wanted to address whether recombination rates were correlated with sequence diversity between founding accessions. Previous studies have resorted to combining XO frequencies derived from several smaller F2 populations into a mean estimated recombination rate value (Kim et al., 2007). We reasoned that differences in recombination patterns between individual populations might become masked when considering a mean rate; the size of our populations was sufficient to allow direct comparisons for all populations individually.
We examined XO distributions and recombination rates across the genome in 17 F2 populations generated by intercrossing 18 genetically distant accessions. Most chromosome pairs counted just one or two XOs (Figure 2). Chromosome pairs with no apparent XOs ranged from 10% (for the longer chromosomes 1 and 5) to 20% (for the shorter chromosomes 2 and 4). The number of XOs in A. thaliana is therefore much lower (both per chromosome and genome-wide) than what has been described in yeast, mice or humans (Broman and Weber, 2000; Broman et al., 2002; Ehrenreich et al., 2010). Although more than one XO may occur between homologs, only a single XO is needed to hold homologs together until the first meiotic division, when they will align at the metaphase plate and each attach to a different spindle pole (Youds and Boulton, 2011). Several mathematical models have been developed to describe how XO positions and numbers are selected (Broman and Weber, 2000; Youds and Boulton, 2011). In a gamma model, double-stranded breaks are distributed randomly along the four-strand bundle, but are resolved into meiotic XOs or non-XOs according to a self-renewal process. A gamma model provided a good fit for the frequencies of XO numbers (Figure 2a, and data not shown), which suggests that double-stranded breaks are generated at the same rate along each chromosome. Subsequently, double-stranded breaks will be resolved into slightly more meiotic XOs on the longer chromosomes, providing an explanation for the distributions observed in Figure 2a.
Recombination hotspots are specific sites (1–2 kb in length) of increased XO formation. In mouse, a genome-wide analysis of likely hotspots revealed that double-stranded breaks during male germ-cell meiosis occur at a consensus sequence that is preferentially occupied by a nucleosome (Smagulova et al., 2011). In A. thaliana, hotspot positions inferred from F2 recombination data do not agree with ones deduced from analysis of patterns of linkage disequilibrium in the global population (Drouaud et al., 2006, 2007; Kim et al., 2007), suggesting that hotspots are accession-specific. Our data support the conclusion that recombination rates vary greatly depending on the cross, even though our marker density was not sufficient for identification of XO hotspots.
In addition to how often and where XOs took place, our analyses of ∼14 000 meioses provided insights into the strength of XO interference in different crosses and into segregation distortion, two phenomena that contribute to shaping the recombination landscape. Almost 1 out of 10 XO events was affected by XO interference, which will cause XO pairs to be separated by a distance greater than expected by chance (Figure 4 and Supplementary Figures 6 and 7). Segregation distortion resulted in the under-representation of alleles over several Mb in over half of our populations, and was not linked to sequence diversity between founding accessions (Figure 1). Distortion in at least three populations is likely to stem from variation in seed dormancy, as the affected regions of the genome are near DOG1, a gene known to have a large effect on this trait in A. thaliana accessions (Bentsink et al., 2006). Segregation distortion driven by selection against strong seed dormancy has been reported in rice (Gu et al., 2008). It is possible that we inadvertently introduced such distortion by manually removing later-germinating seedlings. The remaining examples of segregation distortion we observed are the result of an interaction between specific alleles at two recessive loci, with each parental accession contributing one, a not uncommon occurrence in A. thaliana. Distortion cases associated with selection of genomic regions, or genetic incompatibilities between genomic regions have an important role in shaping the distribution of potential phenotypes observed in F2 and subsequent segregating populations. Our cases of distortion were all caused by recessive alleles, acting alone or through epistatic interaction with another recessive allele, and are likely to be post-zygotic examples of incompatibility between founding accessions. In Silene latifolia, a dioecious plant, the 3:1 sex ratio bias between females and males may be the result of pollen competition between X-bearing and Y-bearing pollen (Taylor and Ingvarsson, 2003). In Arabidopsis lyrata, several cases of segregation distortion have been reported, and most were due to gametic effects (Leppala et al., 2008). In addition, diversity at the self-incompatibility S locus in A. lyrata drives strong distortion between incompatible alleles. Being mostly a selfing species, it is perhaps not surprising that most examples of distortion in A. thaliana are post-zygotic. Interestingly, the distorted regions on chromosome 1 in A. thaliana overlap with distorted regions on A. lyrata chromosomes 1 and 2 (from which A. thaliana chromosome 1 derived; Hu et al., 2011).
