Summary
The noncanonical nuclear factor-κB (NF-κB) signaling pathway mediates activation of the p52/RelB NF-κB complex and, thereby, regulates specific immunological processes. This NF-κB pathway relies on the inducible processing of NF-κB2 precursor protein, p100, as opposed to the degradation of IκBα in the canonical NF-κB pathway. A central signaling component of the noncanonical NF-κB pathway is NF-κB-inducing kinase (NIK), which functions together with a downstream kinase, inhibitor of NF-κB kinase α (IKKα), to induce phosphorylation-dependent ubiquitination and processing of p100. Under normal conditions, NIK is targeted for continuous degradation by a tumor necrosis factor (TNF) receptor-associated factor-3 (TRAF3)-dependent E3 ubiquitin ligase. In response to signals mediated by a subset of TNF receptor superfamily members, NIK becomes stabilized as a result of TRAF3 degradation, leading to the activation of noncanonical NF-κB. This review discusses both the historical perspectives and the recent progress in the regulation and biological function of the noncanonical NF-κB pathway.
Keywords: noncanonical NF-κB, p100 processing, NIK, TRAF3, TRAF2, cIAP
Introduction
The nuclear factor-κB (NF-κB) family of transcription factors regulates diverse biological processes, including many aspects of immunological functions (1, 2). This family is composed of structurally homologous transcription factors, including NF-κB1, NF-κB2, RelA, RelB, and c-Rel, which bind to κB enhancers as homo- or hetero-dimers. NF-κB proteins are normally sequestered in the cytoplasm by specific inhibitory proteins, inhibitors if NF-κB (IκBs), characterized by the presence of ankyrin-repeat structure. The prototypical IκB member is IκBα, which plays a primary role in mediating the activation of the canonical (or classical) NF-κB pathway (3). Several other IκB family members exist and appear to regulate more specific functions of NF-κB. The NF-κB1 and NF-κB2 are translated as precursor proteins, p105 and p100, which also function as IκB-like molecules (4). Their C-terminal portion has structural homology with IκBs and inhibits the nuclear translocation of associated NF-κB members. Proteasome-mediated processing of p105 and p100 not only generate their respective mature proteins, p50 and p52, but also results in the nuclear translocation of sequestered NF-κB members (4).
The signaling pathways that mediate NF-κB activation can be classified into canonical and noncanonical (or alternative) pathways. The canonical pathway responds to numerous stimuli, including ligands for antigen receptors, cytokine receptors, pattern-recognition receptors, etc. (3). The different signals converge to an IκB kinase (IKK) complex, composed of catalytic (IKKα and IKKβ) and regulatory (IKKγ) subunits. Upon activation, IKK phosphorylates IκBα at two N-terminal serines, triggering its ubiquitination and proteasomal degradation; this leads to the nuclear translocation of commonly seen NF-κB complexes, predominantly p50/RelA and p50/c-Rel dimers. Activation of the noncanonical NF-κB pathway involves different signaling molecules and leads to the predominant activation of the p52/RelB dimer (5). This pathway is based on processing of the NF-κB2 precursor protein, p100. In contrast to the constitutive and contranslational processing of p105 (6), the processing of p100 is a signal-induced and posttranslational event (7). Since p100 preferentially interacts with RelB (8), the processing of p100 not only generates p52 but also causes p52/RelB nuclear translocation (5). During the past decade, tremendous progress has been made in elucidating the signaling mechanisms and biological functions of the noncanonical NF-κB pathway. This review provides both the historical perspective and the recent progress of this area of NF-κB research.
Personal and historical narrative
My career in NF-κB research started when I was a graduate student in Stockholm University, where I found the potential involvement of NF-κB-like transcription factors in the regulation of antibacterial genes in insects (9). During my postdoctoral training in Warner Greene’s laboratory at the Gladstone Institute of Virology and Immunology, my colleagues and I discovered the inducible degradation of IκBα as a primary mechanism of NF-κB activation (10). Soon after this discovery, I became interested in the IκB-like protein p100, because of our finding that a large proportion of RelA is sequestered by p100 in tumor necrosis factor-α (TNF-α)-stimulated cells even when IκBα is depleted (11). Since TNF-α did not stimulate this pool of NF-κB, the question arose as to what kind of signals might induce the degradation/processing of p100.
After setting up my laboratory in Penn State University College of Medicine, I continued my effort to address this intriguing question. It became clear at that time that p100 is barely processed in most cell types; however, active processing of p100 occurs in B cells, since these lymphocytes produce abundant p52. These prior findings allowed us to postulate that p100 processing might be induced by a signal that is triggered only in specific cell types, such as B cells. Driven by this hypothesis, two of my former postdoctoral fellows, Gutian Xiao and Edward W. Harhaj, performed extensive biochemical studies to test which of the known NF-κB-activating kinases might induce the processing of p100 in transfected cells. After screening a panel of kinases, Gutian identified the NF-κB-inducing kinase (NIK) as the only kinase capable of inducing productive processing of p100 (7). Although NIK was initially thought to mediate NF-κB activation by proinflammatory cytokines (12), NIK mutation in alymphoplasia (Aly) mice or NIK knockout (KO) surprisingly had no obvious effect on NF-κB activation by TNF-α and interleukin-1 (IL-1) (13, 14). However, we found that the NIK mutation in Aly mice completely blocked the processing of p100 (7). NIK induces p100 site-specific phosphorylation, thereby triggering p100 polyubiquitination by SCFβTrCP E3 ligase and proteasomal processing (7, 15, 16). These initial studies confirmed our hypothesis that p100 processing is a signal-regulated process and identified NIK as a crucial component of this pathway.
When our initial work was published (7), Michael Karin’s laboratory found that mice expressing an IKKα mutant (IKK-AA), with serine-to-alanine substitutions at its phosphorylation sites, displayed phenotypic similarities to the Aly mice. Collaboration was initiated between our laboratories to examine the role of IKKα in NIK-induced p100 processing. Interestingly, we found that IKKα, but not IKKβ, is required for NIK-induced p100 processing in embryonic fibroblasts (MEFs), and our collaborators in the Karin laboratory also demonstrated a blockade of p100 processing in B cells of the IKKα-AA mice (17). Although our initial work revealed that NIK complex isolated from mammalian cells phosphorylated the C-terminus of p100 (7), recombinant NIK produced in insect cells was unable to do so (17). Moreover, recombinant IKKα phosphorylated p100, suggesting that NIK induces p100 phosphorylation via its associated IKKα. Our work also suggested that IKKα and IKKβ preferentially phosphorylate p100 and IκBα, respectively. In another study, we showed that NIK-induced p100 processing does not need IKKγ, the IKK-regulatory subunit required for canonical NF-κB pathway (18). These findings further emphasized the involvement of a noncanonical NF-κB pathway, in the induction of p100 processing.
Identification of signaling receptors that trigger the noncanonical NF-κB pathway was guided by the phenotypic similarities between the Aly mice and the mice that are deficient in certain TNF receptor (TNFR) superfamily members. Since Aly and lymphotoxin-β receptor (LTβR) KO mice shared similar defect in secondary lymphoid organ development, we tested whether LTβR has NIK-stimulating activity. Indeed, transfected LTβR potently stimulated NIK-mediated p100 processing (7). Subsequent work provided definitive evidence that the LTβR signaling pathway induces p100 processing in a NIK- and IKKα-dependent manner (19). Around the same time, several other groups reported the induction of p100 processing by BAFF (B-cell activating factor belonging to TNF family) receptor (BAFFR) and CD40 (20–22). Later on, RANK (receptor activator for NF-κB), TNFR2, and several other TNFR superfamily members were added to the list of noncanonical NF-κB-stimulatory receptors (23–28).
