Abstract
Drug resistance poses a significant challenge in antifungal therapy since resistance has been found for all known classes of antifungal drugs. The discovery of compounds that can act synergistically with antifungal drugs is an important strategy to overcome resistance. For such combination therapies to be effective, it is critical to understand the molecular basis for the synergism by examining the cellular effects exerted by the combined drugs. Genomic profiling technologies developed in the model yeast Saccharomyces cerevisiae have been successfully used to investigate antifungal combinations. This review discusses how these technologies have been used not only to identify synergistic mechanisms but also to predict drug synergies. It also discusses how genome-wide genetic interaction studies have been combined with drug–target information to differentiate between antifungal drug synergies that are target-specific versus those that are non-specific. The investigation of the mechanism of action of antifungal synergies will undoubtedly advance the development of optimal and safe combination therapies for the treatment of drug-resistant fungal infections.
Keywords: drug synergy mechanism, chemogenomic profiling, genetic interaction
Introduction
Currently available antifungal drugs belong to four major drug classes: polyenes (e.g., amphotericin B), azoles (e.g., fluconazole), echinocandins (e.g., caspofungin), and pyrimidine analogs (e.g., 5-fluorocytosine; reviewed in Petrikkos and Skiada, 2007; Pasqualotto and Denning, 2008; Denning and Hope, 2010). These drug classes disrupt membrane integrity, ergosterol biosynthesis, cell wall function, and DNA synthesis respectively. While they are used extensively in the clinic, the drugs within each class suffer from various limitations including toxicity (polyenes), resistance (azoles), narrow spectrum of activity (echinocandins), and lack of efficacy as a single agent (5-fluorocytosine). Drug resistance in particular has been a major concern for the fungistatic azole drugs and also more recently for echinocandins such as caspofungin (reviewed in Sanglard, 2002; Maertens, 2004; Perlin, 2007). In fact, drug resistance has been found for all known drug classes and poses a significant challenge in antifungal therapy (reviewed in Pemán et al., 2009; Mukherjee and Wang, 2010). One of the ways to overcome drug resistance is by administering combination therapy. Combination treatments offer several advantages including the ability to (i) reduce doses, and therefore reduce toxicity, (ii) increase spectrum of activity, and (iii) convert a fungistatic drug to a fungicidal drug, thereby facilitating clearance of the pathogen and decreasing the chance for the development of resistance (reviewed in Johnson et al., 2004; Mukherjee et al., 2005). Several studies, including work from our group, have shown the ability of various compounds, many of which are natural products, to improve the activity of azole antifungal drugs (e.g., Jacob et al., 2003; Onyewu et al., 2003; Niimi et al., 2004; Li et al., 2006; Digirolamo et al., 2009; Pfaller et al., 2009; Ahmad et al., 2010; Gamarra et al., 2010; Sharma et al., 2010; Wei et al., 2011). However, for the development of an effective drug combination regimen, it is important to understand the molecular mechanism behind the combined drug effect. Such an understanding will not only be invaluable in predicting adverse drug interactions and toxic side-effects, but will also be useful in developing chemical probes for exploring biological pathways targeted by the drug combination. This review focuses on chemogenomic profiling and genetic interaction technologies developed in the model yeast Saccharomyces cerevisiae, and discusses the applications of these technologies in predicting drug synergies, elucidating drug synergy mechanisms, and differentiating between specific and non-specific drug synergies.
