Abstract
Contact sites in interaction between light-activated rhodopsin and transducin (T) have been investigated by using a chemically preactivated crosslinking reagent, N-succinimidyl 3-(2-pyridyldithio)propionate. The 3 propionyl-N-succinimidyl group in the reagent was attached by a disulfide exchange reaction to rhodopsin mutants containing single reactive cysteine groups in the cytoplasmic loops. Complex formation between the derivatized rhodopsin mutants and T was carried out by illumination at λ > 495 nm. Subsequent increase in pH (from 6 to 7.5 or higher) of the complex resulted in crosslinking of rhodopsin to the Tα subunit. Crosslinking to Tα was demonstrated for the rhodopsin mutants K141C, S240C, and K248C, and the crosslinked sites in Tα were identified for the rhodopsin mutant S240C. The peptides carrying the crosslinking moiety were isolated from the trypsin-digested peptide mixture, and their identification was carried out by matrix-assisted laser desorption ionization–time of flight mass spectrometry. The main site of crosslinking is within the peptide sequence, Leu-19–Arg-28 at the N-terminal region of Tα. The total results show that both the N and the C termini of Tα are in close vicinity to the third cytoplasmic loop of rhodopsin in the complex between rhodopsin and T.
Keywords: cysteine mutants, disulfide exchange, cytoplasmic domain, matrix-assisted laser desorption ionization–time of flight, avidin-biotin affinity chromatography
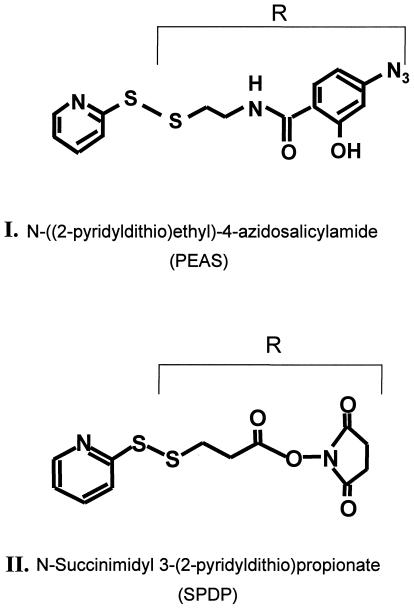
Interactions of rhodopsin with transducin (T) and rhodopsin kinase are the first steps in the two biochemical cascades that are initiated on light activation in visual signal transduction (1). Because both Tαβγ and rhodopsin kinase are known to bind to the cytoplasmic domain of light-activated rhodopsin, it is important to investigate the actual contact sites in the molecular complexes formed between rhodopsin and the above two proteins. With this long-range aim, we have initiated applications of the broad covalent crosslinking approach. We (2) reported on an initial study in which we used a photoactivatable crosslinking agent (Fig. 1 I). Here we report on a study that uses a crosslinking reagent (Fig. 1 II) carrying an activated carboxyl group. An electrophilic attack by the carbonyl group on unprotonated amino groups in suitable proximity will result in crosslinking by formation of amide linkages. Therefore, only a pH change will be required in the crosslinking strategy to deprotonate the relevant amino groups, usually the ɛ-amino groups in lysine residues.
Figure 1.
Structure of the two crosslinking reagents, I and II, used in the work described here and in ref. 2. In both reagents, the moiety R is transferred by a disulfide exchange reaction to the sulfhydryl group of a monocysteine rhodopsin mutant. R in I contains the photoactivatable azidophenyl group whereas R in II contains the activated carboxyl group, by derivatization with the N-oxysuccinimido group.
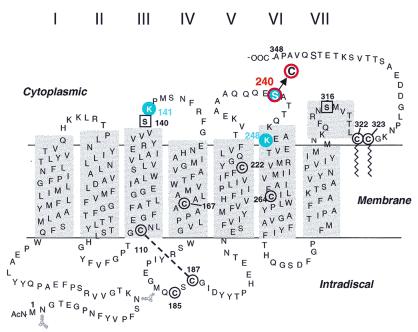
As in the accompanying paper (2), rhodopsin mutants containing single reactive cysteines at predetermined positions in the cytoplasmic domain (Fig. 2) form the starting points for study of interaction with T. The crosslinking moiety, R in Fig. 1 II, is transferred to the rhodopsin mutants by a disulfide exchange reaction with the reactive pyridyl thio group in Fig. 2 II (step 1 in Fig. 3). Three mutants of rhodopsin containing single reactive cysteine residues, K141C, S240C, and K248C (Fig. 2), were selected, and all were demonstrated to undergo crosslinking with T. In this initial study, we have focused mainly on development of the total methodology for identification of the crosslinked peptide sequences, and only work with the mutant S240C was taken to completion.