Soon after the discovery of the noncanonical NF-κB pathway, it became obvious that this new pathway differs from the canonical pathway in some major signaling properties. In particular, canonical NF-κB activation is rapid and independent of protein synthesis, whereas the noncanonical NF-κB activation is slow and dependent on protein synthesis. However, elucidating the mechanism of noncanonical NF-κB regulation turned out to be a tough task, mainly because the endogenous level of NIK was too low to be detected by biochemical approaches, such as immunoblotting (IB) and in vitro kinase assays. After extensive efforts, we realized that the low steady level of NIK is actually a mechanism of its regulation (29). NIK undergoes continuous degradation in resting cells but becomes stabilized and accumulated in response to CD40 and BAFFR signals (29).
To understand the mechanism underlying NIK degradation, we performed yeast two-hybrid screening using NIK as bait, which led to the identification of TRAF3 as a major NIK-binding partner (29). Interestingly, the binding of TRAF3 to NIK functions to target NIK for ubiquitin-dependent degradation. Moreover, induction of p100 processing by CD40 and BAFFR is mediated through degradation of TRAF3 and accumulation of NIK (29). Since recombinant TRAF3 did not ubiquitinate NIK in vitro, we hypothesized that TRAF3 may function as a critical component of a ubiquitin ligase mediating NIK ubiquitination in vivo (29). Indeed, recent studies suggest that TRAF3 functions as the substrate-binding subunit of a NIK ubiquitin ligase composed of TRAF3, TRAF2, and cIAP1 (or cIAP2) (here after named cIAP1/2) (30–33). Collectively, these studies have elucidated a unique signaling mechanism that regulates noncanonical NF-κB activation.
Mechanism of p100 processing
When expressed in mammalian cells, p100 is barely converted to p52 (7), a finding that explains why endogenous NF-κB2 exists predominantly as the precursor, p100 (34). However, the active production of p52 in specific cell types, such as B cells, suggested the existence of an inducible mechanism that mediates the processing of p100. It is now clear that the processing of p100 is suppressed by negative regulatory domains and stimulated via its site-specific phosphorylation (7).
Negative regulatory domains
The lack of constitutive p100 processing is due to the suppressive action of its C-terminal portion (7, 35), which contains a processing-inhibitory domain (PID) and an ankyrin repeat domain (ARD), both contributing to the suppression of processing (7, 36). The PID covers predominantly a death domain (DD) structure, and mutation of the conserved residues within the DD abolishes its processing-suppression function (7). When the structure of both DD and ARD is disrupted or deleted, the constitutive processing of the resulting p100 mutants becomes robust (36).
Although how the DD and ARD suppress p100 processing is yet to be examined by structural studies, one potential mechanism is that these C-terminal domains are involved in the formation of a tight structure of p100 that prevents its processing. In this regard, a notable feature of the constitutive processing of p100 C-terminal truncation (p100ΔC) mutants is the requirement of their nuclear translocation (36). Since ARD is known to mask the nuclear localization sequence (NLS) present in the N-terminal Rel-homology domain (RHD) of p100, it is likely that ARD, with the help of the DD, suppresses p100 processing via inhibition of its nuclear translocation. The requirement of nuclear translocation in p100 constitutive processing suggests that some important regulators may be localized in the nucleus. In this regard, it is well-recognized that a number of E3 ubiquitin ligases are predominantly localized in the nucleus (37–41), although it is unknown whether any of these nuclear E3s is involved in p100ΔC processing. Another potential mechanism was suggested by an interesting study showing that DNA-binding is required for p100 constitutive processing (42). Association with promoter of target genes, via the κB enhancer, may facilitate the recruitment of p100ΔC mutants to the proteasome, where they are initially cleaved, by an endoprotease, into p52 and a C-terminal fragment followed by degradation of the C-terminal fragment. The DNA-binding of p52 may also serve as a mechanism that prevents its degradation, since the DNA-bound p52 is more stable, at least in vitro (42).
Suppression of p100 processing by its negative regulatory domains is critical for preventing abnormal function of NF-κB2, which is suggested by the finding that nfκb2 gene is involved in chromosomal translocations in a subset of lymphomas (43). The rearranged nfκb2 genes encode C-terminally truncated p100 mutants lacking the negative regulatory domains and, thus, undergoing constitutive processing (7). Although the oncogenic potential of over-produced p52 is still under debate (42, 44), it is likely that the loss of IκB-like function of p100 along with over-production of p52 contributes to the development of lymphoma.
p100 phosphorylation
The inducible processing of p100 is triggered through its site-specific phosphorylation. p100 has a C-terminal NIK-responsive element that contains a sequence similar to the phosphorylation site of IκBα (7, 15). Mutation of the conserved serine residues (Ser-866 and Ser-870) abolishes the induction of p100 processing by both NIK and receptor signals (7, 15). Both in vitro kinase assays and phospho-IB assays confirmed that these two residues function as phosphorylation sites of p100 (7, 16). We initially found that a NIK complex isolated from mammalian cells was able to phosphorylate p100 (7) but subsequently identified IKKα as a NIK-associated kinase that directly phosphorylates p100 (17). In vitro, IKKα predominantly phosphorylates a serine (Ser-872) located near the conserved phosphorylation site of p100 (45). However, phosphorylation of p100 in cells expressing either NIK or IKKα occurs at serines 866 and 870 (16). This result may reflect the difference between in vitro and in vivo actions of IKKα, although it is also possible that the physiological function of IKKα may involve a modulating factor(s). In any case, serine 872 is largely dispensable for the inducible processing of p100 (16). In vitro, IKKα also phosphorylates several other serines located in the N-terminal RHD of p100 (45). Combined mutation of these serines interferes with NIK-induced p100 processing, but the putative N-terminal phosphorylation sites of p100 are yet to be confirmed by phospho-IB or mass spectrometry.
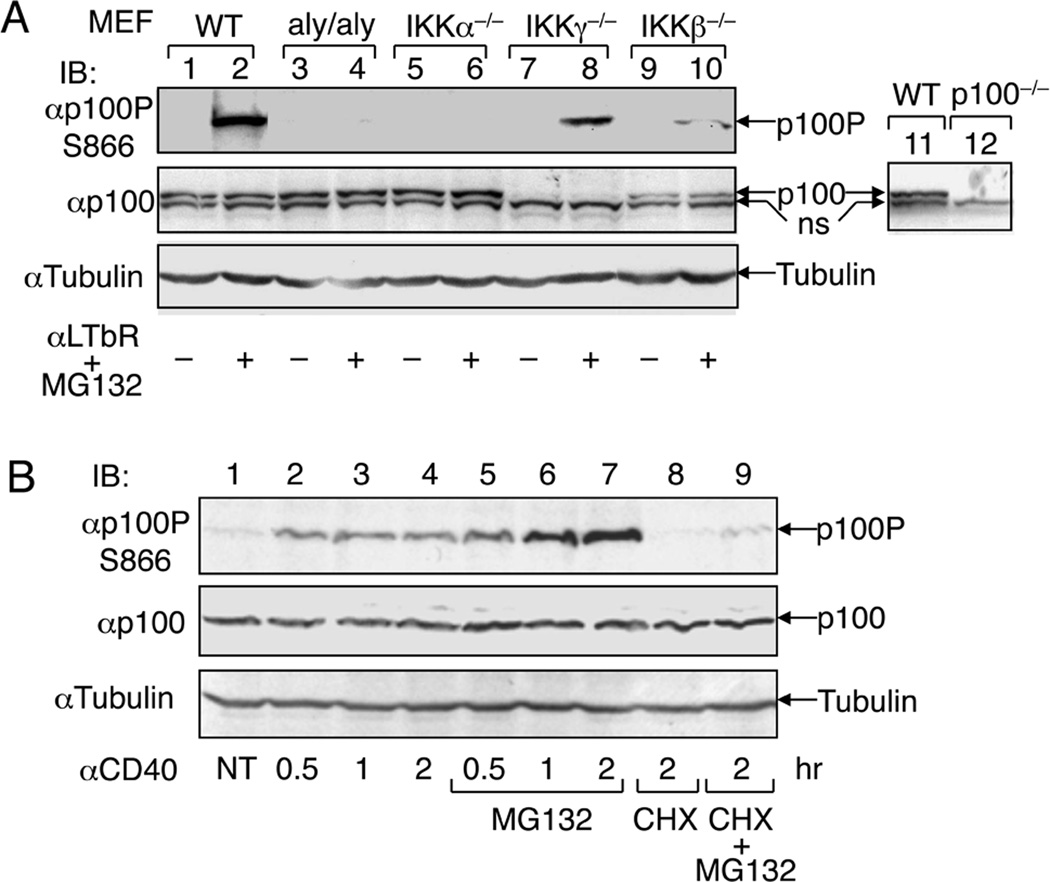
Signal-induced phosphorylation of p100 at serines 866 and 870 has been confirmed by IB using phospho-specific anti-p100 antibodies (16). The induction of p100 phosphorylation is dependent on NIK and IKKα but does not require IKKβ and IKKγ (Fig. 1A). Furthermore, as seen with the processing of p100, the signal-induced p100 phosphorylation is dependent on protein synthesis (16)(Fig. 1B). This latter finding is consistent with the observation that activation of noncanonical NF-κB involves signal-induced NIK stabilization and de novo protein synthesis (29).