Predicting Antifungal Synergies
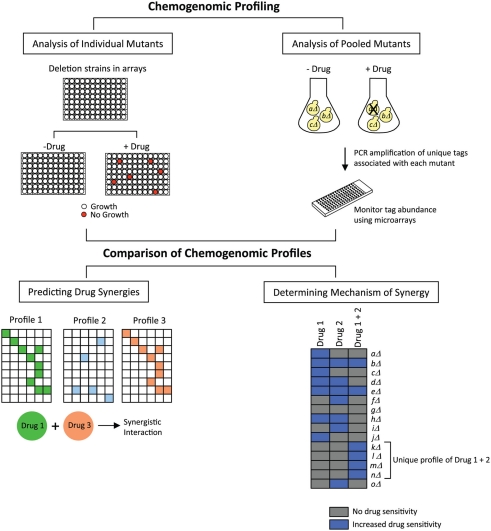
Jansen et al. (2009) have developed a bioinformatics-driven approach that predicts synergy between antifungal drug combinations. Their approach makes use of chemogenomic profiling, a technology in which the cellular response to inhibitory compounds is measured by evaluating the fitness of a library of genome-wide deletion mutant strains. Strains with reduced fitness in the presence of a compound are identified, and the genes deleted in those strains provide information on cellular pathways targeted by the compound as well as pathways that are required for conferring sensitivity to that compound. Given the availability of several whole-genome mutant collections in the model yeast S. cerevisiae, a large number of chemogenomic profiles have been generated for a variety of compounds in this organism (reviewed in Agarwal et al., 2008; Hoon et al., 2008b; Ho et al., 2011). These profiles have been generated using different methods, two of which are shown in Figure 1. In the first method, growth of each individual mutant is monitored in the presence or absence of a drug in a microplate format. In the second method, pooled mutants are analyzed in a single culture, in the presence or absence of a drug. Survival of the mutant strains is assessed by PCR amplification of unique tags associated with each mutant, and tag abundance is measured using a DNA microarray carrying complementary sequences to the tags. The ratio of signals between drug-treated and untreated samples gives an indication of the relative fitness of each mutant.
Figure 1.
Predicting drug synergies and determining synergy mechanisms. Chemogenomic profiling is conducted by screening drugs against collections of yeast deletion mutants. For analyzing individual mutants, the collection is arrayed in microplates, and the growth of each strain is examined in the presence or absence of the drug. For analyzing pooled mutants, the collection is grown in a single culture, with or without drug, and growth is measured by monitoring the abundance of unique tags associated with each mutant, using DNA microarrays consisting of tag complements. Two drugs with similar chemogenomic profiles would be predicted to exhibit a synergistic interaction (for simplicity, exactly matching profiles are depicted). If the combination of two synergizing drugs generates a unique chemogenomic profile of sensitive mutants, then genes deleted in those mutants provide information on the drug synergy mechanism.
In a large study conducted by Parsons et al. (2006), chemogenomic profiles were generated for 82 different compounds, and it was shown that compounds with similar chemogenomic profiles had similarities in their modes of action. To extend this concept to synergistic compounds, Jansen et al. (2009) investigated whether such profiles can be used to predict synergy by assessing if compound pairs with similar profiles are more likely to be synergistic (Figure 1). The authors compiled chemogenomic profiles from the literature for ∼1300 genome-wide screens conducted with a broad range of compounds, and defined a set of hypersensitive genes for each compound. The authors also generated a chemogenomic profile for the antifungal drug fluconazole, and gene deletions that were lethal in the presence of fluconazole were identified. Then, in order to determine if the chemogenomic profile of any given compound was similar to that of fluconazole, pairwise profile comparisons were made to assess correlations between the hypersensitive gene sets. This analysis identified eight compounds that were predicted to be synergistic with fluconazole. These eight compounds were then tested experimentally in dose–matrix response assays, and five compounds were indeed found to be synergistic with fluconazole. Interestingly, it was also found that compounds predicted to be synergistic with fluconazole were also predicted to be synergistic with each other. These were also tested in dose–matrix assays, and six compound pairs were identified that were synergistic, four of which had not been previously reported. To extend their studies to a fungal pathogen, the authors also analyzed the drug pairs in Candida albicans, and found that eight synergistic combinations identified in S. cerevisiae were also synergistic in C. albicans. Finally, one of the novel synergistic combinations (fluconazole + wortmannin) was also tested in a fluconazole-resistant clinical isolate of C. albicans. This combination was strongly synergistic in the clinical isolate – it not only inhibited cell growth in liquid broth, but it was also fungicidal since the cells were not viable in a recovery assay on agar plates. Thus, this method successfully predicted antifungal synergies, and also identified novel drug relationships. In addition, S. cerevisiae resources were effective in identifying synergies in C. albicans. This method will serve as a powerful tool in the discovery of new synergistic drugs and in evaluating the potential of combination therapies.
