Abstract
Interaction of light-activated rhodopsin with transducin (T) is the first event in visual signal transduction. We use covalent crosslinking approaches to map the contact sites in interaction between the two proteins. Here we use a photoactivatable reagent, N-[(2-pyridyldithio)-ethyl], 4-azido salicylamide. The reagent is attached to the SH group of cytoplasmic monocysteine rhodopsin mutants by a disulfide-exchange reaction with the pyridylthio group, and the derivatized rhodopsin then is complexed with T by illumination at λ >495 nm. Subsequent irradiation of the complex at λ310 nm generates covalent crosslinks between the two proteins. Crosslinking was demonstrated between T and a number of single cysteine rhodopsin mutants. However, sites of crosslinks were investigated in detail only between T and the rhodopsin mutant S240C (cytoplasmic loop V-VI). Crosslinking occurred predominantly with Tα. For identification of the sites of crosslinks in Tα, the strategy used involved: (i) derivatization of all of the free cysteines in the crosslinked proteins with N-ethylmaleimide; (ii) reduction of the disulfide bond linking the two proteins and isolation of all of the Tα species carrying the crosslinked moiety with a free SH group; (iii) adduct formation of the latter with the N-maleimide moiety of the reagent, maleimido-butyryl-biocytin, containing a biotinyl group; (iv) trypsin degradation of the resulting Tα derivatives and isolation of Tα peptides carrying maleimido-butyryl-biocytin by avidin-agarose chromatography; and (v) identification of the isolated peptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. We found that crosslinking occurred mainly to two C-terminal peptides in Tα containing the amino acid sequences 310–313 and 342–345.
Keywords: cysteine mutants, disulfide exchange, cytoplasmic domain, avidin-biotin affinity
Light activation of rhodopsin initiates two biochemical cascades, one leading to sensitization (amplification) and the other to desensitization (quenching). Binding and activation of transducin (T) and rhodopsin kinase are, respectively, the first events in the two cascades (1). Competition between T and rhodopsin kinase for interaction with the light-activated rhodopsin and its phosphorylated form is central to the progression of the two cascades. Therefore, understanding the requirements of the two proteins for binding to rhodopsin and the contact sites in the complexes formed is of much interest. Ideally, three-dimensional structures of the complexes between the proteins are needed, but such structural analysis lies in the future. Alternative approaches have provided some insights into the sites in rhodopsin required for binding of T. Thus, peptides corresponding to the amino acid sequences in the different cytoplasmic loops of rhodopsin inhibit T activation (2), and similarly mutagenic studies (3–5) have indicated that a considerable portion of the cytoplasmic domain of light-activated rhodopsin is involved in binding to T. Some information also has been obtained on the regions of T that contact rhodopsin in the complex. Thus, the effects of anti-Tα antibody binding (6, 7), ADP-ribosylation of the α subunit of transducin (Tα), and peptide competition for binding (8) all indicated that the C-terminal region of Tα is a major site of interaction with rhodopsin. More recently, involvement of amino acid sequence 330–350 in the C-terminal region of Tα in binding to rhodopsin received further support by alanine scanning, chimeric receptor studies, and peptide-binding studies (9–11). Additional evidence suggested that the region around residue R310 in Tα also is likely to be involved (9, 12, 13). However, further insights into the contact sites in complex formation between light-activated rhodopsin and T are desirable. An earlier paper described a crosslinking approach to determine the contact sites between the two proteins by the use of a photoactivatable carbene-generating reagent (14). This basic approach has been investigated further in this and an accompanying paper (16). In work herein reported, we use a nitrene-generating photoactivatable reagent for covalent crosslinking of the above two proteins. Photoactivatable crosslinking reagents pioneered by Westheimer and coworkers (17) in exploring three-dimensional proximity relationships have been found to have extensive applications (18–20). The reagents generate highly reactive short-lived electron-deficient carbene or nitrene intermediates after photolysis that react with neighboring amino acids. For identification of the sites of crosslinking, radioactively labeled crosslinking reagents have been used in the past; however, mass spectrometry now offers a more facile method for this purpose.§