Fig. 1. In vivo phosphorylation of p100.
(A) The indicated MEF cells were either not treated (−) or stimulated for 2 h with anti-murine LTβR (1 µg/ml) together with the proteasome inhibitor MG132 (25 µM). IB assays were performed to detect the phosphorylated p100 (p100P) and total p100 (p100) as well as the loading control tubulin using the indicated antibodies. Lane 11 and 12 show the cross-reactivity of the anti-p100 antibody with a non-specific protein (ns), as demonstrated by using the nfκb2 KO (p100−/−) MEFs. (B) M12 B cells expressing human CD40 (M12-CD40) were either not treated (NT) or stimulated for the indicated times with anti-human CD40. Where indicated, the cells were stimulated in the presence of MG132 and/or cycloheximide (CHX, 5 µg/ml). IB was performed using the indicated antibodies. This figure is adapted from Liang et al. (16) with permission.
p100 ubiquitination
The NIK-induced p100 processing is preceded by its polyubiquitination, a molecular event that is dependent on serines 866 and 870 (7). The sequence of the p100 phosphorylation site is similar to the consensus binding sequence of βTrCP (15). Indeed, p100 is bound by βTrCP in a manner that is dependent on NIK-induced p100 phosphorylation (15). Studies using synthetic peptides covering the phosphorylation site of p100 further confirmed that phosphorylation of serines 866 and 870 creates a βTrCP binding site at the C-terminus of p100 (16). As with IκBα degradation, the inducible ubiquitination and processing of p100 are critically dependent on βTrCP, thus establishing SCFβTrCP as a ubiquitin ligase mediating the inducible processing of p100. The ubiquitin acceptor site of p100 was later on mapped to a lysine residue (K856) that is located upstream of the phosphorylation site (46). Of note, the SCFβTrCP-mediated p100 ubiquitination is not required for the constitutive processing of p100ΔC mutants. These p100 mutants do not bind βTrCP nor requires βTrCP for processing (15).
How is the ubiquitinated p100 targeted to the proteasome for limited degradation (processing) is poorly understood. Nevertheless, a yeast two-hybrid screening study identified the binding of a proteasome regulatory subunit, S9 (also called Psmd11), to the C-terminal region of p100 (15). The binding of S9 to p100 requires NIK-induced p100 ubiquitination. In addition, the DD of p100 is critical for its association with S9. It is likely that ubiquitination of p100 facilitates the binding of S9 to the DD of p100. S9 knockdown inhibits NIK-induced processing of p100 but has no effect on the constitutive processing of p100ΔC mutants. Further studies of how the binding of S9 to p100 DD contributes to the inducible processing of p100 may shed light on the post-ubiquitination steps of p100 processing.
Kinases regulating the noncanonical NF-κB pathway
A central signaling component of the noncanonical NF-κB pathway is NIK (5), a mitogen-associated protein 3 kinase (MAP3K) originally thought to mediate NF-κB activation by cytokines, including TNF-α and IL-1 (12). Although overexpressed NIK activates canonical NF-κB, it is dispensable for NF-κB activation by TNF-α and IL-1 under physiological conditions (13, 14). In contrast, NIK is essential for the induction of p100 processing, as first revealed using the Aly mice (7). Furthermore, transiently expressed NIK but not several other kinases potently stimulates the processing of p100 (7). All of the known cellular stimuli of noncanonical NF-κB pathway rely on NIK for inducing p100 processing, although some viral oncoproteins seem to induce p100 processing in a NIK-independent manner (47).
Biochemical and genetic evidence establishes IKKα as a kinase that mediates NIK-induced p100 phosphorylation and processing (17). This finding, together with a prior observation that NIK phosphorylates IKKα (48), suggests that IKKα functions as a downstream kinase of NIK in the noncanonical NF-κB signaling pathway. One puzzle, though, is that IKKα activation does not seem to be sufficient for the induction of p100 processing. IKKα can be activated by various signals, but only the NIK-dependent signals induce p100 processing. Furthermore, when transiently expressed in mammalian cells, IKKα is much less effective than NIK in the induction of p100 processing (7, 45). It seems likely that NIK may induce p100 processing via both activation of IKKα and additional mechanisms. One potential mechanism is that NIK promotes the binding of IKKα to p100 and that the formation of a NIK/IKKα/p100 complex may be critical for p100 phosphorylation and processing (45). It is also possible that NIK may activate a new factor that induces p100 processing in synergy with IKKα.
Although NIK is essential for induction of p100 processing by cellular signals, some viral oncoproteins seem to induce p100 processing in a NIK-independent manner. One example is the oncoprotein Tax encoded by human T-cell leukemia virus type 1 (HTLV1) (18). Tax activates both the canonical and noncanonical NF-κB pathway by targeting the IKK complexes, and this physical association is mediated through direct binding of Tax to the IKK regulatory subunit IKKγ (49). Tax-induced p100 processing is insensitive to dominant-negative mutants of NIK but is dependent on IKKγ, indicating a mechanism that differs from the cellular pathway (18). Tax seems to mimic NIK in that it physically interacts with both p100 and IKKα (via IKKγ) and, thereby, recruits p100 to IKKα (18). Similar findings have been made with the viral FLICE-like inhibitory protein (vFLIP) of Kaposi’s sarcoma-associated herpes virus (KSHV) (50). Like Tax, vFLIP interacts with p100 and IKKα and induces the assembly of the p100/IKKα complex. While these findings suggest that Tax and vFLIP may mimic the action of NIK, the question is how these viral proteins activate the catalytic activity of IKKα. One possibility is that induction of IKKγ ubiquitination and IKKα aggregation by Tax, and possibly by vFLIP, may promote IKKα activation. On the other hand, the involvement of a different IKKα-activating kinase is also possible.
Mechanisms regulating NIK function
Signal-induced activation of protein kinases is usually mediated through their posttranslational modifications, predominantly phosphorylation. However, NIK is unique in that its function is primarily controlled through steady level of expression. The expression level of NIK is kept low under normal conditions but is drastically elevated in response to stimulation via specific receptors, such as CD40 and BAFFR (29). This signaling mechanism is dependent on dynamic ubiquitination and proteasome degradation events that mediate negative or positive regulation of NIK.
