In the 25 years since its discovery, the nuclear factor-κB (NF-κB) family of transcription factors has been the subject of intensive research in virtually all areas of biomedical science. Research into the function and regulation of this single transcription factor family has been at the forefront of studies in immunology, cancer biology, and molecular pathology, and has progressed hand in hand with conceptual and technological advances in molecular biology, genetic engineering, systems biology, and computational biology. The progress made in elucidating the triggers of NF-κB activation, the components of the NF-κB pathway, and characterization of the physiological and pathophysiological functions of NF-κB have been remarkable. This anniversary therefore represents an excellent opportunity to revisit the work that forms the foundation upon which so much effort and progress has been built. The articles in this volume of Immunological Reviews cover much of what is known about the NF-κB pathway and its basic function. Yet each review also draws attention to the myriad and often surprisingly fundamental issues that remain unresolved. Finally, in marking this important landmark, most reviewers have shared a personal and historical narrative documenting their connection to their area of study in NF-κB. These accounts offer a unique view of the early days of NF-κB research, when so much of the fundamental underpinnings of the pathway were laid down in short order, and provide particular insight into how this field was enriched through the expertise of scientific immigrants from disparate areas of biology, further highlighting the amazing diversity of biological systems influenced by this single transcription factor family.
In this introductory article, we lay down some of the most basic and fundamental concepts in order to assist the uninitiated in gaining access to the more detailed discussions that characterize the rest of this issue. Thus, this represents a severely abridged introduction to the NF-κB signaling pathway and its biological function. Where appropriate, we point the reader towards reviews elsewhere in this issue from which they may gain more detailed insight and historical context to the various facets of NF-κB. However, in keeping with the format of this issue, I (S.G.) begin by offering an account of my own involvement with NF-κB.
Personal and historical narrative
I (S.G.) joined David Baltimore’s laboratory in January 1989 after having completed my graduate studies with Dr. Umadas Maitra at Albert Einstein College of Medicine. As a graduate student, I had biochemically purified and characterized eukaryotic translation initiation factors. When I arrived in David’s laboratory, NF-κB was one of the most exciting areas of research in the laboratory (of course along with other exciting areas such as RAG proteins, helix-loop-helix proteins, etc.). Following the discovery of NF-κB by Ranjan Sen in 1986 (1), a number of postdoctoral fellows in the laboratory were pursuing the biology of this transcription factor (Michael Lenardo, Patrick Baeuerle, Gary Nabel, and Jaqueline Pierce). A major breakthrough in understanding how NF-κB is regulated, in particular its inducibility in response to signals such as phorbol-esters, was the discovery by Patrick Baeuerle of the latent, cytoplasmic state of NF-κB in unstimulated cells, followed by the identification of the inhibitor of NF-κB (IκB) 2). Despite the rapid elucidation of the mechanism of NF-κB regulation, further progress was difficult without knowing the identity of the genes encoding these proteins. Therefore, efforts were underway in David’s laboratory, as well as in many others, to clone the NF-κB genes. However, for reasons that were unclear at the time, traditional approaches using binding to oligonucleotide columns yielded amounts of purified protein that were insufficient for protein sequencing. To take a fresh approach at the problem, I decided to try and purify NF-κB and IκB from tissue extracts. To choose the right organ, I made extracts of all mouse organs and analyzed the presence of NF-κB DNA-binding activity in detergent (deoxycholate)-treated extracts to reveal the total amount of NF-κB. This simple analysis revealed that the strongest signal was in lungs, probably because of the presence of resident macrophages. To scale up the purification, I decided to use rabbit lungs, which could be purchased from Pel-Freez, a company that otherwise sold rabbit meat to supermarkets. I began by first purifying the NF-κB:IκB complex, dissociating the two components, and then purifying them individually. The IκB activity was assayed by its ability to inhibit NF-κB DNA-binding activity, and I soon found that IκBα was a 37 kDa protein that could inhibit DNA-binding by NF-κB (3). At that time one of the best characterized inducers of NF-κB activity were phorbol-esters, which were known to function by activating protein kinase C (PKC). I therefore purified PKCs and showed that they could phosphorylate IκBα and that the phosphorylated IκB could no longer inhibit NF-κB DNA-binding (3). It seemed at that time to be a clear explanation of how IκB might be regulated, but as we know now, is not how NF-κB is activated in cells. While we have not revisited these early experiments, it is likely that phosphorylation of IκB somehow alters its conformation, thereby making it unable to block NF-κB DNA-binding.