Strategy for Covalent Crosslinking of T to Light-Activated Rhodopsin and Mass Spectrometric Identification of the Sites of Crosslinking in T
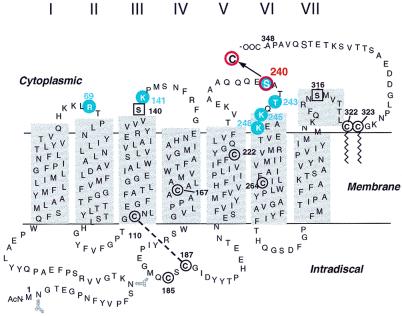
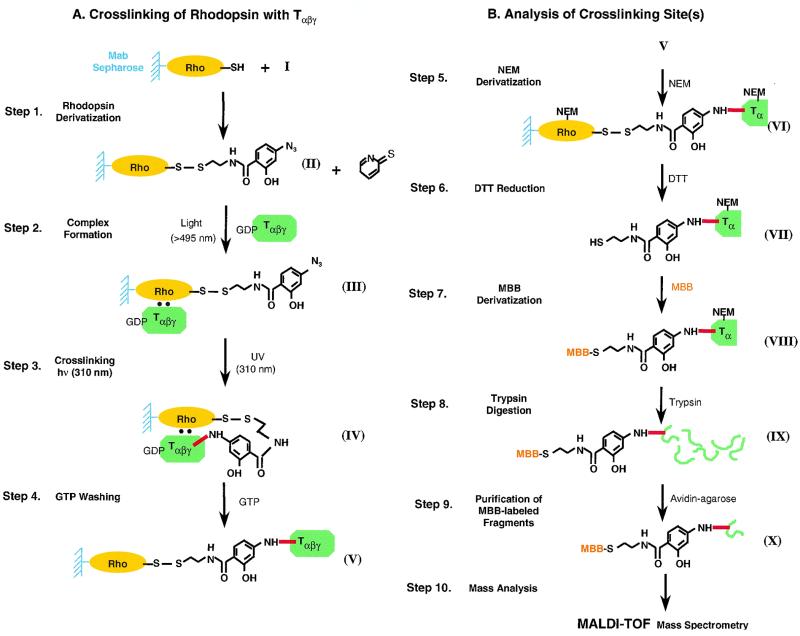
Monocysteine mutants of rhodopsin on the cytoplasmic face (Fig. 1) form the starting points in attachment of the crosslinking agents. Thus, the site of contact with rhodopsin itself is defined at the outset by the position of the cysteine group on the cytoplasmic face. Structure of the crosslinking reagent used here is shown in Fig. 2 (21, 22). The steps used in the strategy for covalent crosslinking are shown in Fig. 3A. The pyridine-thio group, a reactive leaving group, is displaced readily by the SH groups of rhodopsin cysteine mutants, and the disulfide-exchange reaction results in transfer of the group R (Fig. 2) to rhodopsin (Fig. 3A, II, Step 1). This step and several of the subsequent steps are carried out conveniently while the rhodopsin mono-cysteine mutants are bound to Sepharose beads via the rho-1D4-antibody. Rhodopsin derivatives (Fig. 3A, II) are irradiated at λ >495 nm in the presence of GDP-bound transducin (GDP/Tαβγ) to form complex III between the two proteins. Step 3 involves photolysis of the azido-phenolic group at λ310 nm. The photolysis is carried out efficiently by using the monochromatic light from a fluorometer. Subsequent washes with GTP (step 4) selectively release Tβγ as well as the excess of unreacted GDP/Tαβγ. The crosslinked portion of T is retained as complex V. The strategy for subsequent identification of the sites of crosslinks in Tα is shown in Fig. 3B. Isolation of Tα containing the crosslinked moiety donated by reagent I (Fig. 1) aimed at the use of the SH group present in the moiety for linkage to the maleaimido group in maleimido-butyryl-biocytin (MBB; Fig. 4) for subsequent use of biotin-avidin affinity chromatography. Therefore, first, the SH groups in all the cysteines in rhodopsin and Tα were derivatized with N-ethylmaleimide (NEM; step 5, Fig. 3B). This reaction was done at pH 6 to avoid any exchange reaction of the disulfide group present in the crosslinked proteins. The NEM-derivatized product (Fig. 3B, VI) was treated next with DTT (step 6) to release VII. In step 7, the SH group in VII now was derivatized with MBB to form VIII. Subsequent trypsin digestion (step 8) would form a mixture of peptides (Fig. 3B, IX). Only the MBB-derivatized peptides containing the crosslinks to Tα would be expected to bind to avidin-agarose in step 9. The MBB-labeled peptides thus selected (X) were subjected to matrix-assisted laser desorption ionization/time-of-flight mass spectrometry (MALDI/TOF; step 10). Development of the above strategy for the objective of crosslinking and subsequent identification of the sites of crosslinking in T has been the major goal in this initial investigation. Although crosslinking was demonstrated to occur between T and several monocysteine mutants of rhodopsin (Fig. 1), the total methodology leading to the identification of crosslinking sites was completed only for the rhodopsin mutant S240C.
Figure 1.
A secondary structure model of rhodopsin showing positions of the single cysteine-containing mutants that were tested for attachment of R (I) and showed photocrosslinking with heterotrimeric Tαβγ (circled in blue). Mutant S240C (circled in red) was taken through all steps in the strategy described in Fig. 3.
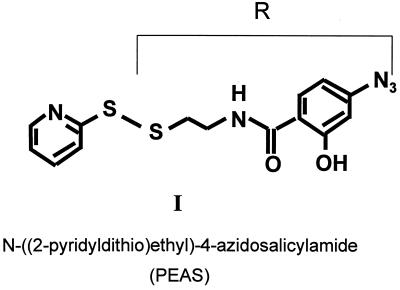
Figure 2.
The photoactivatable reagent N-[(2-pyridyldithio)ethyl]-4-azido-salicylamide (I). The group R is transferred to the SH group of cysteine residue in a rhodopsin mutant by a disulfide-exchange reaction with the reactive pyridylthio group in I.