Determining the Mechanism Behind Antifungal Synergies
In addition to predicting drug synergies, chemogenomic profiles can also be used to evaluate the underlying mechanisms involved in the effects produced by synergistic drug combinations. As shown in Figure 1, for any given synergistic drug pair, chemogenomic profiling can be conducted on cells treated with each drug separately and also on cells treated with both drugs simultaneously. If a unique profile is obtained in the presence of both drugs, then gene deletions represented in that profile provide information on the pathways affected or targeted by the drug combination. An excellent example of the application of this strategy is the study by Hoon et al. (2008a) in which the authors investigated the synergistic effects of the phosphatase inhibitors cantharidin and calyculin A. Chemogenomic profiling was performed in the presence of diluent only, cantharidin, calyculin A, and a cocktail of cantharidin and calyculin A. Comparison of the profiles obtained indicated that fewer deletion strains were sensitive to the cocktail, and more importantly, additional new strains were detected as susceptible to the cocktail treatment that were not susceptible to the single drug treatments. The pathways represented in these gene deletions included golgi vesicle transport, chromatin remodeling, and mRNA polyadenylation. Thus, this study identified new pathways required for buffering the effects of the combined treatment of cantharidin and calyculin A.
Another example in which chemogenomic profiling was used to discover synergistic drug mechanisms is the study by Spitzer et al. (2011). In this study, six compounds were identified that showed synergistic interaction with the antifungal drug fluconazole. To understand the molecular basis for this synergy, the authors generated chemogenomic profiles for fluconazole and also for each compound separately. Strains with deletions in genes required for ergosterol biosynthesis (the pathway targeted by azole antifungals), vesicle-mediated transport, and membrane organization were sensitive to fluconazole alone. Of the six compounds analyzed, five inhibited the growth of strains carrying deletions in genes involved in membrane function, vesicle trafficking, and lipid biosynthesis, suggesting that these five compounds cause general membrane perturbation. Thus, these compounds may exert their synergistic effect on fluconazole by altering its import or export, or they may impair the import of exogenous ergosterol. The chemogenomic profile of the sixth compound analyzed indicated that it interfered with sphingolipid biosynthesis, thus suggesting an interplay between the ergosterol and sphingolipid biosynthetic pathways. Based on this result, the authors also tested myriocin, another sphingolipid biosynthesis inhibitor, and found that myriocin also showed synergistic activity with fluconazole. Thus, this study shows that understanding the mechanism behind drug synergies can be useful in predicting new synergistic interactions. In addition, by revealing that membrane disrupters and sphingolipid biosynthesis inhibitors can potentiate fluconazole activity, this study opens up new avenues for the discovery of novel strategies for antifungal therapy.
In addition to chemogenomics, other “omics”-based technologies such as transcriptomics, proteomics, and metabolomics that measure mRNAs, proteins, and metabolites respectively, can also be used for mechanistic studies on synergistic compounds. For example, gene expression profiling with the anti-microtubule agents docetaxel and estramustine in prostate cancer cells identified new genes affected by both drugs which were not altered by each drug alone (Li et al., 2005). Proteomic analysis in C. albicans with fluconazole and berberine revealed that their synergistic interaction is related to increased generation of endogenous reactive oxygen species (Xu et al., 2009). Metabolomic profiling to investigate the synergistic anti-leukemic activity of the lipid-lowering drug bezafibrate and the contraceptive steroid medroxyprogesterone acetate identified changes in TCA cycle intermediates, supporting the previously known association of reactive oxygen species in the anti-tumor activities of these two drugs (Tiziani et al., 2009). These studies validate the usefulness of genome-wide profiling approaches in elucidating the mechanisms of drug synergies.
Differentiating between Specific and Non-Specific Drug Synergies
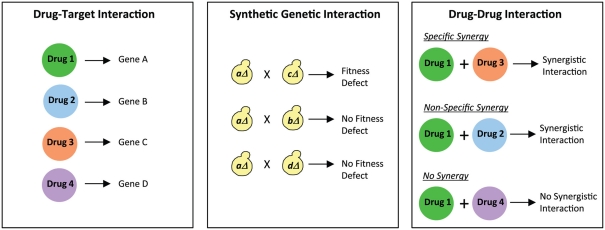
In a recent study (Cokol et al., 2011), the genetic basis of antifungal synergies was explored by evaluating the relationship between synergistic drug interactions and synergistic genetic interactions. In a synergistic genetic interaction, the combination of two genetic perturbations creates a more severe phenotype compared to the single perturbation alone (Tong et al., 2004). Cokol et al. hypothesized that if two genes have a synergistic genetic interaction, then two drugs that target those genes may exhibit a synergistic drug interaction. This prediction would be consistent with the “parallel pathway inhibition model,” according to which two drugs would be synergistic if they inhibit two proteins in parallel pathways (Yeh et al., 2009). For example, terbinafine and rapamycin exhibit drug synergy due to a synergistic genetic interaction between their target genes, ERG1 and TOR1 respectively (Lehár et al., 2007).