From the information collected here, mapping populations may now be simulated that take into account the frequencies of XOs along the chromosomes (Figure 2 and Supplementary Figure 4), their potential locations (Figure 3), as well as other factors that may influence recombination (Figure 4 and Supplementary Figure 6). The accuracy of such simulations is important for making informed experimental design decisions, especially when mapping traits with complex genetic architecture. An impressive proof of concept has been demonstrated in yeast (Ehrenreich et al., 2010), but it remains to be seen how easily this approach can be applied to species with larger genomes such as A. thaliana. In the yeast study, Ehrenreich et al. started with 10 millions haploid cells, each representing harboring 50 XOs, for or a total of 500 millions XOs for a genome of 12.5 Mb. In this study, we grew a little over 7000 plants in 6 months with no phenotypic selection, and the 14 000 meioses in the 17 F2 populations examined here only amounted to 50 000 XOs spread over a 120-Mb genome.
Two possible strategies could be combined to overcome the current limitations of high-resolution deep-sequencing for mapping for genetically complex genetic traits in plants. First, much larger mapping populations than what is currently routinely used are needed in order to accumulate sufficient XOs for a precise estimation of locations of quantitative trait loci. Second, a genome sequence complexity reduction step is performed, whereby only informative regions are sequenced at very high coverage. After library production, informative PCR products (covering known polymorphic regions) would be enriched in the sample by sequence capture using long oligonucleotide probes (Shearer et al., 2010). This critical step could increase fold sequence coverage from the 10s to the many 1000-folds and thus allow each chromosome from a bulked DNA sample to be sequenced, as opposed to a fraction of chromosomes currently sampled (Schneeberger et al., 2009; Schneeberger and Weigel, 2011). The expanding number of genome sequences from Arabidopsis accessions will greatly facilitate the design of fishing probes for deep-sequencing of bulked segregants for mapping simple or complex traits (Weigel and Mott, 2009).
The second goal of our study was to assess the impact of sequence diversity on recombination rates in this species. In 17 large F2 populations we did not find evidence for significant genome-wide correlation between recombination rate and SNP density. There was, however, increased recombination in centromere-adjacent regions, which have higher SNP density (Clark et al., 2007). Differences in recombination rate between homozygous and heterozygous chromosomes have been described (Barth et al., 2001), also suggesting that absence of sequence differences between homologs at meiosis reduces recombination. Interestingly, RIL populations include a similar number of chromosome pairs with no apparent XOs as F2 populations, although RILs have undergone several more meioses than F2 plants (Supplementary Figure 11). Even with 600 markers, XOs were not detected for 10–20% of chromosome pairs of Col-0 × Ler RILs (Singer et al., 2006). These results suggest that recombination between homologs might be suppressed after the initial F1 meiosis, perhaps in part because of the mosaic nature of each homolog. With new sequencing technologies, it should be possible to discover markers even in very closely related strains, which will allow more detailed examination of this important question. New sequencing technologies should also allow a much finer-scale analysis of recombination and sequence variation in F2 populations, as has been described in Drosophila pseudoobscura (Kulathinal et al., 2008).
DATA ARCHIVING
All data used in this study (XO numbers and positions, genetic maps, XO interference) have been deposited at Dryad: doi:10.5061/dryad.v655ns36.
Acknowledgments
We thank Dan Koenig and Tina Hu for help with sliding window analysis scripts in R. Financial support came from postdoctoral fellowships from EMBO (PAS), NIH (KB), the Human Frontiers Science Program (RAEL), Grant FP6 IP AGRON-OMICS (contract LSHG-CT-2006-037704), a Gottfried Wilhelm Leibniz Award of the DFG and the Max Planck Society (DW).
Author contributions: PAS, KB, RAEL, LY, NW, and JF: Conceived and designed the experiments.
PAS, KB, RAEL and LY: Performed the experiments.
PAS: Analyzed the data.