Figure 3.
Steps in strategy for photocrosslinking (A) and subsequent analysis of crosslinked sites in Tαβγ (B). See text for details.
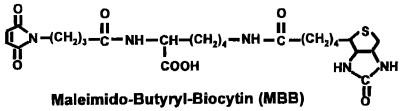
Figure 4.
Structure of MBB, an SH-specific reagent containing a biotinyl group.
Materials and Methods
Materials.
The reagent I, N-[(2-pyridyldithio)-ethyl], 4-azido salicylamide (Fig. 2) was purchased from Molecular Probes; MBB was from Calbiochem, and GTP was from Roche Molecular Biochemicals. GDP/Tαβγ was prepared from bovine rod outer segments as described (23) and was stored in buffer containing 100 mM NaCl, 20 mM Hepes (pH 7.5), 5 mM Mg(OAc)2, 2 mM DTT, and 50% (vol/vol) glycerol. Monoclonal antibody Mab4A against Tα was provided kindly by H. E. Hamm ( Northwestern University Institute of Neuroscience, Evanston, IL). Polyclonal antibodies K-20 against Tα and M-14 against Tβ were purchased from Santa Cruz Biotechnology. Anti-biotin antibody was purchased from New England Biolabs. α-Cyano-4-hydroxycinamic acid, biotin, and monomeric avidin-agarose were purchased from Sigma. Sequencing-grade trypsin was purchased from Promega. C18 zip-tips were from Millipore. Poly(vinylidene difluoride) membrane was purchased from Bio-Rad.
The buffer solutions used are: buffer A, 137 mM NaCl/2.7 mM KCl/1.8 mM KH2PO4/10 mM Na2HPO4 (pH 7.2); buffer B, 5 mM Mes (pH 6.0)/0.05% DM; buffer C, 10 mM Mes (pH 6.0)/100 mM NaCl/2 mM MgCl2/0.012% DM; buffer D, 20 mM Mes (pH 6.0)/50% glycerol/100 mM NaCl/5 mM Mg(OAc)2; buffer E, 5 mM Mes/150 mM NaCl/1 mM EDTA/0.012% DM; buffer F, 10 mM NaPi (pH 7.5)/0.012% DM.
Expression of Rhodopsin Gene Mutants in COS-1 Cells and Purification of Rhodopsin Mutants.
The construction of vectors for the single-cysteine rhodopsin gene mutants, their expression in COS-1 cells, reconstitution of the expressed proteins with 11-cis-retinal, and purification of rhodopsin mutants by using 1D4-immunoaffinity column were all as described (24).
SDS/PAGE and Immunoblotting.
SDS/PAGE was performed by using 10% acrylamide gels. Samples of crosslinked products were reduced immediately after the addition of Laemmli buffer by adding DTT to a final concentration of 10 mM. The samples were incubated at 20°C for half an hour before SDS/PAGE. For immunoblotting, proteins were transferred from the gel to poly(vinylidene difluoride) membrane, and anti-Tα antibody Mab4A or K-20 was used to visualize Tα, anti-Tβ antibody M-14 for Tβ, and anti-biotin antibody for MBB-labeled transducin.
Reaction of Rhodopsin Cysteine Mutants with N-[(2-pyridyldithio)ethyl], 4-azido Salicylamide (I) to Form Disulfide-Bonded Rhodopsin Derivatives (Fig. 3A, Step 1, II).
The reaction with I was carried out while the rhodopsin cysteine mutants were bound to the 1D4-Sepharose beads. The beads containing bound rhodopsin mutants (i.e., mutant S240C, 200 μl) were packed into a 2-ml spin column (Bio-Rad), and the beads were washed first with 30 bed volumes of buffer A followed by a further wash with 15 bed volumes of buffer B. A solution of I in 2 ml of buffer B (200 μM) was added and after a 3-h incubation at room temperature, the beads were washed with 30 bed volumes of buffer B. The washed beads were used for photocrosslinking experiments as below. If required, the derivatized rhodopsin mutants (II) were eluted from the matrix with the same buffer containing C′,C′9 peptide as described (24).
Complex Formation Between Rhodopsin Derivatives (II) and GDP/Tαβγ (Fig. 3, Step II)/Light-Dependent Binding of GDP/Tαβγ to 1D4-Sepharose-Bound Rhodopsin.
1D4-Sepharose resin with bound rhodopsin derivative (Fig. 3, II; 600 pmol) was washed with 10 bed volumes of buffer C; GDP/Tαβγ, from which DTT had been removed by a 1-ml Sephadex G-50 spin column equilibrated with buffer D, then was added to the resin in the dark at a rhodopsin/Tαβγ ratio of 1:2. Half of the mixture was kept in the dark while the other half was bleached with light (>495 nm) to initiate the formation of the rhodopsin–T complex. Excess Tαβγ that was not bound to rhodopsin was washed from the resin with 250 bed volumes of buffer C. The portion of Tαβγ that formed complex with the bound rhodopsin then was eluted either together with rhodopsin with 20 μl of buffer C containing 200 μM of rhodopsin C′,C′9 peptide or alone with 20 μl of buffer C containing 200 μM of GTP. All eluted fractions then were analyzed by reducing SDS/PAGE and immunoblotting using antibody Mab4A or K-20 against Tα.