Figure 2.
A secondary structure model of rhodopsin based on the crystal structure highlighting (in red) the position of the single cysteines introduced in the cytoplasmic domain. The reactive cysteines, Cys-140 and Cys-316, in native rhodopsin were replaced by serine residues (square in black). Cysteines 167, 185, 222, and 264 are not reactive. Cys-110 and Cys-187 form a disulfide bond whereas Cys-322 and Cys-323 carry palmitoyl groups (wiggly lines).
Figure 3.
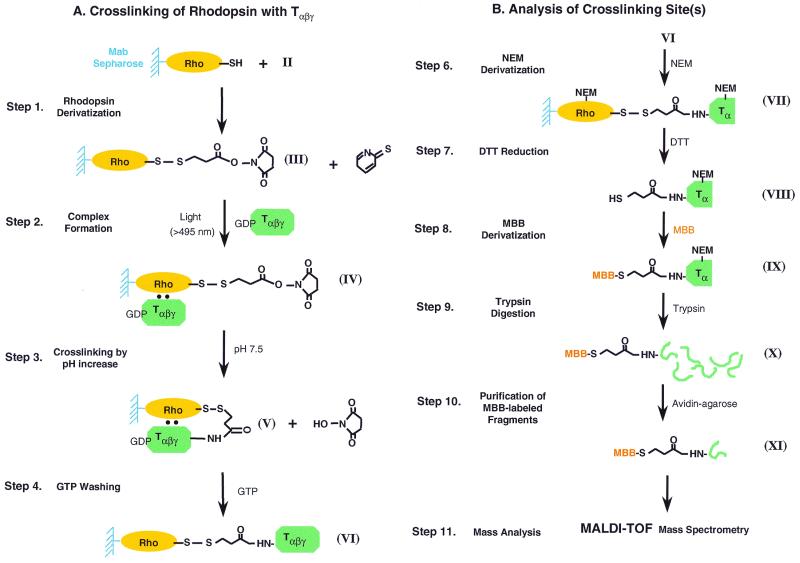
(A) Steps in the strategy for the attachment of R moiety in reagent II to rhodopsin cysteine mutants bound to 1D4-Sepharose beads, complex formation with GDP-Tαβγ on illumination, crosslinking by increasing pH to 7.5 or higher. (B) Steps in the identification of crosslinking site, carrying the HS-CH2-CH2-CO-NH group to a peptide sequence in Tα.
The total steps in the strategy for crosslinking, developed in parallel with that described in ref. 2, are illustrated in Fig. 3. The first two steps in Fig. 3A were carried out at pH 6 or lower because it was important to keep the amino groups, the potential crosslinking sites, as far as possible in protonated form. Only at step 3 was the pH raised to 7.5 or higher to allow crosslinking. The steps for subsequent processing to determine the crosslinking sites are shown in Fig. 3B. In the present study, the main crosslinking site between the rhodopsin mutant S240C and T was established to be at the N terminus of Tα.§
Materials and Methods
Materials.
The reagents used were as described in ref. 2. The reagent N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP) (Fig. 1) was from Pierce. Buffers used were as follows: buffer A, 137 mM NaCl/2.7 mM KCl/1.8 mM KH2PO4/10 mM Na2HPO4, pH 7.2; buffer B, buffer A + 1% dodecylmaltoside (DM); buffer C, buffer A + 0.05% DM; buffer D, 5 mM Mes (pH 6.0); buffer E, buffer D + 0.05% DM; buffer F, buffer E + 100 μM nonapeptide, corresponding to the C-terminal rhodopsin sequences; buffer G, 10 mM NaOAc + 0.05% DM (pH 5.0); buffer H, 10 mM Mes/100 mM NaCl/2 mM MgCl2/0.012% DM, pH 5.5; and buffer I, 10 mM Hepes/100 mM NaCl/2 mM MgCl2/0.012% DM, pH 7.5.
Construction and Expression of Rhodopsin Single Cysteine Mutants.
These were carried out as described in ref. 2.
Purification of the Single Cysteine Rhodopsin Mutants and Derivatization with the R Group in SPDP (Fig. 1 II).