PAS and DW: Wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Balasubramanian S, Schwartz C, Singh A, Warthmann N, Kim MC, Maloof JN, et al. QTL mapping in new Arabidopsis thaliana advanced intercross-recombinant inbred lines. PLoS One. 2009;4:e4318. doi: 10.1371/journal.pone.0004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Melchinger AE, Devezi-Savula B, Lübberstedt T. Influence of genetic background and heterozygosity on meiotic recombination in Arabidopsis thaliana. Genome. 2001;44:971–978. doi: 10.1139/g01-094. [DOI] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Patel D, Le Mette C, Giorgi V, Camilleri C, Bennett MJ, et al. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323:623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- Broman KW, Rowe LB, Churchill GA, Paigen K. Crossover interference in the mouse. Genetics. 2002;160:1123–1131. doi: 10.1093/genetics/160.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Weber JL. Characterization of human crossover interference. Am J Hum Genet. 2000;66:1911–1926. doi: 10.1086/302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Chakravarti A. A graphical representation of genetic and physical maps: the Marey map. Genomics. 1991;11:219–222. doi: 10.1016/0888-7543(91)90123-v. [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Housworth EA, Stahl FW. Crossover interference in Arabidopsis. Genetics. 2002;160:1631–1639. doi: 10.1093/genetics/160.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J, Camilleri C, Bourguignon PY, Canaguier A, Berard A, Vezon D, et al. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination ‘hot spots'. Genome Res. 2006;16:106–114. doi: 10.1101/gr.4319006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J, Mercier R, Chelysheva L, Berard A, Falque M, Martin O, et al. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 2007;3:e106. doi: 10.1371/journal.pgen.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, et al. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss EJ, Stahl FW. A test of a counting model for chiasma interference. Genetics. 1995;139:1201–1209. doi: 10.1093/genetics/139.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XY, Turnipseed EB, Foley ME. The qSD12 locus controls offspring tissue-imposed seed dormancy in rice. Genetics. 2008;179:2263–2273. doi: 10.1534/genetics.108.092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurinke C, van den Boom D, Cantor CR, Koster H. Automated genotyping using the DNA MassArray technology. Methods Mol Biol. 2001;170:103–116. doi: 10.1385/1-59259-234-1:103. [DOI] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Bennett SM, Fitzpatrick CL, Noor MA. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc Natl Acad Sci USA. 2008;105:10051–10056. doi: 10.1073/pnas.0801848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SY, Horn SR, Radford SJ, Housworth EA, Stahl FW, Copenhaver GP. Crossover interference on nucleolus organizing region-bearing chromosomes in Arabidopsis. Genetics. 2005;170:807–812. doi: 10.1534/genetics.104.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppala J, Bechsgaard JS, Schierup MH, Savolainen O. Transmission ratio distortion in Arabidopsis lyrata: effects of population divergence and the S-locus. Heredity. 2008;100:71–78. doi: 10.1038/sj.hdy.6801066. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- O'Neill CM, Morgan C, Kirby J, Tschoep H, Deng PX, Brennan M, et al. Six new recombinant inbred populations for the study of quantitative traits in Arabidopsis thaliana. Theor Appl Genet. 2008;116:623–634. doi: 10.1007/s00122-007-0696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, et al. The scale of population structure in Arabidopsis thaliana. PLoS Genet. 2010;6:e1000843. doi: 10.1371/journal.pgen.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Laitinen RAE, Yant L, Mott R, Weigel D. Genetic architecture of flowering time variation in Arabidopsis thaliana. Genetics. 2011;188:421–433. doi: 10.1534/genetics.111.126607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, et al. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Weigel D. Fast-forward genetics enabled by new sequencing technologies. Trends Plant Sci. 2011;16:282–288. doi: 10.1016/j.tplants.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J, 2nd, Scherer S, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci USA. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Loudet O, Durand S, Berard A, Brunel D, Sennesal FX, et al. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics. 2008;178:2253–2264. doi: 10.1534/genetics.107.083899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, Fan Y, Chang HS, Zhu T, Hazen SP, Briggs SP. A high-resolution map of Arabidopsis recombinant inbred lines by whole-genome exon array hybridization. PLoS Genet. 2006;2:e144. doi: 10.1371/journal.pgen.0020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Taylor DR, Ingvarsson PK. Common features of segregation distortion in plants and animals. Genetica. 2003;117:27–35. doi: 10.1023/a:1022308414864. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Vlad D, Rappaport F, Simon M, Loudet O. Gene transposition causing natural variation for growth in Arabidopsis thaliana. PLoS Genet. 2010;6:e1000945. doi: 10.1371/journal.pgen.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthmann N, Fitz J, Weigel D. MSQT for choosing SNP assays from multiple DNA alignments. Bioinformatics. 2007;23:2784–2787. doi: 10.1093/bioinformatics/btm428. [DOI] [PubMed] [Google Scholar]
- Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009;10:107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Boulton SJ. The choice in meiosis—defining the factors that influence crossover or non-crossover formation. J Cell Sci. 2011;124:501–513. doi: 10.1242/jcs.074427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.