UV-Activation of Azido-Phenyl Group in R (I) for Crosslinking to Tαβγ.
Photolyis was carried out at λ310 nm by using a xenon lamp in a PTI MPIII fluorescence spectrophotometer. A sample tube containing both rhodopsin-bound 1D4-Sepharose beads and Tαβγ was placed in the cuvette holder in the fluorometer, and irradiation at 310 nm as well as by the yellow light (λ >495 nm) from a slide-projector lamp were carried out simultaneously to keep rhodopsin at the meta II state. We found that UV irradiation at 310 nm of rhodopsin meta II caused the red shift of meta II UV absorption. However, such effect was diminished by bleaching the sample with >495 nm light at the same time. The slide projector was placed above the cuvette holder such that both the yellow light and the UV light from the xenon lamp illuminated the sample. Photolysis was performed for 3 min.
Photocrosslinking of Derivatized Rhodopsin Cysteine Mutant Derivatized with R (Fig. 3B, II) to Tαβγ.
Photocrosslinking of derivatized rhodopsin cysteine mutants to Tαβγ was carried out while rhodopsin was bound to 1D4-Sepharose following the procedure in Fig. 3A. Approximately 3 nmol of N-[(2-pyridyldithio)-ethyl], 4-azido salicylamide-labeled rhodopsin S240C was mixed with Tαβγ in buffer C (rhodopsin/Tαβγ = 1:2) in the dark. The samples were bleached with >495 nm light and subsequently irradiated with UV at 310 nm. To remove excess unbound Tαβγ, the resin was washed extensively with 250 bed volumes of buffer C, the portion of Tαβγ that was bound to but not covalently crosslinked to rhodopsin then was eluted with 100 μl of buffer C containing 200 μM GTP for three times followed by a further washing with 15 bed volumes of the same GTP-containing buffer.
Derivatization of SH Groups in Rhodopsin–T Complex with NEM.
After being washed with the GTP-containing buffer, the 1D4-Sepharose-bound rhodopsin–Tα complex (V) was incubated with 40 mM NEM in buffer E for 1 h at room temperature (Fig. 3B, step 5). The resin then was washed with 50 bed volumes of buffer E followed by 10 bed volumes of buffer F. The resulting NEM-derivatized product (Fig. 3, VI) was incubated with 1 mM DTT in 150 μl of the same buffer (vortexing for 20 min at room temperature; Fig. 3B, step 6).
Derivatization of the Single SH Group with MBB (Fig. 3, VII, Step 7).
The single SH group now released in VII was incubated with 5 mM MBB in buffer F to give VIII (Fig. 3B). The latter was analyzed and purified by reducing SDS/PAGE. After the electrophoresis, protein bands corresponding to MBB-derivatized T were sliced out before the gel was stained. The position of the protein band was determined either by the standard protein marker or by staining one of the lanes on which some of the tagged Tα was applied. The gel slices then were macerated and immersed into 300 μl of water, and the diffusive elution of the protein from the gel was allowed overnight.
Trypsin Digestion of Purified Tα Tagged with Crosslinking Moiety (Fig. 3B, Step 8).
VIII (Fig. 3B) eluted from the acrylamide gel was incubated with trypsin at a substrate/trypsin ratio of 50:1 in 50 mM NH4HCO3 (pH 8.0). The digestion was allowed for 9 h at 37°C. The reaction then was terminated by snap-freezing in liquid N2.
Purification of MBB-labeled Trypsin-Digestion Fragments by Avidin-Agarose Affinity Chromatography (Fig. 3B, Step 9).
Biotin (1 mg/ml in buffer A) was used to block the nonexchangeable biotin-binding sites in 40 μl of monomeric avidin-agarose beads. First, agarose resin was washed with 20 bed volumes of 0.1 M glycine-HCl (pH 2.0) and then equilibrated with 20 bed volumes of buffer A. Trypsin-digested peptide fragments (Fig. 3B, IX) were mixed with equilibrated avidin-agarose and mutated for 1 h at room temperature. The resin then was washed with 100 bed volumes of buffer A, and MBB-containing peptides (Fig. 3B, X) that were bound to avidin-agarose were eluted subsequently with biotin (1 mg/ml); four fractions (100 μl each fraction) were collected. To increase the elution efficiency, agarose beads were allowed to sit for 20 min at room temperature in between each fraction.
Mass Spectrometry.
Mass analysis of avidin-agarose purified tryptic fragments of T was performed (step 10) by using MALDI/TOF (PerSeptive Biosystems, Framingham, MA). Before the mass analysis, peptide fragments eluted with biotin were purified further by using C18 zip-tips to remove biotin and excess salt. Purified peptides (1 μl) then were mixed with an equal volume of the matrix α-cyano-4-hydroxycinamic acid (in 50% acetonitrile/0.1% trifluoroacetic acid) and subjected to mass spectrometry. Mass spectra were taken by using the linear mode.