Unlike IκBα, the final steps of purification of the dissociated NF-κB was using affinity purification of κB oligonucleotide-coupled resins. Previous studies had suggested that NF-κB bound with very high affinity to its cognate binding site, and therefore would elute from the resin only at high salt: thus separating bona fide NF-κB from other non-specific DNA-binding proteins. However, my experiments indicated that NF-κB eluted along with many other contaminating DNA-binding proteins. To overcome this problem, I created another column with a mutated κB-site oligonucleotide and went through a sequential purification, first through the wildtype oligo column, where NF-κB eluted at high salt, followed by its purification through the mutant oligo column, where NF-κB now eluted at low salt, but most contaminating DNA-binding proteins continued to elute at high salt. Using this approach, I was able to obtain a large amount of highly purified NF-κB that contained the p50 and p65 subunits. I sent a portion of my purified p50 sample to a protein sequencing facility at another institution, and after 2 months was informed that they had been unable to obtain any sequence. I embarked on a frantic search for another protein sequencing facility and fortunately became aware of Paul Tempst at Harvard, who had recently established his protein sequencing laboratory after completing his postdoctoral training at Lee Hood’s laboratory. Paul agreed to sequence the sample, and in about a month, he gave me nearly 250 amino acids of sequence out of a 440 amino acid protein! I immediately began the process of cloning the gene from a mouse λgt11 cDNA library. Although we were unaware of any specific competition, I felt I had to get the cloning and characterization done as soon as possible. Between March 1990 and July 1990 we succeeded in cloning the 4.5 kB cDNA, completely sequencing it, and demonstrating that it was encoded as a precursor of 105 kDa (4). The mad rush was justified in hindsight, as it turned out Alain Israel’s laboratory had also sequenced the p105 gene and had submitted their paper one day earlier to the same journal (5)!
Besides the sequence of p50, Paul Tempst had also given me sequences from p65 (some of these sequences were published in the p50 paper to show the similarity between the proteins), and another postdoctoral fellow, Garry Nolan, who had joined the laboratory went on to clone p65 (6). However, the situation in the laboratory had changed in the meantime. David had announced earlier in the spring that he would join Rockefeller University to become the President beginning July 1, 1990. While initially I had expected to move with him to New York, the publication of the p50 paper suddenly raised the possibility that I would be able to start my own laboratory far sooner that I had expected. As I had also purified sufficient amounts of IκBα, it was my goal to clone IκB as soon as I had finished my job interviews. Unfortunately Paul Tempst moved from Harvard to Sloan-Kettering that winter, and my purified IκBα sample remained unsequenced in his laboratory until May 1991. Upon receiving the sequences, I geared up to clone IκBα, however Nathan Davis, a student from Henry Bose’s laboratory, with whom I had been collaborating on characterizing the chicken IκB, pp40, informed me that he had succeeded in cloning the pp40 gene. It was immediately clear from the sequence that pp40 was the chicken homolog of rabbit IκBα. We rushed our paper with the mammalian sequence and description of the chicken pp40 clone, and publication of our study (7), along with Al Baldwin’s paper (8) clearly established the nature of IκB proteins.
I began my laboratory at Yale in July 1991 and continued to work on NF-κB. However, this was also the time when the field had exploded, and as a young starting laboratory, I was unable to participate in many of the discoveries other than as an onlooker. The project that I focused on was the purification and cloning of IκBβ, but interestingly the first independent paper from my laboratory was on something quite different. The concept that NF-κB was a key regulator of inflammation was just beginning to be established, and a very talented student in the laboratory, Elizabeth Kopp, became interested in testing whether known anti-inflammatory drugs might act through NF-κB. We were debating between testing glucocorticoids and nonsteroidal anti-inflammatory drugs when we realized that the well-known effect of aspirin on cyclooxygenases, which involved the transfer of acetyl group of aspirin to cyclooxygenase, did not explain the anti-inflammatory effect of salicylates, which lacked the acetyl group. We went on to show that high dose salicylates and aspirin can inhibit NF-κB, and it is now accepted to be the mechanism by which salicylates exert their anti-inflammatory functions (9).
In parallel, we did clone IκBβ and showed that it behaved differently from IκBα, although many details of its regulation of NF-κB remained unclear (10). For a variety of reasons, the mice lacking IκBβ remained uncharacterized, and it was only recently that analysis of these mice revealed the surprising role of IκBβ in both inhibiting and facilitating inflammatory gene expression (11). It was from our efforts to biochemically purify and study IκBβ containing complexes that we realized that regulation of gene expression by NF-κB involved post-translational modifications of NF-κB itself. We have continued to explore the regulation of the transcriptional activity of NF-κB, using a combination of biochemical and genetic approaches (12–14) Finally, we have also explored the potential therapeutic utility of disrupting the IκB kinase (IKK)-NF-κB essential modulator (NEMO) interaction using NBD-peptides, and many labs, including our own, have demonstrated the utility of this strategy in numerous animal models of inflammatory, autoimmune, and oncological disease (15, 16). However, extending this approach to patients remains the ultimate goal, and we are exploring strategies to enhance the efficacy of the NBD-peptides that we hope will allow translation of NBD-peptides to the clinic in the near future.