In the Cokol et al. study, a comprehensive analysis was conducted to establish the overlap between drug synergies and genetic synergies. The authors first compiled a list of 113 known drug–target interactions in S. cerevisiae from the literature, i.e., a list of known drugs and the target genes affected by those drugs was generated (Figure 2). This list was then integrated with known synergistic genetic interactions in this organism. Genetic interaction profiles for approximately 75% of all genes in S. cerevisiae have been generated using the synthetic genetic array (SGA) technology (Costanzo et al., 2010). In SGA analysis (Figure 2), a query mutation is crossed into a set of ∼5000 viable deletion mutants and the resulting double mutants are screened for fitness defects (Tong et al., 2004; Baryshnikova et al., 2010; Costanzo et al., 2010). Using the available information from several SGA studies, Cokol et al. (2011) identified gene pairs that exhibited severe fitness defects, and examined if these genes were present in their list of 113 known drug–target interactions. This analysis generated 211 drug pairs that were predicted to be synergistic. Of these, 38 drug pairs were assessed in dose–matrix response assays, and 14 drug pairs predicted to be synergistic did indeed show significant synergy in the assays. These drug pairs thus displayed “specific synergy” since genetic interaction profiles of their targeted genes suggested that the drugs would target parallel pathways (Figure 2). In contrast, several drug pairs (33% of the drug pairs analyzed) that were not predicted to target synergistic gene products were also found to show synergy, thus representing drugs displaying “non-specific synergy” (Figure 2). A set of six drugs were identified that were classified as promiscuous synergizers that were responsible for 92% of all the synergies discovered. This set included four drugs that target the biosynthesis of ergosterol, a cell membrane lipid, thus showing consistency with the “bioavailability model” of drug synergy in which one drug’s action affects the second drug’s availability in the cell (Zimmermann et al., 2007). The remaining two promiscuous synergizers are not known to disrupt the cell membrane, suggesting that their promiscuity involves a novel mechanism. Interestingly, the predominant form of drug synergy in this study was promiscuous synergy. Therefore, promiscuous drugs that enhance the potency of other drugs would have potential value in driving the search for new synergistic drug combinations. However, from a therapeutic standpoint, drug combinations that are specific to particular pathways would be attractive since they would produce less toxic side-effects. Thus, the ability to differentiate between specific and promiscuous synergizers will clearly play an important role in the discovery of novel combination therapies.
Figure 2.
Differentiating between specific and non-specific drug synergies. Comparing drug–target interactions with synthetic genetic interactions provides information on specific and non-specific drug synergies. For example, Drug 1 and Drug 3 exhibit specific synergy since the genes targeted by them (Gene A and Gene C) genetically interact with each other. On the other hand, Drug 1 and Drug 2 exhibit non-specific synergy since there is no genetic interaction between the genes targeted by them (Gene A and Gene B). Finally, an example is shown of two drugs (Drug 1 and Drug 4) that do not synergize with each other, and also do not show any genetic interaction between the genes they target (Gene A and Gene D).