Results
Light-Induced Binding of GDP/Tαβγ to Rhodopsin Bound to 1D4-Sepharose.
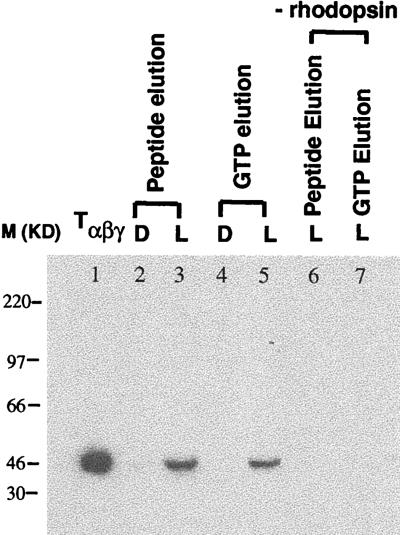
Development of the strategy in Fig. 3 necessitated investigation of conditions to be used essentially at every step. Use of rhodopsin while bound to 1D4-Sepharose offered significant simplification of the procedure at several steps. Therefore in the first experiment, light-dependent binding of GDP/Tαβγ to Sepharose-bound rhodopsin was tested. Rhodopsin thus bound was illuminated (λ >495 nm) in the presence of GDP/Tαβγ. Excess of Tαβγ was removed by washing, and the portion of GDP/Tαβγ complexed with rhodopsin then was eluted from the Sepharose matrix together with rhodopsin in the presence of the epitope peptide. In an alternative experiment, elution with GTP was performed to release Tα (formed by breakdown of GTP/Tα from the matrix). Fig. 5 shows the results obtained with immunoblot analysis of fractions after SDS/PAGE, under reducing conditions, by using anti-Tα antibody Mab4A. Lane 2 contains the eluate with the epitope peptide in the dark, and lane 3 contains the corresponding eluate after illumination of the matrix. Lanes 4 and 5 are the corresponding samples (dark and light) after elution with GTP. Immunoblots of lanes 2 and 3 and 4 and 5 showed clearly that the binding of GDP/Tαβγ to rhodopsin was light dependent. Lane 1 is the control showing the position of Tα, and lanes 6 and 7 are controls (rhodopsin−) for lanes 3 and 5, respectively.
Figure 5.
Light-dependent binding of GDP/Tαβγ to wild-type rhodopsin bound to 1D4-Sepharose. Analysis was performed by SDS/PAGE (reducing). Tαβγ was visualized by immunoblotting using Ab Mab4A against Tα. Samples were treated as described in Materials and Methods. After extensive washing of 1D4 Sepharose beads both in the dark (D) and after illumination (L), Tα and rhodopsin were eluted together from the matrix with the epitope peptide (lane 2, dark; lane 3, after illumination). Tα was released also alone from the matrix by the addition of GTP (lane 4, dark; lane 5, after illumination). Controls are: lane 1, Tαβγ; lanes 6 and 7, controls of lanes 3 and 5 without 1D4-Sepharose-bound rhodopsin.
Next, control experiments were performed to test the effect of binding of rhodopsin to 1D4 Ab on the rate of Tαβγ activation by rhodopsin. The rates of Tαβγ activation by wild-type rhodopsin were compared by using the fluorescence assay (25) in the presence of various concentrations of the antibody 1D4 in solution with the rate in the absence of the antibody. No significant difference in the rates was seen (data not shown). Thus, binding of the antibody to the C terminus of rhodopsin either in solution or when bound to the Sepharose matrix did not affect the binding or activation of Tαβγ.
Crosslinking of Rhodopsin Cysteine Mutant S240C with Tα After Photolysis of Group R (Fig. 1) in II (Fig. 3A) at λ310 nm.
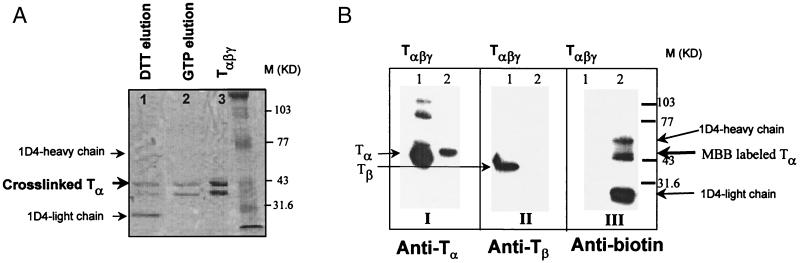
The rhodopsin mutant S24C carrying R (Fig. 3A, II) was complexed with GDP/Tαβγ by light activation, and the resulting III (Fig. 3A) was illuminated at λ310 nm. The matrix then was washed with GTP (Fig. 3A, step 4), followed by treatment of the complex bound to the matrix (Fig. 3A, V) with 10 mM DTT. Both the DTT eluate and the GTP eluate were analyzed (Coomassie blue staining) by SDS/PAGE, and the results are shown in Fig. 6. As judged by the intensity of the protein bands, GTP eluate (Fig. 6A, lane 2) appeared to contain more Tβ than Tα, whereas in the DTT eluate (lane 1), the band corresponding to Tα was more prominent. The DTT eluate (Fig. 6A, lane 1) also showed two additional bands that were identified as 1D4-heavy and -light chains, respectively, resulting from partial reduction of the antibody by DTT.