The transfected COS cells were solubilized in buffer B for 1 h at 4°C in the presence of 0.1 μM PMSF. After centrifugation at 35 K for 30 min, 1D4 Sepharose was added to the supernatants and the mixture was mutated for 4 h at 4°C. The beads were washed with 50 bed vol of buffer C, followed by 50 bed vol of buffer E. Then the beads were incubated with 100 μM SPDP in buffer E for 30 min at room temperature. After further washing of the beads with 30 bed vol of buffer G to remove excess SPDP, the derivatized rhodopsin mutants (III in Fig. 3), were eluted with buffer F. The eluates were immediately frozen in liquid N2 and stored at −80°C.
Chemical Crosslinking Between the R-Derivatized Rhodopsin Mutants and T.
The rhodopsin derivatives (III in Fig. 3) containing the epitope nonapeptide were passed through Sephadex G-50 to remove the nonapeptide and the eluates were incubated with 10 μl of 1D4-Sepharose beads for 20 min in the dark at room temperature. The Sepharose beads with bound rhodopsin derivatives (III in Fig. 3) (50 pmol) were washed with buffer G and then suspended in buffer H containing GDP-Tαβγ (100 pmol). The samples were bleached with yellow light (>495 nm) for 30 s (Fig. 3A, step 2). The Sepharose beads then were first washed with buffer H to remove unbound Tαβγ and then suspended in buffer I and incubated at 10°C for 30 min. The reaction was terminated by adding excess Tris (pH 7.5) to a final concentration of 10 mM (Fig. 3A, step 3). Free T was first eluted by incubation with 200 μM GTP in buffer H (Fig. 3A, step 4). After washing the beads with buffer H, crosslinked product (V in Fig. 3A) was eluted by incubation of the beads with buffer D containing 10 mM DTT for 1 h. DTT- and GTP-eluted fractions were analyzed by 10% SDS/PAGE. The proteins were visualized by Western blotting using anti-Tα, anti-Tβ, and antibiotin antibodies.
Purification of Peptide Fragments of T Carrying Crosslinker Moiety (R).
Large-scale chemical crosslinking reactions were carried out by using 20 or 30 nmol of S240C-rhodospin derivatives (III in Fig. 3A) and 30 or 45 nmol of Tαβγ as described above. Purification of peptide fragments attached to crosslinker were carried out as described (2).
Matrix-Assisted Laser Desorption Ionization–Time of Flight (MALDI-TOF) Mass Spectrometry.
MALDI-TOF mass spectrometry analysis was carried out as described (2).
Results
Light and pH Increase Dependent Crosslinking Between SPDP-Labeled Rhodopsin Mutants and T (III in Fig. 3A).
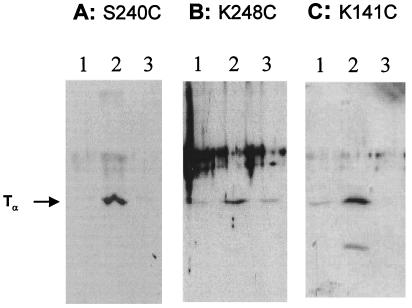
Among 10 cysteine residues in wild-type rhodopsin, only C140 and C316 are the reactive cysteines with sulfhydryl reagents in dark state (3). Single cysteine mutants were constructed with replacement of the two native reactive cysteines to serine residues (C140S and C316S) and introduction of one cysteine at a time at sites K141, S240, and K248 (Fig. 2). In these mutants, the introduced single cysteines are the only reactive residue toward thiol-specific crosslinking reagents. SPDP (Fig. 1) is a heterobifunctional chemical crosslinker containing at one end a 2-pyridyldithio group, which reacts with SH groups, and at the other end N-succinimidyl ester, which reacts with unprotonated amino groups (Fig. 1 II). The R group in reagent II was attached with a single reactive cysteine group in the cytoplasmic loop of rhodopsin mutants S240C, K248C, and K141C in dark state by disulfide exchange reaction (Fig. 3A, step 1). Derivatized rhodopsin mutants that bound on 1D4-Sepharose beads were mixed with T and activated by light (>495 nm) to form a complex with T at pH 5.5 (Fig. 3A, step 2). Then pH was increased to 7.5 to induce crosslinking with amino groups in T presumed to be in proximity (Fig. 3A, step 3). Subsequently, free T, which was bound but not covalently crosslinked to rhodopsin, was eluted with GTP (Fig. 3A, step 4). Crosslinked T then was eluted by DTT reduction of the disulfide linkage. The results of Western blotting analysis of DTT-eluted fraction by anti-Tα antibody are shown in Fig. 4. For each mutant, there are three samples, each prepared under three sets of identical conditions. The first sample was illuminated at λ > 495 nm to activate rhodopsin and form a complex with T at pH 5.5 and then pH was increased to 7.5 (Fig. 4, lanes 2). The second sample was illuminated at λ > 495 nm at pH 5.5 but pH was not increased (Fig. 4, lanes 3). The last sample was maintained in dark and then pH was increased to 7.5 (Fig. 4, lanes 1). Fig. 4A shows the results of SPDP-derivatized mutant S240C. Although there were some background signal in dark and light without a pH increase (samples shown in Fig. 4A, lanes 1 and 3), significant increase in signals dependent on light activation and pH increase was found (Fig. 4A, lane 2). Fig. 4A demonstrates that light activation and pH increase-dependent formation of crosslinked complex between rhodopsin and Tα occurred with the single cysteine rhodopsin mutant S240C. Similarly, light activation and pH increase-dependent signals also were seen in both K248C and K141C (Fig. 4 B and C). Thus all three rhodopsin mutants labeled with SPDP were crosslinked to Tα.