Negative regulation of NIK
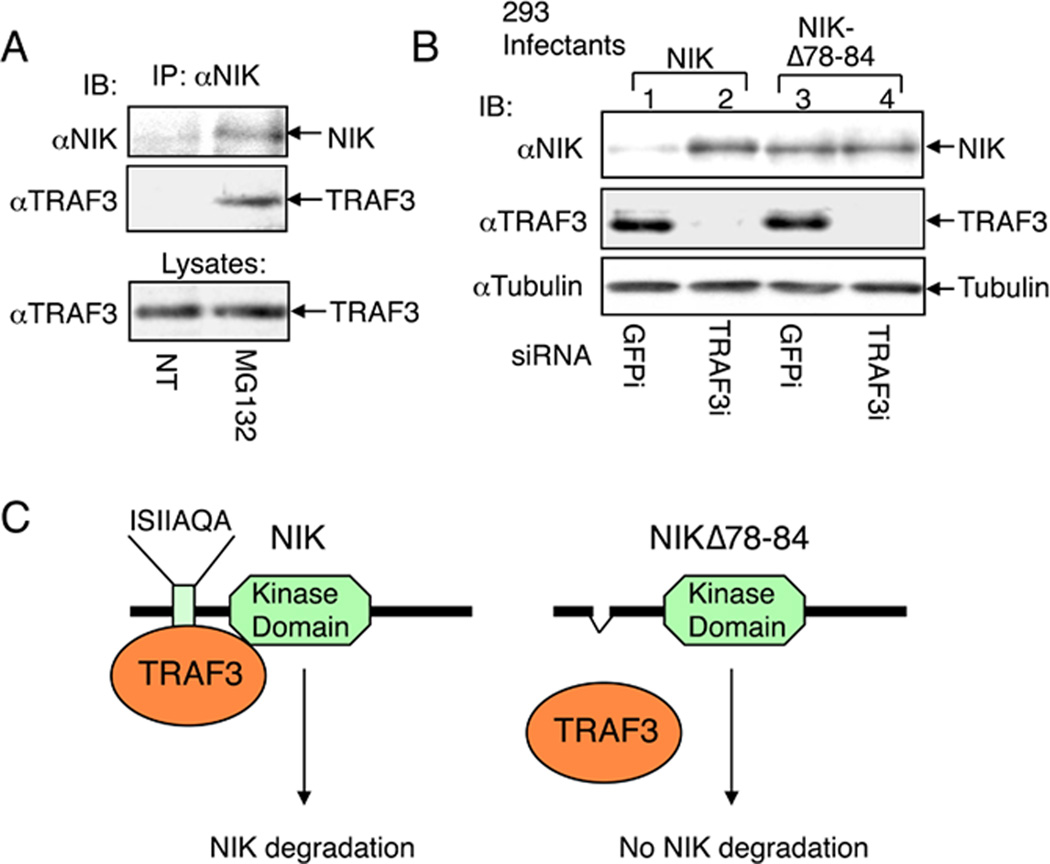
The negative mechanism of NIK regulation was first uncovered in a yeast two hybrid screening experiment that identified TRAF3 as a major NIK-binding protein (29). The strong binding between NIK and TRAF3 also occurs in mammalian cells, under both transfected and endogenous conditions. Although NIK was initially identified as a TRAF2-binding protein (12), the NIK-TRAF2 interaction is substantially weaker than the NIK-TRAF3 binding (29). The NIK-TRAF3 association serves as a key negative mechanism of NIK regulation. It appears that upon de novo synthesis, NIK is immediately bound by TRAF3 and targeted for ubiquitin-proteasome-mediated degradation. Thus, NIK undergoes constant degradation to keep the its steady level extremely low in unstimulated cells, and incubation of cells with a proteasome inhibitor, MG132, leads to the stabilization of NIK within the TRAF3 complex (29) (Fig. 2A). TRAF3 binds to an N-terminal region of NIK containing a core sequence (amino acid 78–84, ISIIAQA) that is not homologous to the previously defined TRAF2/3-binding motif (29). An NIK mutant lacking the TRAF3-binding motif, NIKΔ78–84 (harboring deletion of amino acid 78–84), becomes stabilized due to escape from TRAF3-mediated degradation(29) (Fig. 2B, C). When expressed in B cells of transgenic mice, this stable form of NIK mediates B-cell hyperplasia and severe autoimmunity (51). Expression of the NIK mutant in myeloid cells or osteoclast precursor cells also enhances osteoclast differentiation and function, leading to osteoporosis (52).
Fig. 2. NIK degradation through NIK-TRAF3 association.
(A) M12 B cells were either not treated (NT) or incubated for 2 h with proteasome inhibitor MG132. The NIK protein complex was isolated by IP followed by detection of NIK (top panel) and TRAF3 (middle panel) by IB using anti-NIK and horseradish peroxidase-conjugated anti-TRAF3, respectively. The TRAF3 expression level was detected by direct IB (bottom level). (B) 293 cells were infected with retroviruses encoding NIK or a NIK mutant lacking the core region of TRAF3-binding site (NIKΔ78–84). Stably infected cells were transfected with small-interfering RNA (siRNA) for either the control GFP or TRAF3. Cell lysates were subjected to IB using the indicated antibodies to detect the expression level of NIK (top panel), TRAF3 (panel 2), or control tubulin (panel 3). (C) Schematic picture depicting the regulation of NIK degradation by TRAF3. Wildtype NIK is bound by TRAF3 via an N-terminal domain and targeted for continuous degradation, which explains why NIK is barely detectable unless when the cells are treated with a proteasome inhibitor (A) or TRAF3 siRNA (B). NIKΔ78–84 is not bound by TRAF3 and stable even in the presence of TRAF3. This figure is adapted from Liao et al. (29) with permission.
More recently, NIK was found to be cleaved by APl2-MALT1, a fusion oncoprotein created by recurrent t(11;18)(q21;q21) chromosomal translocations in mucosa-associated lymphoid tissue (MALT) lymphoma (53). In addition to activating canonical NF-κB, APl2-MALT1 induces p100 processing and nuclear translocation of RelB/p52. APl2-MALT1 cleaves NIK at arginine 325, generating an N-terminally truncated NIK mutant, NIK(326–947), lacking the TRAF3-binding site and retaining kinase activity. This NIK mutant is resistant to TRAF3-mediated degradation and, thus, causes deregulated activation of noncanonical NF-κB signaling and enhanced B-cell adhesion and survival. This finding further emphasizes the importance of NIK/TRAF3 physical association in the negative regulation of NIK.
Although TRAF3 mediates NIK ubiquitination and degradation in mammalian cells, purified TRAF3 does not display E3 ubiquitin ligase activity towards NIK, leading to the hypothesis that TRAF3 may not directly ubiquitinate NIK but rather functions as a critical component of a ubiquitin ligase mediating NIK ubiquitination (29). Indeed, several more recent studies identified a NIK ubiquitin ligase, composed of cIAP1/2, TRAF2, and TRAF3 (hereafter named TRAF-cIAP E3 complex), in which TRAF3 functions as the substrate-binding component (30–33). Within the TRAF-cIAP E3 complex, TRAF2 serves as a linker to connect TRAF3 and cIAP1/2, since TRAF3 does not directly bind cIAP1/2 (5, 30, 33); this is why deficiency in either TRAF3 or TRAF2 leads to constitutive activation of the noncanonical NF-κB pathway (29, 54–57). In contrast to TRAF2 and TRAF3, cIAP1 and cIAP2 are functionally redundant, since simultaneous ablation of both cIAP1 and cIAP2 is required for stabilizing NIK (58). Knockin mice expressing an E3-inactive cIAP2 mutant also display aberrant noncanonical NF-κB activation in B cells, because the cIAP2 mutant interferes with the function of endogenous cIAP1 (59).