Figure 6.
SDS/PAGE analysis of products formed in crosslinking of rhodopsin derivative S240C-R and Tαβγ. The photocrosslinking procedure was performed as described in Fig. 3. All samples were treated with 10 mM DTT immediately after the addition of Laemmli SDS sample buffer. The gel was visualized by Coomassie blue staining. (A) After GTP washing (Step 4) to remove uncrosslinked Tαβγ (lane 2), sample V was reduced by DTT and the eluted protein is shown in lane 1. Lane 3 contains 25 pmol of purified Tαβγ as a control. (B) Sample V in Fig. 3 was treated with NEM and eluted with DTT, and the eluate was then derivatized with MBB (Steps 5–7) in Fig. 3B. Sample VIII (Fig. 3B) then was analyzed by Western blotting using antibodies against Tα (I), Tβ (II), and biotinyl moiety (III). Lane 1 in all three cases contained purified Tαβγ as a control.
Characterization of the Derivatization of the SH Group in R (Fig. 2) with MBB.
The strategy for identification of the site of crosslinking in Tα (Fig. 3B) required the selective derivatization of the SH group in R as linked to specific peptides. As a control, the reactivity of the SH group in R was checked. The latter was prepared from I by reduction with an excess of DTT for 30 min at room temperature and treated with an 8-fold excess of MBB (Fig. 4) overnight. The expected adduct with MBB was characterized by MALDI/TOF. The main peak obtained (m/z, 788.16) corresponded to the K+ salt of the adduct (data not shown). The nonadduct form of this product (MH+ = 750.4) also was seen as minor peaks in adduct forms with Na+ and K+ (data not shown).
Derivatization of V (Fig. 3B) with NEM, Reduction of NEM-Derivatized VI with DTT, and Reaction of the Single Cysteine in VII with MBB.
Covalently linked product V from mutant S240C and Tα (Fig. 3A) was treated with NEM (Fig. 3B), and the derivatized VI was reduced with DTT that released the Tα-derivative VII containing a single SH group. The latter then was derivatized with MBB (Fig. 4), and the product was characterized as in Fig. 6B by probing with antibodies against Tα, Tβ, and biotin group. The derivatized VIII contained predominantly the Tα (Fig. 6B, I, lane 2) and not much of the β subunit (II, lane 2). Moreover, it carried biotin moiety as shown by immunoblot analysis by using the anti-biotin antibody (Fig. 6B, III). Also seen in III are the protein bands with sizes matching those of 1D4-heavy and -light chains. These antibody 1D4 chains formed after reduction with DTT evidently were derivatized by MBB together with VIII.
Therefore, to purify VIII from both the antibody components of 1D4 and the excess of free MBB, SDS/PAGE was performed, and the protein bands corresponding to VIII were sliced out from the gel, gel slices were macerated, and VIII was eluted from the gel by passive diffusion. The resulting sample then was digested with trypsin (Fig. 3B, step 8), and MBB-derivatized fragments were purified by chromatography using avidin-agarose (Fig. 3B, step 9).
MALDI/TOF Identification of Sites of Crosslinking to Tα.
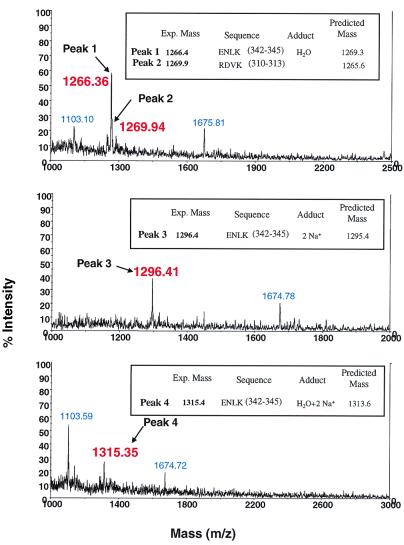
Results of analysis of MBB-labeled fragments (Fig. 3B, X) are shown in Fig. 7. As seen in the three panels, a total of four peaks (with mass shown in red) corresponding to m/z of 1,266.4 (peak 1), 1,269.9 (peak 2), 1,296.4 (peak 3), and 1,315.3 (peak 4) were observed. Their identifications are given in the insets of the individual panels. They correspond to the peptide sequences 310–313 (R DVK) and 342–345 (ENLK; Figs. 7 and 8) in Tα. Two additional peaks (mass highlighted in blue) that were observed were found also in the two controls when Tα alone or trypsin alone was used in the trypsin-digestion experiments (data not shown).
Figure 7.
MALDI analysis of the MBB-labeled tryptic fragments of T2 obtained after avidin affinity purification (Fig. 3B, ). Peaks labeled in red are the identified fragments. Amino acid sequences and the predicted masses of these fragments are indicated in the insets. Unidentified peaks labeled in blue also appeared in the two control samples (trypsin alone and Tα-MBB alone, data not shown).