The NF-κB pathway
The mammalian NF-κB family consists of p65 (RelA), c-Rel, RelB, p50/p105 (NF-κB1), and p52/p100 (NF-κB2). These proteins exist in the resting state as homodimers or heterodimers primarily bound to IκB family proteins. The NF-κB family proteins are characterized by the presence of a conserved N-terminal 300 amino acid Rel homology domain (RHD) that is responsible for dimerization, interaction with IκBs and binding to DNA (reviewed in 17). IκB proteins, which function by constraining the function of NF-κB dimers, are defined by the presence of five to seven C-terminal ankyrin repeats and in mammalian cells contain seven members: IκBα, IκBβ, IκBε, Iκζ, BCL-3, and the precursor proteins p100 and p105 (reviewed in 18). An additional potential IκB family protein, IκBη, has recently been identified (19). The typical IκB proteins function to prevent DNA binding by NF-κB dimers. For example, IκBα or IκBβ bound to p65/p50 or p65/c-Rel dimers mask the nuclear localization sequence (NLS) of p65, which despite the presence of an accessible p50 NLS results in steady state cytoplasmic localization (reviewed in 17). Release of NF-κB dimers from typical IκB proteins depends upon the action of the IκB kinase complex (reviewed in 20). Upon phosphorylation by IKKs, IκB proteins are recognized and ubiquitinated by members of the Skp1-Culin-Roc1/Rbx1/Hrt-1-F-box (SCF or SCRF) family of ubiquitin ligases (reviewed in 21). βTrCP, the receptor subunit of the SCF family ubiquitin ligase machinery, binds directly to the phosphorylated degron sequence (DS*GXXS*) on IκBs. Recognition by the ubiquitin ligase machinery leads to the polyubiquitination and subsequent degradation of IκBs or processing in the case of p100 by the proteasome. Freed NF-κB dimers translocate to the nucleus and bind to specific sequences, termed κB sites, in the promoter or enhancer regions of target genes (reviewed in 22, 23). Activated NF-κB can then be downregulated through multiple mechanisms (reviewed in 24, 25) including the well-characterized feedback pathway whereby newly synthesized IκBα restores steady state cytoplasmic localization of NF-κB dimers. Aspects of this basic signaling scheme and even some parts of the signaling pathways leading to NF-κB activation are well conserved in terms of mechanism and biological functions, perhaps for as much as a billion years of evolutionary history (reviewed in 26).
NF-κB signaling is generally considered to occur through either the canonical or noncanonical pathways. The so called canonical, or classical, pathway of NF-κB activation is triggered by numerous extracellular and intracellular signals, prototypically proinflammatory cytokines such as tumor necrosis factor (TNF) and interleukin-1 (IL-1), Toll-like receptors (TLRs), antigen receptors, as well as various cells stressors. While most of these signaling events emanate from cell surface receptors, various intracellular signals, including recognition of intracellular pathogens (reviewed in 27, 28), as well as more direct cell stresses including reactive oxygen species and DNA double strand breaks (reviewed in 29).
Whether initiated from an extracellular or intracellular source, signaling in the canonical pathways generally occurs through the recruitment of adapter proteins containing homotypic and heterotypic protein:protein interaction motifs such as receptor interacting protein (RIP) homotypic interaction motifs (RHIMs), death domains (DDs), and Toll IL-1 receptor homology domains (TIRs). For example caspase activation and recruitment domain (CARD)-containing proteins are implicated in the formation of signaling platforms in multiple classical NF-κB signaling pathways (reviewed in 30). The resulting oligomeric structures may be crucial for signaling to NF-κB, although this has not been directly proven in vivo for the NF-κB pathway. It is thought that in conjunction with non-degradative ubiquitination of multiple signaling intermediates (reviewed in 27), these complexes serve as platforms for the recruitment and activation of the kinases responsible for phosphorylation of IκB, the IκB kinase (IKK) complex. IKKα, IKKβ, or both complexed with the regulatory protein NEMO (IKKγ) are recruited to receptor/adapter protein complexes and either directly via trans-autophosphorylation or through the induction of an IKK kinase, such as TAK1, are activated. Thus, it is hypothesized that induction of the canonical NF-κB pathway is an aggregative signaling process in which formation of a multiprotein complex leads to kinase activation, either IKK or upstream kinase, through proximity-induced trans-autophosphorylation. The noncanonical pathway may be described by comparison as a dissociative signaling mechanism. Receptor signaling leads to IKKα activation through disruption of a protein complex that mediates constitutive downregulation of the IKKα activating kinase, NF-κB-inducing kinase (NIK). Dissociation of this inhibitory complex upon ligand binding to certain members of the TNFR superfamily, such as B-cell activating factor belonging to the TNF family receptor (BAFFR), CD40, and lymphotoxin βR (LTβR), leads to accumulation of NIK, which phosphorylates IKKα leading to phosphorylation of p100 (reviewed in 31).
Degradation of IκB is a tightly regulated event that is initiated upon specific phosphorylation by activated IKK. The IKK complex has an apparent molecular weight of 700 – 900 kDa and appears to be composed of a heterodimer of IKKα (IKK1) and IKKβ (IKK2), and the regulatory subunit NEMO (reviewed in 20). Limited structural information suggests that this kinase complex is assembled as an elongated dimer of dimers (reviewed in 17). The canonical NF-κB signaling pathway results in NEMO-dependent phosphorylation of IκBα on Ser32 and Ser36 and IκBβ on Ser19 and Ser23. Both IKKβ and IKKα can contribute to canonical pathway activation, although in many cases IKKβ is sufficient (discussed in 20). The noncanonical pathway however depends only on the IKKα subunit by NIK, independent of NEMO. NIK and IKKα then cooperate in phosphorylating p100 and causing its inducible processing to p52 (reviewed in 31). Yet another IKK-dependent pathway is mediated by the induced degradation of the precursor protein p105. IKK-mediated p105 degradation results in activation of the Cot/Tpl2 kinase, resulting in activation of extracellular signal-regulated kinase (ERK) (reviewed in 32). In the 15 years since the identification of the IKK complex, numerous substrates in addition to IκB proteins have been identified. These serve to integrate the NF-κB pathway with other signaling pathways regulating inflammation, cell proliferation, and cell survival (reviewed in 20, 33).