Figure 4.
SDS/PAGE followed by immunoblotting in analysis of crosslinking of rhodopsin mutants, S240C, K248C, andK141C with the R group in reagent II. Crosslinking depends both on light and increase in pH. Lanes 1, eluate from 1D4-Sepharose after reduction with DTT in the dark (Fig. 3A, step 1); lanes 2, sample after illumination (Fig. 3A, step 2) and pH increase (Fig. 3A, step 3); and lanes 3, sample after illumination (Fig. 3A, step 2) only. Immunoblotting was with anti-Tα antibody.
Identification of Crosslinking Sites Between Light-Activated Rhodopsin and T.
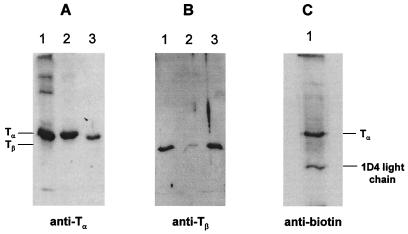
To identify the peptide fragments of T that carries crosslinking moiety, we have established the system to purify the fragments attached to crosslinker. Fig. 3B illustrates the strategy of purification and identification of peptide fragments of T crosslinked to rhodopsin. After crosslinking reaction (Fig. 3 VI), crosslinked complex between rhodopsin and T were treated with NEM to block all free sulfhydryl groups on both proteins (Fig. 3B, step 6). Reduction of disulfide linkage with DTT (Fig. 3B, step 7) eluted Tα containing only one SH group derived from crosslinker. Then this newly generated SH group was biotinylated by maleimido-butyryl-biocytin (MBB) (Fig. 3B, step 8). MBB-derivatized crosslinked T with S240C rhodopsin mutant were analyzed by Western blotting using antibodies against Tα (Fig. 5A), Tβ (Fig. 5B), and biotin (Fig. 5C). MBB-derivatized crosslinked T contained Tα (Fig. 5A, lane 2) but a very small amount of Tβ (Fig. 5B, lane 2). In contrast, GTP-eluted fraction contained both Tα and Tβ subunits (Fig. 5 A and B, lane 3). These results confirmed that crosslinking occurred predominantly with Tα. Similar results were obtained by K248C and K141C (data not shown). MBB-derivatized Tα also was detected by antibiotin antibody (Fig. 5C). This result indicated that the sulfhydryl group in crosslinked Tα was derivatized with MBB. Also seen is the light chain of 1D4 antibody eluted by DTT reduction and subsequently derivatized with MBB (Fig. 5C).
Figure 5.
SDS/PAGE analysis of product formed in crosslinking of rhodopsin S240C-R with T. Protein bands were detected by immunoblotting with antibodies against Tα, Tβ, and biotin. (A and B) Lanes 1, control Tαβγ; lanes 2, eluate from 1D4-Sepharose after reduction with DTT (Fig. 3B, IX); and lanes 3, eluate from 1D4-Sepharose with GTP after reduction with DTT (Fig. 3A, step 4). (C) Eluate from 1D4-Sepharose after reduction with DTT (Fig. 3B, IX).
MBB-derivatized Tα then was extracted from SDS/PAGE and digested with trypsin as described (2) (Fig. 3B, step 9). The peptide fragments that derivatized with MBB were purified by avidin Sepharose (Fig. 3B, step 10) and subjected to MALDI-TOF mass spectrometry.