Figure 8.
Amino acid sequence of Tα. Shaded sequences indicate the secondary structural elements in the C-terminal region (26). The two regions identified from photocrosslinking are highlighted in red. Sequence 310–313 (R DVK), is located between the α4 and β6 regions of Tα, and the sequence 342–345 is near the C terminus.
Discussion
The study of protein–protein interactions by probing covalent crosslinking between proximal sites in the proteins is a time-honored approach (17–20). Thus, in the 1960s, it provided extensive information on proximity relationships between structural proteins in the ribosomal subunits (27). Research using this approach continues to find applications in studies of a variety of important questions. Two specific and elegant recent applications are in studies of multisubunit protein–DNA interactions (e.g., refs. 28 and 29) and in studies of dynamics of protein transport across the eukaryotic endoplasmic reticulum (30). Previously, the general approach has been applied in this laboratory to studies of membrane protein–phospholipids interactions (31–34) and in probing the retinal binding pockets in both bacteriorhodopsin (35) and rhodopsin (36). In this report and in an accompanying paper (16), we have developed a general strategy partly based on earlier work (14) that first defines the location of a crosslinking reagent on the cytoplasmic face of rhodopsin and subsequent crosslinking that aims at identifying the sites in T that are proximal.
Previous work on the interaction of T with light-activated rhodopsin by several groups has indicated that the interaction involves mainly the α subunit. The total evidence suggests that the C-terminal region of Tα is a major site of interaction. The strategy now developed has provided more precise information on the contact sites in rhodopsin and T interaction. The photoactivatable crosslinking group attached at position 240 in rhodopsin crosslinked mainly to two C-terminal sequences, 310–313 [between α-helix 4 and β-sheet 6 in crystal structure (26)] and 342–345. Both regions containing the above sequences were postulated previously to be involved in binding to rhodopsin (9–13). Our results have defined more specifically the amino acid sequences in Tα that are proximal to the third cytoplasmic loop of rhodopsin in the interaction between the two proteins.
The major aim of the present work has been to develop a general strategy for studies of protein–protein interactions that uses the covalent-crosslinking approach and includes mass spectrometric analysis of the crosslinked peptide sequences. The latter is far superior to the labor-intensive radioactive-labeling methods used previously. Further work on mapping of the contact sites can take a number of directions. There is an infinite variety in chemical concepts that have been used in design of crosslinking agents previously, and undoubtedly these can be developed further. In every crosslinking reagent, the spacer length between the two ends carrying the functional groups can be systematically varied. Then, there are the two main classes of photoactivatable groups, those generating carbenes and those forming nitrenes as the reactive intermediates. Only one of the simplest conceivable nitrene-generating reagent (I) has been investigated in the present work. Finally, it is noted that more than 100 rhodopsin mutants containing single reactive cysteines that span all of the cytoplasmic face of rhodopsin are available from our previous structural studies. Availability of these mutants and versatility in crosslinking reagents should enable detailed mapping of interactions of rhodopsin with the proteins involved in visual signal transduction.
Acknowledgments
We thank Professor U. L. RajBhandary (Biology, Massachusetts Institute of Technology) for valuable discussions. Enthusiastic assistance of Ms. Judy Carlin in the preparation of the manuscript is acknowledged. This work was supported by National Institutes of Health Grant GM28289 and National Eye Institute Grant EY11717 (H.G.K.). Y.I. was supported by Banyu Tsukuba Research Institute, Tsukuba 300-2611, Japan during a leave of absence. K.C. was supported by the Massachusetts Institute of Technology Department of Chemistry Cancer Training Grant CA 09112.