Following degradation of IκB, the released NF-κB is able to bind promoter and enhancer regions containing κB sites with the consensus sequence GGGRNNYYCC (N - any base, R – purine, and Y – pyrimidine), although κB sites are now well demonstrated with considerably more degenerate sequences (reviewed in 34). The crystal structures of NF-κB dimers bound to the κB enhancer reveal how both immunoglobulin (Ig)-like domains that make up the RHD contact the DNA of the κB site. The NH2-terminal Ig-like domain is primarily responsible for sequence specificity of NF-κB, while hydrophobic residues within the COOH-terminal domain form the dimerization interface (35–40). RelB, c-Rel, and p65 contain a transactivation domain (TAD) located towards the C-terminus that is necessary for transactivation by these proteins. Homodimers of p52 and p50 lack TADs and hence have no intrinsic ability to drive transcription. In fact, binding of p52 or p50 homodimers to κB sites of resting cells leads to repression of gene expression (12). Atypical IκB proteins capable of binding to DNA-bound NF-κB dimers can either augment this repressive function or, as in the case of IκBζ and BCL-3, induce transcriptional activation (reviewed in 18). The TADs on p65, c-Rel, and RelB promote transcription in part through the recruitment of transcriptional co-activators, such as the histone acetyltransferases PCAF, CBP, or p300 (reviewed in 22). Recruitment of transcriptional co-activators is modulated through post-translational modifications (PTMs) targeting NF-κB proteins. For p65, alone, PTMs including phosphorylation, ubiquitination, methylation, acetylation, nitrosylation, and sulfhydration have been described (reviewed in 22, 41). A comprehensive understanding of the interplay and significance of the various PTMs targeting p65, alone, seems several years away, and work on other Rel proteins is less developed. As a result, our understanding of truly fundamental aspects of transcriptional regulation by NF-κB is in its infancy. The reviews in this volume by Natoli (22) and Smale (34) highlight some of the questions that lie ahead in trying to decipher the regulation of gene programs, both their initiation and termination, by NF-κB.
Biological roles of NF-κB
Various aspects of the functions of NF-κB, IκB, and IKK proteins are reviewed in detail in this issue. Kaileh and Sen (42) and Gerondakis et al. (43) provide detailed reviews of NF-κB in hematopoietic cell development and function. Pasparakis (44) reviews the considerable progress in the past decade in understanding the role of NF-κB in epithelial biology and highlights some of the many paradoxes suggested by epithelial NF-κB knockout models. Staudt and colleagues (45) and Karin and colleagues (46) provide detailed analysis of NF-κB in lymphoma and inflammation associated malignancy. Chan and Greene (28) review the interplay of NF-κB and virology, particularly the give and take between NF-κB and HTLV1 and HIV1. Here we provide a brief overview of some of the biological functions of these proteins. All of the mammalian Rel, IκB, and IKK family members have been genetically targeted in mice. As many of these knockouts, particularly of NF-κB and IκB proteins, predate routine use of tissue specific cre-lox systems, there has been a push in recent years to generate new conditional knockouts. These efforts have shed additional light on some of the tissue-specific roles of NF-κB pathway components, although additional work is needed in this area. The role of individual pathway components in the development and response of hematopoietic cells (reviewed in 42, 43) as well as in epithelial tissues (reviewed in 44) is reviewed in depth in this volume. The reviews from the laboratories of Verma (47) and Scheidereit (48) also provide detailed discussions of the IKK complex and IκB family knockouts.
The first genetic studies targeting the NF-κB pathway revealed the crucial role of p65 in providing protection from apoptosis during TNF signaling (reviewed in 33), a recurrent finding in NF-κB pathway knockouts that was recapitulated in IKKβ and NEMO knockout animals (reviewed in 20). Mice lacking p65 (Rela−/−) exhibit lethality due to liver apoptosis at gestational day 15–16 (49). The observed liver apoptosis was shown to be the result of TNF signaling in the developing liver, as crossing mice lacking p65 to either Tnf−/− (48) or Tnfrsf1a−/− (50) mice rescues the embryonic lethality and revealed the role of p65 in innate immune responses (51). In contrast, p50 (Nfkb1−/−) deficient mice do not show developmental defects (52), demonstrating p50 is not essential for induction of anti-apoptotic genes but instead exhibit defective humoral immune responses. B cells lacking p50/p105 do not respond efficiently to lipopolysaccharide (LPS), underscoring the role of the classical p65/p50 heterodimer in TLR signaling pathways. Mice lacking p52 (Nfkb2−/−) have defects in their splenic architecture and Peyer’s patch development (53–55), emphasizing the importance of the noncanonical signaling pathway in lyphmorganogenesis. The p52 knockout phenotype is consistent with a role for p52 in LTβR signaling, particularly with regard to the development of secondary lymphoid organs (56, 57).
The c-Rel protein forms homodimers and heterodimers with p50 and p65. Although c-Rel-deficient mice fail to generate a productive humoral immune response and exhibit defects in both T- and B-cell responses (58), there is evidence to suggest that under some circumstances there is functional redundancy between c-Rel and p65. As a result, mice lacking the c-Rel TAD, which cannot induce gene expression but can form dimers, display a more severe phenotype. These c-Rel mutants have hypoplastic bone marrow and hyperplasia of secondary lymphoid organs, suggesting c-Rel functions in activation of B-cell class switching (59, 60). Loss of c-Rel in conjunction with p50 loss also suggests a role for c-Rel in dendritic cell survival (61). Recent work also demonstrates an essential role for c-Rel in the development of regulatory T cells (reviewed in 43).