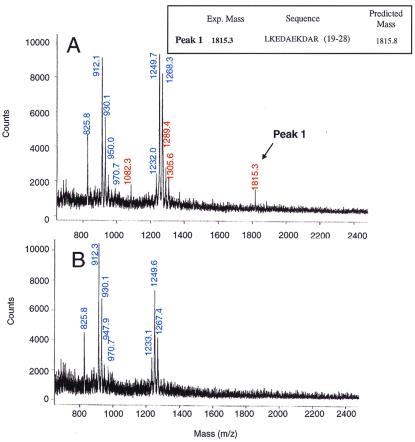
Experiments were carried out by using 20 and 30 nmol of S240C-SPDP rhodopsin. Fig. 6 shows tryptical mass spectra of crosslinked Tα (A) and trypsin alone control (B). Seven signals, 825.8, 912.3, 930.1, 947.9, 970.7, 1233.1, 1249.6, and 1267.4, were detected in trypsin alone control (Fig. 6B). These signals also were seen in mass spectrum of crosslinked samples (Fig. 6A). In addition, four peaks, with m/z values of 1082.3, 1289.4, 1305.6, and 1815.3, appeared only in crosslinked samples (Fig. 6A). These signals most likely are derived from crosslinked Tα, and one of them (m/z = 1815.3) was identified as the fragment from Leu-19 to Arg-28 located near the N-terminal region of Tα (Fig. 7). This result suggests that residue 240 in rhodopsin is very close to the N-terminal region of Tα in the complex formed by these two proteins. However, we have so far not been able to identify the other three fragments seen above.
Figure 6.
(A) MALDI-TOF mass spectrum of peptide fragments containing MBB-β-thio-propionyloxyamido-Tα obtained after purification on avidin-agarose of trypsin-digested peptide mixture. (B) Mass spectrum of a control sample in which trypsin alone was subjected to the trypsin digestion conditions used in A. Mass (m/z) lines (blue) originate from trypsin autolysis whereas the mass (m/z) values (red) originated crosslinked Tα.
Figure 7.
Amino acid sequence of Tα. The fragment identified by mass spectrum (Fig. 6) is underlined. This fragment (Leu-19–Arg-28, red) identified is near N-terminal region of the Tα. Also shown in blue are the two crosslinked peptide sequences at the C terminus of Tα identified in ref. 2.
Discussion
With the long-range aim of mapping contact sites in complexes formed between light-activated rhodopsin and the key proteins involved in visual transduction, we have taken a broad covalent crosslinking approach. We (2) reported on the development of a strategy for the study of interactions between rhodopsin and T by using a photoactivatable reagent. Essentially the same strategy has been used in this paper except for the important difference that the crosslinking reagent (Fig. 1 II) uses a different principle. It contains a preactivated carboxyl group that is expected to attack unprotonated amino groups in proximity to form amide linkages. Thus, the only requirement is to increase the pH of the medium at a suitable step (Fig. 3A, step 3) to deprotonate the ɛ amino groups of lysine residues. Thus, this approach becomes specific almost exclusively for amide crosslinks with lysines.
In the present work, crosslinking between T and the three rhodopsin mutants, K141C in cytoplasmic loop III-IV, S240C and K248C in loop V and VI, was demonstrated. Crosslinking depended both on light and pH increase. In all cases, crosslinking occurred mainly to Tα. Thus, the results show the involvement of the cytoplasmic loop between helices III and IV and between helices V and VI. These findings are in agreement with previous results (4–9).
The total procedure leading to identification of the site of crosslinking was taken to completion only for the rhodopsin mutant S240C. Mass spectrometric analysis showed that its crosslinking occurred mainly to the N-terminal region of Tα. Although a number of previous reports (10–13), as well as the work described in ref. 2, all have indicated that the C-terminal region of Tα is important for its interaction with rhodopsin, previous work has indicated that the N-terminal region of Tα also could be involved in the binding. It has been shown that peptide with sequence Glu-8–Ala-23 can inhibit the binding of T to rhodopsin (13). Onrust et al. (12) also reported that substitution of Leu-19, Lys-20, Glu-21, and Asp-22 in Tα decreased the binding. Thus, there is clear evidence for the involvement of the N-terminal region in Tα in interaction with rhodopsin.