Concluding Remarks
Chemogenomic profiling and genetic interaction studies have proven to be powerful technologies in investigating the molecular and genetic basis of synergistic drug relationships. However, it should be noted that just like any other whole-genome technology, these technologies suffer from a few drawbacks. For example, one of the weaknesses of SGA technology is that since it primarily involves crosses between two haploid mutants, it generally interrogates only non-essential genes. For chemogenomic profiling, analysis of individual mutants in microplate format requires large quantities of drug, which can be a limitation when studying natural products. While chemogenomic profiling of pooled mutants, in which tag abundance is monitored using DNA microarrays, makes use of smaller quantities of drug, the disadvantage is the high cost, the need for DNA microarray facilities, and potential errors in some tags making them undetectable upon hybridization. With regards to using chemogenomic profiling in predicting drug synergies, it is worth noting that an identical chemogenomic profile for two drugs does not automatically guarantee that there will be synergy. If two drugs target the same pathway, synergy may not be likely to occur – for example, sulfamethoxazole and trimethoprim do not show any synergy in Escherichia coli even though both drugs target the same folate metabolism pathway (reviewed in Jia et al., 2009). In spite of these limitations, the various examples discussed in this review do indeed demonstrate that chemogenomic profiling and SGA technologies have led to greater mechanistic understanding of synergistic drug interactions and have been useful in predicting drug synergies, and also in distinguishing specific synergies from non-specific ones. The availability of similar technologies in fungal pathogens such as C. albicans and Cryptococcus neoformans will of great value in studying drug combinations in clinically relevant isolates. Given the ever-growing problem of drug resistance, continued investigations in drug synergy mechanisms are greatly needed for the discovery and development of new combination therapies for treating fungal infections.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research conducted by the authors in this area is supported in part by R01 AI27094 (to Alice M. Clark) from the National Institute of Allergy and Infectious Diseases, and the Agricultural Research Service Specific Cooperative Agreement No. 58-6408-2-0009 from the United States Department of Agriculture.
References
- Agarwal A. K., Xu T., Jacob M. R., Feng Q., Li X. C., Walker L. A., Clark A. M. (2008). Genomic and genetic approaches for the identification of antifungal drug targets. Infect. Disord. Drug Targets 8, 2–15 [DOI] [PubMed] [Google Scholar]
- Ahmad A., Khan A., Khan L. A., Manzoor N. (2010). In vitro synergy of eugenol and methyleugenol with fluconazole against clinical Candida isolates. J. Med. Microbiol. 59, 1178–1184 10.1099/jmm.0.020693-0 [DOI] [PubMed] [Google Scholar]
- Baryshnikova A., Costanzo M., Kim Y., Ding H., Koh J., Toufighi K., Youn J. Y., Ou J., San Luis B. J., Bandyopadhyay S., Hibbs M., Hess D., Gingras A. C., Bader G. D., Troyanskaya O. G., Brown G. W., Andrews B., Boone C., Myers C. L. (2010). Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat. Methods 7, 1017–1024 10.1038/nmeth.1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M., Chua H. N., Tasan M., Mutlu B., Weinstein Z. B., Suzuki Y., Nergiz M. E., Costanzo M., Baryshnikova A., Giaever G., Nislow C., Myers C. L., Andrews B. J., Boone C., Roth F. P. (2011). Systematic exploration of synergistic drug pairs. Mol. Syst. Biol. 7, 544. 10.1038/msb.2011.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., Vandersluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., Van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010). The genetic landscape of a cell. Science 327, 425–431 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D. W., Hope W. W. (2010). Therapy for fungal diseases: opportunities and priorities. Trends Microbiol. 18, 195–204 10.1016/j.tim.2010.02.004 [DOI] [PubMed] [Google Scholar]