RelB is unique in that it does not homodimerize and undergoes limited heterodimerization. RelB selectively heterodimerizes with p100, p52, and p50 (62, 63) and apparently does not interact with any IκB molecules other than p100 (64). Because RelB/p52 heterodimers do not bind other IκBs, they are generally thought to exhibit constitutive nuclear localization (62, 63, 65, 66). However, RelB:p52 dimers may be regulated by p100 by p105 in typical IκB-like p100 and p105 complexes (67, 68). Although RelB may heterodimerize with p65 under certain circumstances, the physiological role of this heterodimer is not clear (reviewed in 17). Loss of RelB results in multi-organ as well as severe deficits in adaptive immunity (69). Mice lacking RelB have defective cellular immune responses due to defects in thymic medullary cells and dendritic cell function (70). This phenotype is exaggerated when p50 is also knocked out, indicating that p50 and RelB cooperate in the regulation of genes that limit inflammation (71). RelB−/− B cells, although crippled in their proliferative response, undergo normal IgM secretion and class switching in response to various stimuli (72). The importance of RelB in lymphoid organogenesis is well established (56). RelB knockout animals lack Peyer’s patches (55, 73), likely secondary to defects in LTβR signaling. Noncanonical signaling to RelB is also likely responsible for the defective thymic medullary epithelial cell development, as this process depends upon CD40 and RANKL (74). While the role of the noncanonical pathway has been best developed in the context of lymphorganogenesis, the canonical pathway is also activated by these same TNFR family members and clearly also contributes to lymphoid organ development and organization (57). Thus, not only does NF-κB play multiple essential roles in the specific developmental steps of development and activation (42, 43), it is also crucial for the development of the specialized tissues within which lymphocyte development and activation occur.
Transcriptional specificity is partially regulated by the ability of specific NF-κB dimers to preferentially associate with certain members of the IκB family. For example, the classical NF-κB heterodimer (p65/p50) is predominantly, though not exclusively, regulated by IκBα. Therefore, IκBα-deficient mice display increased but not constitutive p65/p50 DNA binding activity, demonstrating that additional mechanisms regulate the activity of this dimer (47, 75). IκBα is known to participate in a feedback loop where newly synthesized IκBα inhibits the activity of NF-κB; although how this removes NF-κB from κB sites, if it in fact does, is not known (discussed in 22). Regardless of the precise mechanism, in the absence of IκBα, the termination of NF-κB activation in response to TNF is significantly delayed. In fact, if one places IκBβ, which otherwise does not participate in the downregulation of NF-κB activity, under control of the IκBα promoter, the normal kinetics of NF-κB activation and downregulation are restored (76). This is not to say that IκBα and IκBβ are interchangeable; indeed, they clearly are not. Nevertheless, analysis of cell lines in which multiple isoforms of IκBs have been knocked out indicates that a significant portion of the functional characteristics of IκBα, IκBβ, and IκBε are the result of temporal differences in their degradation and differential transcriptional regulation of their promoters (77). The intrinsic properties of the typical IκB proteins, separate from their transcriptional regulation, that contribute to their differential functions remain to be identified.
Like NF-κB family proteins, our understanding of the functions of individual IκBs remains somewhat cursory. At the signaling level, there has been considerable progress in parsing out how individual IκBs shape the transcriptional response. A notable portion of this work has been the result of efforts in mathematical modeling of the NF-κB pathway, and is reviewed by Basak et al. (78). However, while knockouts have shed particular light on the unexpected functions of atypical IκB proteins, there remains much to be learned about the distinct biological roles of IκBs. In particular, the ability of IκB proteins to selectively augment and inhibit NF-κB transcriptional programs, to selectively repress and promote the formation of different NF-κB dimers, and the capacity of to mediate crosstalk with other signaling pathways need to be investigated in vivo to more fully understand the functions of the IκB family (discussed in 18).