How the activated form of NIK is regulated is less well studied. A recent study suggests a role for IKKα in mediating feedback regulation of NIK (60). Upon activation by NIK, IKKα not only phosphorylates p100 to trigger p100 processing but also phosphorylates NIK. The IKKα-mediated NIK phosphorylation targets activated NIK for degradation, presumably serving as a mechanism to prevent accumulation of too much NIK in cells exposed to noncanonical NF-κB stimuli. This feedback degradation pathway does not require the TRAF-cIAP E3 complex, although the detailed underlying mechanism has not been defined (60, 61).
Mechanism of NIK activation
In contrast to the activation of canonical NF-κB by diverse receptor signals, only a specific subset of TNFR superfamily members mediates induction of noncanonical NF-κB signaling; these include LTβR (19), CD40 (21), BAFFR (20, 22), RANK (23), TNFR2 (24, 26), CD27 (25), etc. A hallmark of noncanonical NF-κB activation is the slow kinetics and dependence of de novo protein synthesis (16, 20). This feature can now be explained by the finding that NIK activation is a slow process that involves signal-induced TRAF3 degradation and NIK accumulation, the latter of which is likely a result of NIK stabilization and de novo synthesis (29) (Fig. 3).
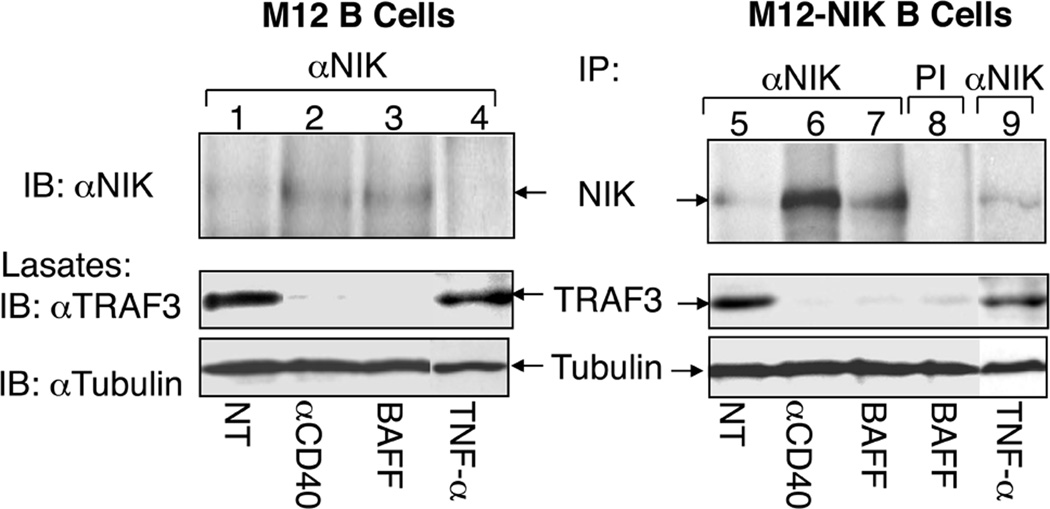
Fig. 3. Signal-induced NIK activation involves TRAF3 degradation and NIK accumulation.
M12 cells and M12-NIK cells (M12 cells stably infected with retroviruses encoding NIK) were either not treated (NT) or incubated for 2 h with anti-CD40, BAFF, or TNF-α. Whole-cell lysates were subjected to IP using anti-NIK (lanes 1–7 and 9) or a preimmune serum (lane 8) followed by detection of the enriched NIK protein by IB (top panel). The expression of TRAF3 (middle panel) and control tubulin (bottom panel) was detected by direct IB. This figure is adapted from Liao et al. (29) with permission.
Strong evidence suggests that NIK activation is predominantly mediated through its protein accumulation. First, exogenously expressed NIK is notoriously active in the induction of p100 processing (7). Second, NIK accumulation in TRAF3 knockdown or TRAF3 KO cells is sufficient for triggering the processing of p100 (29, 54). Third, cIAP1/2 degradation by antagonists induces NIK accumulation and activation of noncanonical NF-κB pathway (31, 32). In fact, genetic deficiency in any component of the NIK ubiquitin ligase complex, including TRAF2, TRAF3, cIAP1/2, leads to the accumulation of NIK and activation of noncanonical NF-κB pathway (29, 54–58). Based on the finding that NIK activation is also associated with its activation-loop phosphorylation (62), it is conceivable that NIK may undergo autophosphorylation along with the elevation of its expression level. However, at least for some receptors, such as CD27, the activation of NIK also involves its recruitment into the receptor complex (25). The receptor recruitment of NIK may increase its local concentration, thereby facilitating its trans-phosphorylation and catalytic activation.
The function of cIAP1/2 in NF-κB signaling is complex. In addition to mediating K48 ubiquitination and degradation of NIK in unstimulated cells, cIAP1/2 is responsible for the inducible degradation of TRAF3 in response to receptor signals, contributing to both constitutive NIK degradation and signal-induced NIK activation (30). Moreover, cIAP1/2 also possess K63 type of E3 activity towards receptor interacting protein 1 (RIP1), which is important for mediating activation of canonical NF-κB and cell survival (63, 64). Consistently, double ablation of cIAP1 and cIAP2 in B cells attenuates CD40-mediated activation of canonical NF-κB and MAPK pathways, in addition to causing constitutive activation of noncanonical NF-κB (58). Therefore, the cIAP1 and cIAP2 double KO mice display both BAFF-independent B-cell survival (due to constitutive noncanonical NF-κB activation) and impaired formation of germinal centers (due to attenuated canonical NF-κB signaling)(58). TRAF2 also plays dual roles in the regulation of noncanonical NF-κB signaling. While mediating constitutive degradation of NIK in untreated cells, TRAF2 is required for signal-induced degradation of TRAF3 (30, 65), thus contributing to inducible NIK stabilization. In addition to serving as an adapter of TRAF3 and cIAP1/2, TRAF2 mediates the recruitment of cIAP1/2 to the receptors and induces K63 ubiquitination of cIAP1/2, a modification that stimulates the K48 E3 ligase activity of the cIAP1/2 towards TRAF3 (30). How K63 ubiquitination of cIAP1/2 activates their K48 E3 function is unknown. One intriguing possibility is that the K63 ubiquitin chain may facilitate the association of cIAP1/2 with a specific E2 mediating K48 polyubiquitin chain conjugation.
It is likely that activation of NIK or noncanonical NF-κB is subject to regulation by additional factors that modulate the steps involved in cIAP1/2 activation or TRAF3 ubiquitination and degradation. In this regard, TRAF1 positively regulates noncanonical NF-κB activation by promoting TRAF3 degradation and NIK stabilization (66). TRAF1 directly interacts with BAFFR, providing a potential mechanism by which this TRAF member regulates BAFF-induced noncanonical NF-κB activation. However, since TRAF1 also promotes noncanonical NF-κB activation independently of BAFFR, additional mechanisms must exist. One other mechanism is suggested by a recent structural study demonstrating that TRAF1 binds to TRAF2 to form a ternary complex that associates with cIAP2 with elevated affinity (67). Therefore, TRAF1 may promote TRAF3 degradation and thus NIK stabilization by modulating the TRAF2/cIAP1/2 complex. Another newly identified positive regulator of noncanonical NF-κB pathway is MALT1, which is important for the induction of p100 processing and p52/RelB nuclear expression by BAFFR in B cells (68). MALT1 physically associates with TRAF3 and may function as a scaffold to promote TRAF3 ubiquitination by cIAP1/2 (68). Bcl10 (B-cell lymphoma 10), a partner of MALT1, may also have a role in mediating noncanonical NF-κB activation (69, 70), although it is unknown whether it functions together with MALT1.