- Digirolamo J. A., Li X. C., Jacob M. R., Clark A. M., Ferreira D. (2009). Reversal of fluconazole resistance by sulfated sterols from the marine sponge Topsentia sp. J. Nat. Prod. 72, 1524–1528 10.1021/np900177m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarra S., Rocha E. M., Zhang Y. Q., Park S., Rao R., Perlin D. S. (2010). Mechanism of the synergistic effect of amiodarone and fluconazole in Candida albicans. Antimicrob. Agents Chemother. 54, 1753–1761 10.1128/AAC.01728-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. H., Piotrowski J., Dixon S. J., Baryshnikova A., Costanzo M., Boone C. (2011). Combining functional genomics and chemical biology to identify targets of bioactive compounds. Curr. Opin. Chem. Biol. 15, 66–78 10.1016/j.cbpa.2010.10.023 [DOI] [PubMed] [Google Scholar]
- Hoon S., Smith A. M., Wallace I. M., Suresh S., Miranda M., Fung E., Proctor M., Shokat K. M., Zhang C., Davis R. W., Giaever G., St Onge R. P., Nislow C. (2008a). An integrated platform of genomic assays reveals small-molecule bioactivities. Nat. Chem. Biol. 4, 498–506 10.1038/nchembio1008-632 [DOI] [PubMed] [Google Scholar]
- Hoon S., St Onge R. P., Giaever G., Nislow C. (2008b). Yeast chemical genomics and drug discovery: an update. Trends Pharmacol. Sci. 29, 499–504 10.1016/j.tips.2008.07.006 [DOI] [PubMed] [Google Scholar]
- Jacob M. R., Hossain C. F., Mohammed K. A., Smillie T. J., Clark A. M., Walker L. A., Nagle D. G. (2003). Reversal of fluconazole resistance in multidrug efflux-resistant fungi by the Dysidea arenaria sponge sterol 9α,11α-epoxycholest-7-ene-3β,5α,6α,19-tetrol 6-acetate. J. Nat. Prod. 66, 1618–1622 10.1021/np030317n [DOI] [PubMed] [Google Scholar]
- Jansen G., Lee A. Y., Epp E., Fredette A., Surprenant J., Harcus D., Scott M., Tan E., Nishimura T., Whiteway M., Hallett M., Thomas D. Y. (2009). Chemogenomic profiling predicts antifungal synergies. Mol. Syst. Biol. 5, 338. 10.1038/msb.2009.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J., Zhu F., Ma X., Cao Z., Li Y., Chen Y. Z. (2009). Mechanisms of drug combinations: interaction and network perspectives. Nat. Rev. Drug Discov. 8, 111–128 10.1038/nrd2922-c1 [DOI] [PubMed] [Google Scholar]
- Johnson M. D., Macdougall C., Ostrosky-Zeichner L., Perfect J. R., Rex J. H. (2004). Combination antifungal therapy. Antimicrob. Agents Chemother. 48, 693–715 10.1128/AAC.48.3.693-715.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehár J., Zimmermann G. R., Krueger A. S., Molnar R. A., Ledell J. T., Heilbut A. M., Short G. F., III, Giusti L. C., Nolan G. P., Magid O. A., Lee M. S., Borisy A. A., Stockwell B. R., Keith C. T. (2007). Chemical combination effects predict connectivity in biological systems. Mol. Syst. Biol. 3, 80. 10.1038/msb4100116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. C., Jacob M. R., Ding Y., Agarwal A. K., Smillie T. J., Khan S. I., Nagle D. G., Ferreira D., Clark A. M. (2006). Capisterones A and B, which enhance fluconazole activity in Saccharomyces cerevisiae, from the marine green alga Penicillus capitatus. J. Nat. Prod. 69, 542–546 10.1021/np0600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hong X., Hussain M., Sarkar S. H., Li R., Sarkar F. H. (2005). Gene expression profiling revealed novel molecular targets of docetaxel and estramustine combination treatment in prostate cancer cells. Mol. Cancer Ther. 4, 389–398 10.1158/1535-7163.MCT-05-0184 [DOI] [PubMed] [Google Scholar]
- Maertens J. A. (2004). History of the development of azole derivatives. Clin. Microbiol. Infect. 10(Suppl. 1), 1–10 10.1111/j.1470-9465.2004.00861.x [DOI] [PubMed] [Google Scholar]
- Mukherjee P. K., Sheehan D. J., Hitchcock C. A., Ghannoum M. A. (2005). Combination treatment of invasive fungal infections. Clin. Microbiol. Rev. 18, 163–194 10.1128/CMR.18.1.163-194.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P. K., Wang M. (2010). “Antifungal drug resistance: significance and mechanisms,” in Antifungal Therapy, eds Ghannoum M. A., Perfect J. R. (New York: Informa Healthcare; ), 63–86 [Google Scholar]