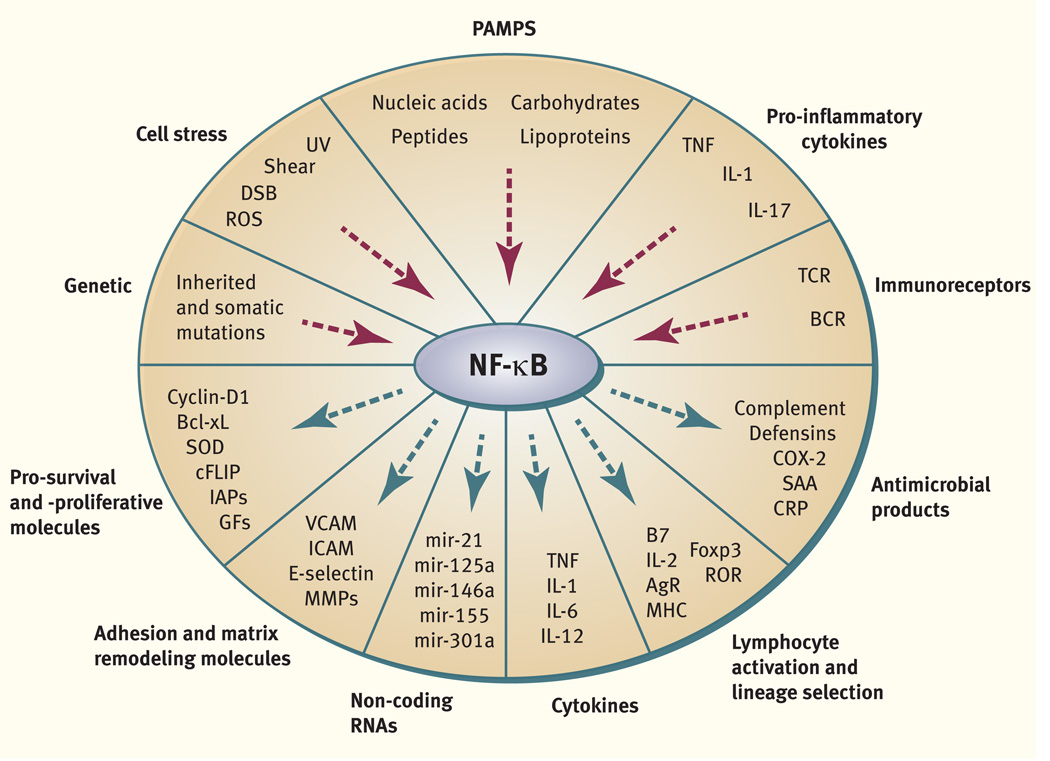
The brief discussion above is by no means intended to be comprehensive. Instead, it serves to highlight a few basic aspects of NF-κB biology, as a regulator of anti-apoptotic genes and crucial determinant of immune system development and function. By mediating signals from a diverse array of physiological and environmental stimuli to an expansive set of target genes (Fig. 1), NF-κB can contribute to a vast swath of normal physiology and pathobiology. For comprehensive discussion of these and many other aspects of NF-κB regulation and biology, we leave the reader to the works that follow in this volume of Immunological Reviews.
Fig. 1. NF-κB is a central regulator of the transcriptional responses to a wide variety of physiological and environmental stimuli.
Signals ranging from pro-inflammatory cytokines to stresses including reactive oxygen species (ROS), ultraviolet light (UV) and DNA double strand breaks (DSB), to engagement of antigen receptors lead to activation of NF-κB. Several inherited and somatically acquired mutations can also lead to activation of NF-κB. NF-κB transcriptional programs induce a wide variety of genes (http://www.nf-kb.org). These transcriptional programs have far-reaching biological consequences and include survival factors, growth factors, cytokines, chemokines, numerous core mediators of adaptive and innate immunity, as well as micro-RNAs.
Perspectives: conclusions and outstanding questions
Reviews on NF-κB are produced in a disproportionate number, and many have appeared in this anniversary year. Given this plethora of material, we would like to call special attention to the two unique aspects of the articles in this issue of Immunological Reviews. As each new review in the field is written, space and formatting constraints dictate that older findings be covered in less detail in order to accommodate new work. As such, not only does the story of how this field developed become lost but also nuances in the data that often have important biological consequences are often, necessarily, ignored. Therefore, the personal and historical narratives included in this volume are especially valuable. Second, this process of reflecting back on the history of the field has also led many of the authors to discuss truly fundamental questions that remain unanswered. The outstanding questions discussed in the reviews in this volume highlight how much remains to be done to understand the regulation and function of NF-κB. The mechanisms of IKK activation, IκB function, transcriptional activation and termination all are inadequately understood. Yet perhaps the most resonating of these unresolved issues, touched upon in several of the reviews in this issue, is the unfulfilled promise of NF-κB as a therapeutic target. Where these past 25 years have seen remarkable progress in the identification of NF-κB pathway components and functions, the research in the coming decades must strive to make equivalent progress in elucidating mechanistic detail and leveraging this knowledge for the development of therapies that can selectively target the myriad diseases characterized by NF-κB dysregulation.
Acknowledgements
We would like to thank the contributors to this volume, who despite numerous other commitments were able to generate the articles that follow this introduction. Work in the authors’ laboratories is supported by grants from the NIH (including AI033443, AI068977 and AI093985 to S.G. and ARRA P30 AR058886, M.S.H.)