Model of NIK regulation
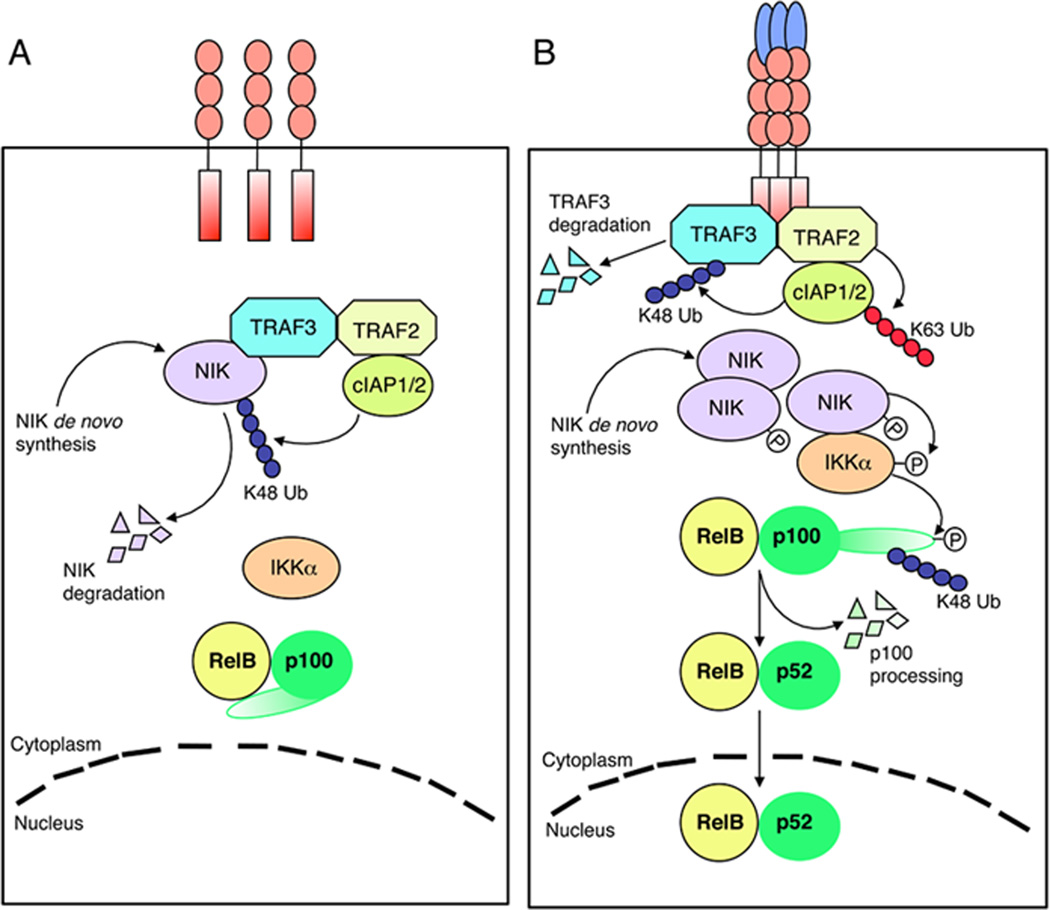
Based on our initial observation and the recent findings discussed above, a model of NIK regulation has emerged (Fig. 4). In cells that are not exposed to noncanonical NF-κB inducers, newly synthesized NIK is rapidly bound by TRAF3 and targeted to the TRAF-cIAP E3 ubiquitin ligase complex for K48-polyubiquitination and proteasomal degradation (Fig. 4A). Within the TRAF-cIAP complex, TRAF2 functions as an adapter of TRAF3 and cIAP1/2, whereas TRAF3 mediates substrate binding. The continuous degradation of NIK keeps its level extremely low to prevent induction of p100 processing under normal conditions. In cells stimulated by noncanonical NF-κB inducers, the TRAF-cIAP E3 complex is recruited to the receptors (Fig. 4B). Within the receptor complex, cIAP1/2 is activated by TRAF2-mediated K63 ubiquitination, and the activated cIAP1/2 then targets TRAF3 for K48 ubiquitination and degradation. In the absence of sufficient TRAF3, de novo synthesized NIK is accumulated and activated via trans-phosphorylation. NIK then activates IKKα, leading to p100 processing and nuclear translocation of RelB/p52. For some receptors, the activation of NIK may also involve its recruitment into the receptor complex, a mechanism that may promote NIK activation through increase of its local concentration.
Fig. 4. A model of noncanonical NF-κB regulation.
(A) Under normal conditions, de novo synthesized NIK is immediately bound by TRAF3 and recruited to the TRAF-cIAP E3 ubiquitin ligase complex via TRAF3-TRAF2 dimerization. cIAP1/2 catalyzes K48 ubiquitination of NIK, targeting NIK for degradation in the proteasome. The continuous degradation of NIK prevents NIK accumulation or activation of noncanonical NF-κB signaling. (B) Ligand engagement of the noncanonical NF-κB-stimulatory receptors induces recruitment of the TRAF-cIAP E3 components to the receptor complex. Probably due to its aggregation, TRAF2 becomes activated and mediates K63 ubiquitination of cIAP1/2, a modification that stimulates the K48-specific E3 activity of cIAP1/2. cIAP1/2 then conjugates K48 polyubiquitin chains to TRAF3, resulting in TRAF3 degradation. In the absence of TRAF3, the de novo synthesized NIK is stabilized and accumulated to higher levels; this allows NIK to induce IKKα-dependent p100 phosphorylation and ubiquitination, leading to the nuclear translocation of the generated RelB/p52 heterodimer.
Interplay between canonical and noncanonical pathways
The biological function of NF-κB often involves crosstalk within the family or with other families of transcription factors. In particular, the noncanonical and canonical NF-κB pathways have extensive interplays at different levels, including both upstream signaling crosstalk and nuclear interactions via formation of diverse NF-κB dimers. Although the two pathways usually cooperate in their biological functions, negative interplays have also been identified. The positive and negative interplays between the two NF-κB pathways may serve to modulate the kinetics and magnitude of expression of NF-κB target genes.
Indirect crosstalk via gene induction
A well-recognized mechanism of crosstalk between the canonical and noncanonical NF-κB pathways is the induction of nfκb2 gene expression by the canonical pathway; this positively regulates noncanonical NF-κB signaling by providing more NF-κB2 precursor, p100, for p52 generation (19). However, in cells stimulated with inducers that activate only the canonical NF-κB pathway, such as TNF-α, the accumulated p100 functions as an IκB to inhibit both the noncanonical NF-κB member RelB and the canonical NF-κB member RelA (11, 71, 72). The canonical NF-κB pathway also induces the expression of RelB, which may promote or inhibit the activation of noncanonical pathway. In human umbilical vein endothelial cells (HVEC), the TNF-induced canonical NF-κB suppresses CXCL12 gene induction by LTβR-mediated noncanonical NF-κB pathway (73). It appears that the TNF-α-induced excess amount of RelB represses the transactivation of CXCL12 gene by the noncanonical NF-κB. How precisely RelB exerts this negative function is yet to be defined, but the balance between RelB and p52 may be a determining factor. This possibility is also suggested by a recent study using human monocytes (74). During the differentiation of human monocytes to macrophages, IKKα steady level increases due to the downregulation of IKKα-specific microRNAs, which in turn leads to heightened production of p52. Due to the low level of RelB in resting macrophages, the excessive p52 production represses the expression of NF-κB target genes. However, induction of RelB by proinflammatory stimuli converts the role of p52 from negative to positive, leading to elevated NF-κB target gene expression (74).
A recent study provides an example for how noncanonical NF-κB pathway influences canonical NF-κB signaling by modulating expression of a signaling factor, WW domain–containing oxidoreductase (WWOX) (75). WWOX is a tumor suppressor encoded by a gene located at a chromosomal common fragile site and is affected in multiple cancers (76). Although WWOX may function through different pathways (77), it serves as an inhibitor of canonical NF-κB activation stimulated by the Tax oncoprotein of the human T-cell leukemia virus type 1 (HTLV1). In HTLV1-infected cells, Tax suppresses the expression of WWOX via activation of the noncanonical NF-κB pathway, thereby promoting activation of canonical NF-κB (75).