Abbreviations
- T
transducin
- Tα
the α subunit of T
- Tβ
the β subunit of T
- Tαβγ
heterotrimeric T
- MBB
maleimido-butyryl-biocytin
- GDP/T
GDP-bound transducin
- NEM
N-ethylmaleimide
- MALDI/TOF
matrix-assisted laser desorption ionization/time-of-flight mass spectrometry
Footnotes
References
- 1.Khorana H G. J Biol Chem. 1992;267:1–4. [PubMed] [Google Scholar]
- 2.Konig B, Arendt A, McDowell J H, Kahlert M, Hargrave P A, Hofmann K P. Proc Natl Acad Sci USA. 1989;86:6878–6882. doi: 10.1073/pnas.86.18.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke R H, Sakmar T P, Graham R M, Khorana H G. J Biol Chem. 1992;267:14767–14774. [PubMed] [Google Scholar]
- 4.Yang K, Farrens D L, Hubbell W L, Khorana H G. Biochemistry. 1996;35:12464–12469. doi: 10.1021/bi960848t. [DOI] [PubMed] [Google Scholar]
- 5.Cai K, Klein-Seetharaman J, Farrens D, Cheng Z, Altenbach C A, Hubbell W L, Khorana H G. Biochemistry. 1999;38:7925–7930. doi: 10.1021/bi9900119. [DOI] [PubMed] [Google Scholar]
- 6.Mazzoni M R, Malinski J A, Hamm H E. J Biol Chem. 1991;266:14072–14081. [PubMed] [Google Scholar]
- 7.Hamm H E, Deretic D, Arendt A, Hargrave P A, Koenig B, Hofmann K P. Science. 1988;241:832–835. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 8.Ramdas L, Disher R M, Wensel T G. Biochemistry. 1991;30:11637–11645. doi: 10.1021/bi00114a005. [DOI] [PubMed] [Google Scholar]
- 9.Onrust R, Herzmark P, Chi P, Garcia P D, Lichtarge O, Kingsley C, Bourne H R. Science. 1997;275:381–384. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 10.Natochin M, Muradov K G, McEntaffer R L, Artemyev N O. J Biol Chem. 2000;275:2669–2675. doi: 10.1074/jbc.275.4.2669. [DOI] [PubMed] [Google Scholar]
- 11.Ernst O P, Meyer C K, Marin E P, Henklein P, Fu W Y, Sakmar T P, Hofmann K P. J Biol Chem. 2000;275:1937–1943. doi: 10.1074/jbc.275.3.1937. [DOI] [PubMed] [Google Scholar]
- 12.Mazzoni M R, Hamm H E. J Biol Chem. 1996;271:30034–30040. doi: 10.1074/jbc.271.47.30034. [DOI] [PubMed] [Google Scholar]
- 13.Natochin M, Granovsky A E, Muradov K G, Artemyev N O. J Biol Chem. 1999;274:7865–7869. doi: 10.1074/jbc.274.12.7865. [DOI] [PubMed] [Google Scholar]
- 14.Resek J R, Farrens D L, Khorana H G. Proc Natl Acad Sci USA. 1994;91:7643–7647. doi: 10.1073/pnas.91.16.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwa J, Klein-Seetharaman J, Khorana H G. Proc Natl Acad Sci USA. 2001;98:4872–4876. doi: 10.1073/pnas.061632798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh Y, Cai K, Khorana H G. Proc Natl Acad Sci USA. 2001;98:4883–4887. doi: 10.1073/pnas.051632998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Thornton E R, Westheimer F H. J Biol Chem. 1962;237:3006–3008. [PubMed] [Google Scholar]
- 18.Peters K, Richards F M. Annu Rev Biochem. 1977;46:523–551. doi: 10.1146/annurev.bi.46.070177.002515. [DOI] [PubMed] [Google Scholar]
- 19.Bayley H. Laboratory Techniques in Biochemistry and Molecular Biology: Photogenerated Reagents in Biochemistry and Molecular Biology. Amsterdam: Elsevier; 1983. [Google Scholar]
- 20.Brunner J. Annu Rev Biochem. 1993;62:483–514. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Ebright Y W, Ebright R H. Science. 1994;265:90–92. doi: 10.1126/science.8016656. [DOI] [PubMed] [Google Scholar]
- 22.Trieschmann L, Martin B, Bustin M. Proc Natl Acad Sci USA. 1998;95:5468–5473. doi: 10.1073/pnas.95.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baehr W, Morita E A, Swanson R J, Applebury M L. J Biol Chem. 1982;257:6452–6460. [PubMed] [Google Scholar]
- 24.Cai K, Langen R, Hubbell W L, Khorana H G. Proc Natl Acad Sci USA. 1997;94:14267–14272. doi: 10.1073/pnas.94.26.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fahmy K, Jager F, Becki M, Zvyaga T A, Sakmar T P, Siebert F. Proc Natl Acad Sci USA. 1993;90:10206–10210. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambright D G, Noel J P, Hamm H E, Sigler P B. Nature (London) 1994;369:621–628. doi: 10.1038/369621a0. [DOI] [PubMed] [Google Scholar]
- 27.Nomura M, Tissieres A, Lengyel P, editors. Ribosomes. Plainview, NY: Cold Spring Harbor Lab. Press; 1974. [Google Scholar]
- 28.Bartholomew B, Durkovich D, Kassavetis G A, Geiduschek E P. Mol Cell Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst S A. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 30.Rapoport T A, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 31.Gupta C M, Radhakrishnan R, Gerber G E, Olsen W L, Quay S C, Khorana H G. Proc Natl Acad Sci USA. 1979;76:2595–2599. doi: 10.1073/pnas.76.6.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross A H, Radhakrishnan R, Robson R J, Khorana H G. J Biol Chem. 1981;257:4152–4161. [PubMed] [Google Scholar]
- 33.Takagaki Y, Radhakrishnan R, Gupta C M, Khorana H G. J Biol Chem. 1983;258:9128–9135. [PubMed] [Google Scholar]
- 34.Takagaki T, Radhakrishnan R, Wirtz K W A, Khorana H G. J Biol Chem. 1983;258:9136–9162. [PubMed] [Google Scholar]
- 35.Huang K-S, Radhakrishnan R, Bayley H, Khorana H G. J Biol Chem. 1982;257:13616–13623. [PubMed] [Google Scholar]
- 36.Nakayama T, Khorana H G. J Biol Chem. 1990;265:15762–15769. [PubMed] [Google Scholar]