References
- 1.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- 2.Baeuerle PA, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh S, Baltimore D. Activation in vitro of NF-kappa B by phosphorylation of its inhibitor I kappa B. Nature. 1990;344:678–682. doi: 10.1038/344678a0. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh S, Gifford AM, Riviere LR, Tempst P, Nolan GP, Baltimore D. Cloning of the p50 DNA binding subunit of NF-kappa B: homology to rel and dorsal. Cell. 1990;62:1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]
- 5.Kieran M, et al. The DNA binding subunit of NF-kappa B is identical to factor KBF1 and homologous to the rel oncogene product. Cell. 1990;62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- 6.Nolan GP, Ghosh S, Liou HC, Tempst P, Baltimore D. DNA binding and I kappa B inhibition of the cloned p65 subunit of NF-kappa B, a rel-related polypeptide. Cell. 1991;64:961–969. doi: 10.1016/0092-8674(91)90320-x. [DOI] [PubMed] [Google Scholar]
- 7.Davis N, Ghosh S, Simmons DL, Tempst P, Liou HC, Baltimore D, Bose HR., Jr Rel-associated pp40: an inhibitor of the rel family of transcription factors. Science. 1991;253:1268–1271. doi: 10.1126/science.1891714. [DOI] [PubMed] [Google Scholar]
- 8.Haskill S, et al. Characterization of an immediate-early gene induced in adherent monocytes that encodes I kappa B-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 9.Kopp E, Ghosh S. Inhibition of NF-kappa B by sodium salicylate and aspirin. Science. 1994;265:956–959. doi: 10.1126/science.8052854. [DOI] [PubMed] [Google Scholar]
- 10.Thompson JE, Phillips RJ, Erdjument-Bromage H, Tempst P, Ghosh S. I kappa B-beta regulates the persistent response in a biphasic activation of NF-kappa B. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 11.Rao P, et al. IkappaBbeta acts to inhibit and activate gene expression during the inflammatory response. Nature. 2010;466:1115–1119. doi: 10.1038/nature09283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 13.Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-kappaB is regulated by the IkappaB- associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 14.Dong J, Jimi E, Zhong H, Hayden MS, Ghosh S. Repression of gene expression by unphosphorylated NF-kappaB p65 through epigenetic mechanisms. Genes Dev. 2008;22:1159–1173. doi: 10.1101/gad.1657408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May MJ, D'Acquisto F, Madge LA, Glockner J, Pober JS, Ghosh S. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science. 2000;289:1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 16.Jimi E, et al. Selective inhibition of NF-kappa B blocks osteoclastogenesis and prevents inflammatory bone destruction in vivo. Nat Med. 2004;10:617–624. doi: 10.1038/nm1054. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh G, Wang WY-F, Huang D-B, Fusco A. NF-κB regulation: lessons from structures. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinz M, Arslan SC, Scheidereit C. It takes two to tango: IκBs, the multifunctional partners of NF-κB. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01102.x. [DOI] [PubMed] [Google Scholar]
- 19.Yamauchi S, Ito H, Miyajima A. IkappaBeta, a nuclear IkappaB protein, positively regulates the NF-kappaB-mediated expression of proinflammatory cytokines. Proc Natl Acad Sci USA. 2010;107:11924–11929. doi: 10.1073/pnas.0913179107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lui F, Xia Y, Parker AS, Verma IM. IKK biology. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanarek N, Ben-Neriah Y. Regulation of NF-κB by ubiquitination and degradation of the IκBs. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01098.x. [DOI] [PubMed] [Google Scholar]
- 22.Natoli G. NF-κB and chromatin: ten years on the path from basic mechanisms to candidate drugs. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01103.x. [DOI] [PubMed] [Google Scholar]
- 23.Smale ST. Dimer-specific regulatory mechanisms within the NF-κB family of transcription factors. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 24.Harhaj WE, Dixit VM. Regulation of NF-κB by deubiquitinases. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-κB. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- 26.Gilmore TD, Wolenski FS. NF-κB: where did it come from and why? Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01096.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan J, Greene WC. Dynamic roles for NF-B and HTLV-I and HIV-1 pretroviral pathogenesis. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01094.x. [DOI] [PubMed] [Google Scholar]
- 29.McCool KW, Miyamoto S. DNA damage-dependent NF-κB activation: NEMO turns nuclear signaling inside out. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang C, Lin X. Regulation of NF-κB by the CARD proteins. Immnol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun S-C. The noncanonical NF-κB pathway. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gantke T, Sriskantharajah S, Sadowski M, Ley SC. IkB kinase regulation of the TLP-2/ERK MAPK pathway. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01104.x. [DOI] [PubMed] [Google Scholar]
- 33.Baldwin AS. Regulation of cell death and autophagy by IKK and NF-κB: critical mechanisms in immune function and cancer. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01095.x. [DOI] [PubMed] [Google Scholar]
- 34.Smale ST. Dimer-specific regulator mechanisms within the NF-B family of transcription factors. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh G, van Duyne G, Ghosh S, Sigler PB. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 36.Muller CW, Rey FA, Sodeoka M, Verdine GL, Harrison SC. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature. 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 37.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF- kappaB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 38.Chen YQ, Ghosh S, Ghosh G. A novel DNA recognition mode by the NF-kappa B p65 homodimer. Nat Struct Biol. 1998;5:67–73. doi: 10.1038/nsb0198-67. [DOI] [PubMed] [Google Scholar]
- 39.Huxford T, Huang DB, Malek S, Ghosh G. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 40.Huang DB, Chen YQ, Ruetsche M, Phelps CB, Ghosh G. X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2. Structure. 2001;9:669–678. doi: 10.1016/s0969-2126(01)00635-9. [DOI] [PubMed] [Google Scholar]