NF-κB subunit dimerization
The five NF-κB members can form various homo- and hetero-dimers with very few exceptions (78). For example, RelB functions as dimers with both p52 and the canonical NF-κB member p50. Furthermore, p52 can dimerize with RelA as well as with a coactivator protein, Bcl3, whose expression is induced by the canonical NF-κB pathway (79). While NF-κB dimerization typically mediates functional cooperation between the two NF-κB pathways, this mechanism may also mediate negative crosstalk. For example, in TNF-α stimulated cells, RelA functions as a repressor of RelB (80). Binding of RelA to RelB prevents the DNA binding activity of RelB. Therefore, although RelB moves to the nucleus in response to TNF-α stimulation, it does not engage DNA and remains inactive in the nucleus. The p65-mediated RelB suppression has also been detected in human dendritic cells (DCs) stimulated with the ligand of a C-type lectin, dectin-1 (81). Dectin-1 stimulates the nuclear translocation of both RelA and RelB, but the DNA-binding activity of RelB is repressed by RelA due to RelA/RelB dimer formation mediated by the Raf-1 signaling pathway. Via RelB suppression, the dectin-1-stimulated Raf-1 pathway promotes the expression of genes encoding IL-12 family members and, thereby, facilitates T-cell differentiation into T-helper 1 (Th1) and Th17 cells.
RelA and RelB also cooperate at the level of chromotin remodeling, as seen in the regulation of the GM-CSF (granulocyte-macrophage colony-stimulating factor) gene promoter (82). LTβR-activated RelB and TNF-α-activated RelA are independently recruited to the GM-CSF promoter, where they synergize in the induction of chromatin remodeling. Consequently, the LTβR signal primes NIH 3T3 cells for GM-CSF induction by TNF-α. The noncanonical and canonical NF-κB pathways, stimulated through LTβR and TNFR, also cooperate in the induction of the CXCL13 (also called BLC) gene in a LN stromal cell line, although the underlying mechanism is unclear (83).
Crosstalk via upstream signaling
An example of upstream signaling crosstalk between the canonical and noncanonical NF-κB pathways is provided by recent studies that RIP1 suppresses TNFR1-mediated activation of noncanonical NF-κB pathway (84, 85). Under normal conditions, TNFR1 activates the canonical but not the noncanonical NF-κB pathway. In fact, TNFR1 has an intrinsic activity to activate the noncanonical NF-κB pathway, but this activity is suppressed by a mechanism mediated by the adapter kinase RIP-1. RIP-1 inhibits TNF-α-stimulated degradation of TRAF2 and cIAP1 in a manner that does not require the kinase activity of RIP-1 (85) and may involve inhibition of the recruitment of TRAF2 to TNFR1 (84). Thus, although TNFR1 does not mediate TRAF2 degradation under normal conditions, genetic deficiency of RIP1 renders MEFs and other cell types sensitive to TNF-α-induced degradation of TRAF2 and cIAP1, leading to the induction of p100 processing. These findings suggest that the recruitment of RIP1 to TNFR1, during TNF-α stimulation, serves to both mediate activation of the canonical NF-κB pathway and inhibition of the noncanonical NF-κB pathway. However, it is currently unknown under what physiological or pathological conditions does TNFR1 mediates induction of the noncanonical NF-κB pathway.
Canonical NF-κB activation by NIK
The physiological function of NIK is primarily the activation of the noncanonical NF-κB pathway. However, under certain conditions, NIK may also contribute to the activation of canonical NF-κB (25). This crosstalk becomes prominent when NIK is aberrantly accumulated, as seen in TRAF3-deficient cells (86). Similarly, NIK accumulation in multiple myeloma and diffuse large B-cell lymphomas contributes to the activation of both noncanonical and canonical NF-κB pathways (87, 88). Another example is provided by the study of the oncogenic RET/PTC3 fusion protein (also called RP3), which is generated by chromosomal translocation that occurs in papillary thyroid carcinomas (PTCs) (89). RP3 activates canonical NF-κB, which is independent of IKKβ but is dependent on IKKγ, IKKα, and NIK. It appears that RP3 stabilizes NIK, presumably allowing the accumulated NIK to activate the canonical NF-κB pathway. Although the mechanism underlying this function of RP3 is unclear, RP3 does not induce the degradation of TRAF3 or TRAF2, indicating a mechanism that is independent of the TRAF-cIAP ubiquitin ligase. However, it remains to be examined whether RP3 induces the degradation of cIAP1 and cIAP2, which would also lead to NIK stabilization.
Biological roles of the noncanonical NF-κB pathway
The noncanonical and canonical NF-κB pathways typically function as a coordinative system. However, based on animal model studies, it is apparent that the function of noncanonical pathway is far more selective than that of the canonical pathway. One important factor that contributes to the functional selectivity of the noncanonical NF-κB pathway is its specific response to a small group of receptors, such as LTβR, BAFFR, CD40, RANK, etc. Indeed, the major biological functions of the noncanonical NF-κB pathway can be assigned to the different activating receptors. Well-known functions of the noncanonical NF-κB pathway include secondary lymphoid organogenesis and architecture organization, thymic epithelial cell (TEC) differentiation, B-cell maturation and survival, DC maturation, and bone metabolism. In addition, recent studies suggest a role for the noncanonical NF-κB pathway in regulating T-cell differentiation. Since most of these functions have been comprehensively discussed in recent reviews (79, 90–92), the following section focuses on the role of the noncanonical NF-κB pathway in regulating lymphoid organ development and T-cell differentiation.
Secondary lymphoid organ development and architecture organization
Secondary lymphoid organs are anatomical sites that provide a microenvironment for the maintenance and activation of lymphocytes and, thus, are important for initiation of adaptive immune responses (93). In addition, the lymphoid organs also play a role in mediating peripheral immune tolerance (94). The development of secondary lymphoid organs, particularly lymph nodes (LNs) and Peyer’s patches (PPs), involves interaction between hematopoietic cell-derived lymphoid tissue inducer (LTi) cells and the mesenchymal cell-derived stromal organizer cells (95). The stromal organizer cells express LTβR that is critical for transducing signals required for secondary lymphoid organogenesis. LTβR has two major ligands, LTα1β2 and LIGHT (homologous to lymphotoxins, inducible expression, competes with herpes simplex virus glycoprotein D for HVEM, a receptor expressed on T lymphocytes), the former of which has a dominant role in LTβR stimulation during lymphoid organ development (93). Engagement of LTβR by its ligand stimulates the stromal organizer cells to express chemokines, including CXCL13, CCL21 (also called SLC), and CCL19 (also called ELC) and cell adhesion molecules, such as vascular cell adhesion molecule 1 (VCAM1), intercellular adhesion molecule 1 (ICAM1), and mucosal addressin cell adhesion molecule 1 (MADCAM1), which mediate attraction and retention of LTi cells at the site of developing LNs and PPs as well as the recruitment and segregation of lymphocytes in mature secondary lymphoid organs (95, 96). Thus, mice with genetic deficiency in LTβR or LTα lack both LNs and PPs and have disorganized splenic architecture (97–102).
Studies using various mouse models have demonstrated a critical role for NF-κB pathways in mediating the function of stromal cells during lymphoid organogenesis. The noncanonical pathway, in particular, has a vital role in this biological process, although the canonical pathway is also importantly involved (2, 103). Aly mice, in which NIK mutation causes a complete blockade of noncanonical NF-κB signaling (7), lack all LNs and PPs and have disorganized splenic and thymic architectures (104, 105). This phenotype is similar to mice deficient in LTβR, suggesting a role for the noncanonical NF-κB pathway in mediating LTβR signaling in stromal organizer cells. Indeed, an initial in vitro study revealed that overexpressed LTβR stimulates NIK-mediated p100 processing (7), and the critical role of NIK and IKKα in LTβR signaling was definitively elucidated using MEFs and lymphoid stromal cells derived from Aly and IKKα-AA mice (19, 106). Ligation of LTβR induces robust processing of p100 in a NIK- and IKKα-dependent manner. It appears that the noncanonical NF-κB pathway is critical for LTβR-induced expression of several chemokines, whereas the canonical NF-κB pathway mediates the induction of specific cell adhesion molecules (such as VCAM1) and proinflammatory factors (19).