- Niimi K., Harding D. R., Parshot R., King A., Lun D. J., Decottignies A., Niimi M., Lin S., Cannon R. D., Goffeau A., Monk B. C. (2004). Chemosensitization of fluconazole resistance in Saccharomyces cerevisiae and pathogenic fungi by a D-octapeptide derivative. Antimicrob. Agents Chemother. 48, 1256–1271 10.1128/AAC.48.4.1256-1271.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyewu C., Blankenship J. R., Del Poeta M., Heitman J. (2003). Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob. Agents Chemother. 47, 956–964 10.1128/AAC.47.3.956-964.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons A. B., Lopez A., Givoni I. E., Williams D. E., Gray C. A., Porter J., Chua G., Sopko R., Brost R. L., Ho C. H., Wang J., Ketela T., Brenner C., Brill J. A., Fernandez G. E., Lorenz T. C., Payne G. S., Ishihara S., Ohya Y., Andrews B., Hughes T. R., Frey B. J., Graham T. R., Andersen R. J., Boone C. (2006). Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126, 611–625 10.1016/j.cell.2006.06.040 [DOI] [PubMed] [Google Scholar]
- Pasqualotto A. C., Denning D. W. (2008). New and emerging treatments for fungal infections. J. Antimicrob. Chemother. 61(Suppl. 1), i19–i30 10.1093/jac/dkm428 [DOI] [PubMed] [Google Scholar]
- Pemán J., Cantón E., Espinel-Ingroff A. (2009). Antifungal drug resistance mechanisms. Expert Rev. Anti. Infect. Ther. 7, 453–460 10.1586/eri.09.18 [DOI] [PubMed] [Google Scholar]
- Perlin D. S. (2007). Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10, 121–130 10.1016/j.drup.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrikkos G., Skiada A. (2007). Recent advances in antifungal chemotherapy. Int. J. Antimicrob. Agents 30, 108–117 10.1016/j.ijantimicag.2007.05.008 [DOI] [PubMed] [Google Scholar]
- Pfaller M. A., Messer S. A., Georgopapadakou N., Martell L. A., Besterman J. M., Diekema D. J. (2009). Activity of MGCD290, a Hos2 histone deacetylase inhibitor, in combination with azole antifungals against opportunistic fungal pathogens. J. Clin. Microbiol. 47, 3797–3804 10.1128/JCM.00618-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D. (2002). Resistance of human fungal pathogens to antifungal drugs. Curr. Opin. Microbiol. 5, 379–385 10.1016/S1369-5274(02)00344-2 [DOI] [PubMed] [Google Scholar]
- Sharma M., Manoharlal R., Negi A. S., Prasad R. (2010). Synergistic anticandidal activity of pure polyphenol curcumin I in combination with azoles and polyenes generates reactive oxygen species leading to apoptosis. FEMS Yeast Res. 10, 570–578 [DOI] [PubMed] [Google Scholar]
- Spitzer M., Griffiths E., Blakely K. M., Wildenhain J., Ejim L., Rossi L., De Pascale G., Curak J., Brown E., Tyers M., Wright G. D. (2011). Cross-species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol. Syst. Biol. 7, 499. 10.1038/msb.2011.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiziani S., Lodi A., Khanim F. L., Viant M. R., Bunce C. M., Günther U. L. (2009). Metabolomic profiling of drug responses in acute myeloid leukaemia cell lines. PLoS ONE 4, e4251. 10.1371/journal.pone.0004251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., Chen Y., Cheng X., Chua G., Friesen H., Goldberg D. S., Haynes J., Humphries C., He G., Hussein S., Ke L., Krogan N., Li Z., Levinson J. N., Lu H., Menard P., Munyana C., Parsons A. B., Ryan O., Tonikian R., Roberts T., Sdicu A. M., Shapiro J., Sheikh B., Suter B., Wong S. L., Zhang L. V., Zhu H., Burd C. G., Munro S., Sander C., Rine J., Greenblatt J., Peter M., Bretscher A., Bell G., Roth F. P., Brown G. W., Andrews B., Bussey H., Boone C. (2004). Global mapping of the yeast genetic interaction network. Science 303, 808–813 10.1126/science.1091317 [DOI] [PubMed] [Google Scholar]
- Wei G. X., Xu X., Wu C. D. (2011). In vitro synergism between berberine and miconazole against planktonic and biofilm Candida cultures. Arch. Oral Biol. 56, 565–572 10.1016/j.archoralbio.2010.11.021 [DOI] [PubMed] [Google Scholar]
- Xu Y., Wang Y., Yan L., Liang R. M., Dai B. D., Tang R. J., Gao P. H., Jiang Y. Y. (2009). Proteomic analysis reveals a synergistic mechanism of fluconazole and berberine against fluconazole-resistant Candida albicans: endogenous ROS augmentation. J. Proteome Res. 8, 5296–5304 10.1021/pr900251d [DOI] [PubMed] [Google Scholar]
- Yeh P. J., Hegreness M. J., Aiden A. P., Kishony R. (2009). Drug interactions and the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7, 460–466 10.1038/nrmicro2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G. R., Lehár J., Keith C. T. (2007). Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov. Today 12, 34–42 10.1016/j.drudis.2006.11.008 [DOI] [PubMed] [Google Scholar]