- 41.Diaz-Meco MT, Moscat J. The atypical PKCs in inflammation: NF-κB and beyond. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaileh M, Sen R. NF-κB function in B lymphocytes. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01106.x. [DOI] [PubMed] [Google Scholar]
- 43.Gerondakis S, et al. NF-B subunit specificity in hemopoiesis. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2011.01090.x. [DOI] [PubMed] [Google Scholar]
- 44.Pasparakis M. Role of NF-κB in epithelial biology. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01109.x. [DOI] [PubMed] [Google Scholar]
- 45.Lim K-H, Yang Y, Staudt LM. Pathogenetic importance and therapeutic implications of NF-κB in lymphoid malignancies. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiDonato JA, Mercurio F, Karin M. NF-κB and the link between inflammation and cancer. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 47.Beg AA, Sha WC, Bronson RT, Baltimore D. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 48.Doi TS, Marino MW, Takahashi T, Yoshida T, Sakakura T, Old LJ, Obata Y. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proc Natl Acad Sci USA. 1999;96:2994–2999. doi: 10.1073/pnas.96.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 50.Alcamo E, et al. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-kappa B in leukocyte recruitment. J Immunol. 2001;167:1592–1600. doi: 10.4049/jimmunol.167.3.1592. [DOI] [PubMed] [Google Scholar]
- 51.Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y. NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J Exp Med. 1997;185:953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 53.Caamano JH, et al. Nuclear factor (NF)-kappa B2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J Exp Med. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franzoso G, et al. Mice deficient in nuclear factor (NF)-kappa B/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J Exp Med. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paxian S, et al. Abnormal organogenesis of Peyer's patches in mice deficient for NF-kappaB1, NF-kappaB2, and Bcl-3. Gastroenterology. 2002;122:1853–1868. doi: 10.1053/gast.2002.33651. [DOI] [PubMed] [Google Scholar]
- 56.Mebius RE. Organogenesis of lymphoid tissues. Nat Rev Immunol. 2003;3:292–303. doi: 10.1038/nri1054. [DOI] [PubMed] [Google Scholar]
- 57.Hayden MS, Ghosh S. NF-kappaB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 59.Zelazowski P, Carrasco D, Rosas FR, Moorman MA, Bravo R, Snapper CM. B cells genetically deficient in the c-Rel transactivation domain have selective defects in germline CH transcription and Ig class switching. J Immunol. 1997;159:3133–3139. [PubMed] [Google Scholar]
- 60.Carrasco D, et al. Multiple hemopoietic defects and lymphoid hyperplasia in mice lacking the transcriptional activation domain of the c-Rel protein. J Exp Med. 1998;187:973–984. doi: 10.1084/jem.187.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kriehuber E, et al. Balance between NF-kappaB and JNK/AP-1 activity controls dendritic cell life and death. Blood. 2005;106:175–183. doi: 10.1182/blood-2004-08-3072. [DOI] [PubMed] [Google Scholar]
- 62.Ryseck RP, Bull P, Takamiya M, Bours V, Siebenlist U, Dobrzanski P, Bravo R. RelB, a new Rel family transcription activator that can interact with p50-NF-kappa B. Mol Cell Biol. 1992;12:674–684. doi: 10.1128/mcb.12.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobrzanski P, Ryseck RP, Bravo R. Both N- and C-terminal domains of RelB are required for full transactivation: role of the N-terminal leucine zipper-like motif. Mol Cell Biol. 1993;13:1572–1582. doi: 10.1128/mcb.13.3.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV. RelB cellular regulation and transcriptional activity are regulated by p100. J Biol Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- 65.Dobrzanski P, Ryseck RP, Bravo R. Differential interactions of Rel-NF-kappa B complexes with I kappa B alpha determine pools of constitutive and inducible NF-kappa B activity. EMBO J. 1994;13:4608–4616. doi: 10.1002/j.1460-2075.1994.tb06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lernbecher T, Kistler B, Wirth T. Two distinct mechanisms contribute to the constitutive activation of RelB in lymphoid cells. EMBO J. 1994;13:4060–4069. doi: 10.1002/j.1460-2075.1994.tb06723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Basak S, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Savinova OV, Hoffmann A, Ghosh G. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes. Mol Cell. 2009;34:591–602. doi: 10.1016/j.molcel.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weih F, et al. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 70.Burkly L, et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- 71.Weih F, Durham SK, Barton DS, Sha WC, Baltimore D, Bravo R. p50-NF-kappaB complexes partially compensate for the absence of RelB: severely increased pathology in p50(−/−)relB(−/−) double-knockout mice. J Exp Med. 1997;185:1359–1370. doi: 10.1084/jem.185.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Snapper CM, et al. B cells lacking RelB are defective in proliferative responses, but undergo normal B cell maturation to Ig secretion and Ig class switching. J Exp Med. 1996;184:1537–1541. doi: 10.1084/jem.184.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yilmaz ZB, Weih DS, Sivakumar V, Weih F. RelB is required for Peyer's patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 2003;22:121–130. doi: 10.1093/emboj/cdg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akiyama T, et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 2008;29:423–437. doi: 10.1016/j.immuni.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 75.Klement JF, et al. IkappaBalpha deficiency results in a sustained NF-kappaB response and severe widespread dermatitis in mice. Mol Cell Biol. 1996;16:2341–2349. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng JD, Ryseck RP, Attar RM, Dambach D, Bravo R. Functional redundancy of the nuclear factor kappa B inhibitors I kappa B alpha and I kappa B beta. J Exp Med. 1998;188:1055–1062. doi: 10.1084/jem.188.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 78.Basak S, Behar M, Hoffmann A. Lessons from mathematical modeling the NFκB pathway. Immunol Rev. 2012;246 doi: 10.1111/j.1600-065X.2011.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]