In the meantime, the crystal structure of Tαβγ has been solved (14). Examination of the structure shows that the N-terminal peptide sequence of Tα, Leu-19–Arg-28, now shown to be involved in crosslinking, is exposed to the surface and is close to the C-terminal region of Tα. Furthermore, the C-terminal sequences, Glu-342–Phe-350 and Arg-310–Lys-313, shown to be crosslinked to Tα in ref. 2 (also highlighted in blue in Fig. 7) and the N-terminal sequence, Leu-19–Arg-28, actually face the same side and are relatively close to each other. Thus, our results strongly support the conclusion that both the N-terminal and C-terminal regions of Tα are involved in binding to the third cytoplasmic loop of rhodopsin.
The extent of crosslinking using reagent II was several-fold lower than that obtained with reagent I (2). However, it is highly significant that the information obtained was complementary. Reagent II has the specificity for crosslinking only with the unprotonated amino groups and thus has a unique feature. Future work with the present approach should include investigation of at least two parameters, the manner of prior activation of the carboxyl group in the reagent and the length of the carbon chain (spacers) between the dithiopyridyl group and the activated carboxyl group. Efficiency of crosslinking and the sites where crosslinking can occur will be expected to vary widely with these parameters. These comparative studies would be important for mapping the sites of interactions between the proteins. The total mapping would benefit greatly from the current availability of the very large number of monocysteine mutants of rhodopsin spanning the entire cytoplasmic domain of rhodopsin (15).
Acknowledgments
We are grateful to Professor U. L. RajBhandary (Massachusetts Institute of Technology) for valuable discussions. Enthusiastic assistance of Ms. Judy Carlin in the preparation of the manuscript is acknowledged. This work was supported by National Institutes of Health Grant GM28289 and National Eye Institute Grant EY11717 (H.G.K.). Y.I. was supported by Banyu Tsukuba Research Institute, Tsukuba, Japan during a leave of absence.
Abbreviations
- SPDP
N-succinimidyl 3-(2-pyridyldithio)propionate
- T
transducin
- MBB
maleimido-butyryl-biocytin
- MALDI-TOF
matrix-assisted laser desorption ionization–time of flight
- DM
dodecylmaltoside
Footnotes
References
- 1.Khorana H G. J Biol Chem. 1992;267:1–4. [PubMed] [Google Scholar]
- 2.Cai K, Itoh Y, Khorana H G. Proc Natl Acad Sci USA. 2001;98:4877–4882. doi: 10.1073/pnas.051632898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Findlay J B C, Barclay P L, Bett M, Davidson M, Pappin D S L, Thompson P. Vision Res. 1984;24:1301–1308. doi: 10.1016/0042-6989(84)90312-2. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn H, Hargrave P A. Biochemistry. 1981;20:2410–2417. doi: 10.1021/bi00512a007. [DOI] [PubMed] [Google Scholar]
- 5.Konig B, Arendt A, McDowell J H, Kahelrt M, Hargrave P A, Hofmann K P. Proc Natl Acad Sci USA. 1989;86:6878–6882. doi: 10.1073/pnas.86.18.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franke R R, Konig B, Sakmar T P, Khorana H G, Hofmann H P. Science. 1989;250:123–125. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- 7.Franke R R, Konig B, Sakmar T P, Graham R M, Khorana H G. J Biol Chem. 1992;267:14767–14774. [PubMed] [Google Scholar]
- 8.Yang K, Farrens D L, Hubbell W L, Khorana H G. Biochemistry. 1995;35:12464–12469. doi: 10.1021/bi960848t. [DOI] [PubMed] [Google Scholar]
- 9.Ridge K D, Zhang C, Khorana H G. Biochemistry. 1995;34:8804–8811. doi: 10.1021/bi00027a032. [DOI] [PubMed] [Google Scholar]
- 10.Garcia P D, Onrust R, Bell S M, Sakmar T P, Bourne H R. EMBO J. 1995;14:4460–4469. doi: 10.1002/j.1460-2075.1995.tb00125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozawa S, Weiss E R. J Biol Chem. 1995;270:31052–31058. doi: 10.1074/jbc.270.52.31052. [DOI] [PubMed] [Google Scholar]
- 12.Onrust R, Herzmark P, Chi P, Garcia P D, Lichtarge O, Kingsley C, Bourne H R. Science. 1997;275:381–385. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 13.Hamm H E, Deretic D, Arendt A, Hargrave P A, Koenig B, Hofmann K P. Science. 1988;241:832–835. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 14.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. Nature (London) 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 15.Khorana H G. J Biomol Struct Dyn. 2000;11:1–16. doi: 10.1080/07391102.2000.10506598. [DOI] [PubMed] [Google Scholar]