In addition to NIK, its downstream noncanonical NF-κB signaling components, including IKKα, NF-κB2, and RelB, are required for secondary lymphoid organ development (17, 72, 107, 108). Genetic deficiency of any of these molecules abolishes the development of PPs and also results in different degrees of defects in LN development (93, 103). The level of LN defect is correlated with the severity of the signaling blockade in noncanonical NF-κB activation. Compared to Aly and NIK KO mice, the nfκb2 KO mice have milder defects in LN development (109, 110). This is obviously because the nfκb2 deficiency, despite the lack of p52, does not block activation of RelB, and the RelB/p50 dimer may partially compensate the function of RelB/p52. Indeed, mice with double ablation of nfκb1 and nfκb2 have severe defect in LN development, which resembles the phenotype of the Aly and NIK KO mice (110). Similarly, mutant mice expressing a processing-defective p100 mutant (Lym1) lack all of the peripheral LNs and PPs and have shrunk mesenteric LNs (111). In contrast to the nfκb2 KO mice, the Lym1 mice are defective in both p52 production and RelB nuclear translocation. Moreover, the impaired p100 processing in Lym1 mice also inhibits LTβR-mediated nuclear translocation of RelA (111), a prototypical canonical NF-κB member that is also required for secondary lymphoid organ development (112).
The NF-κB pathways are also important for the architecture organization of the secondary lymphoid organs, which has been extensively characterized for the white pulp of the spleen (103, 113). The splenic white pulp, as well as the LN, is compartmentalized into T- and B-cell zones, which is important for efficient immune responses (113). Within the B-cell zones (or B-cell follicles), clonal expansion of antigen-activated B cells forms germinal centers (GCs) that support antibody isotype switching, affinity maturation, and other B-cell differentiation events. Formation and maintenance of the T/B-cell zones is regulated by specific chemokines, including CXCL13, CCL19, and CCL21, which attract B cells to the B-cell follicles (CXCL13) and T cells and DCs to the T-cell zone (CCL19 and CCL21) (113). Production of these homing chemokines is induced by the LTβR signals in stromal cells and critically involves the noncanonical NF-κB pathway (103). As such, noncanonical NF-κB regulates both the development of LNs and PPs and the formation of spleen architecture.
CD4+ T-cell differentiation
The canonical NF-κB pathway is well known for its role in regulating the activation and differentiation of T cells (79). In contrast, the involvement of the noncanonical NF-κB pathway in this aspect of immune function is less well studied. Nevertheless, it was noticed about a decade ago that T cells derived from the Aly mice had impaired proliferation and IL-2 production in response to stimulation by anti-CD3 and anti-CD28 (114). A similar phenotype was seen with T cells derived from NIK KO and RelB KO mice (115). However, the hyporesponsive phenotype of these mutant T cells appeared to be due to the presence of immunosuppressive CD25−Foxp3−CD44+ memory CD4+ T cells (115). Purified naive Aly T cells do not show proliferative defect but rather display a hyper-responsive phenotype. The mechanism by which Aly memory CD4+ T cells mediate immunosuppression is unclear, but these cells express high levels of CD25, despite their attenuated IL-2 expression, suggesting the possible involvement of CD25-mediated IL-2 consumption (115).
A role for NIK in regulating CD4+ T-cell differentiation is suggested by a recent finding that NIK is required for formation of Th17 cells (116), a subset of CD4+ effector T cells that produce IL-17 family of cytokines and are involved in autoimmunity (117). In an autoimmunity model, the NIK KO mice are refractory to the induction of experimental autoimmune encephalitis (EAE), coupled with impaired Th17 cell differentiation (116). In vitro experiments suggest that NIK deficiency diminishes the synergistic effect of the TCR and IL-6 receptor (IL-6R) in the activation of signal transducer and activator of transcription 3 (STAT3), a transcription factor important for Th17 cell differentiation. However, it remains to be further examined whether this function of NIK involves the induction of p100 processing or simply an NF-κB-independent function of NIK.
The noncanonical NF-κB pathway is also required for the generation of T follicular helper (Tfh) cells, a subset of CD4+ effector T cells mediating B-cell activation and antibody responses (118). Tfh generation requires both the T-cell receptor (TCR) signal and costimulatory signals, particularly the one mediated by the inducible costimulator (ICOS) (119). Although ICOS ligand (ICOSL) expression can be induced in DCs and fibroblasts, B cells characteristically express a high level of ICOSL and provide a crucial supporting function in Tfh cell differentiation (120, 121). The ICOSL expression in B cells is regulated by the noncanonical NF-κB pathway, which is chronically activated in peripheral B cells by the homeostatic inducer BAFF (118). The promoter region of ICOSL contains a κB-like sequence that is predominantly bound by p52, RelB, and p50 in BAFF-stimulated primary B cells and a B-cell line. Since both p52 and p50 form dimers with RelB, these findings suggest that the noncanonical NF-κB pathway may regulate Tfh cell induction via mediating ICOSL expression in B cells. The BAFF-induced signal in B cells also promotes the development of regulatory T cells (Tregs), although the underlying molecular mechanism is unclear (122).
Noncanonical NF-κB signaling in DCs also has an important role in mediating T-cell differentiation, which was suggested by a study using the Aly mice (123). It is thought that the noncanonical NF-κB inhibits proinflammatory cytokine induction and mediates expression of the anti-inflammatory enzyme indoleamine 2,3-dioxygenase (IDO), which together support the production of Treg-like immunosuppressive cells (124). Activation of noncanonical NF-κB in DCs is primarily mediated by engagement of CD40 by its ligand, CD40L, on T cells. In addition, a recent study suggests a role for a C-type lectin, dectin-1, in the activation of noncanonical NF-κB pathway in DCs, which in turn impacts on the CD4+ T-cell development (124). Dectin-1 mediates activation of both canonical and noncanonical NF-κB, but the latter is normally suppressed by a Raf-1 signaling axis downstream of the dectin-1. DC stimulation by the dectin-1 ligand curdlan induces the expression of IL-12, IL-23, and IL-1β, cytokines that promote Th1 and Th17 cell differentiation. However, inhibition of Raf-1 causes activation of noncanonical NF-κB, which represses the induction of these cytokines, leading to a shift of CD4+ T-cell differentiation towards the Th2 subset. It is important to note that the function of noncanonical NF-κB in DCs may be complex, since this pathway has also been shown to mediate costimulation of T cells that promote Th1 and Th17 cell differentiation in the EAE model (125).
γδ Th17 cell differentiation
A recent study demonstrates an essential role for both RelA and RelB in the development of γδ Th17 cells (126). While RelA appears to regulate expression of the LTβR ligands, LTα, LTβ, and LIGHT on accessory thymocytes, RelB has a dominant role in mediating the LTβR signaling within the γδ Th17 precursor cells. This cell intrinsic pathway mediates induction of RORγt and RORα4, transcription factors regulating the expression of the Th17 signature cytokine IL-17. As with RelB, p52 is required for γδ Th17 differentiation. These findings suggest a role for the noncanonical NF-κB pathway in the regulation of innate immune Th17 responses. Since γδ Th17 cells mediate neutrophil recruitment during bacterial infections, this novel function of noncanonical NF-κB pathway is important for early phase of host response to bacterial infections (126).
Perspectives: conclusions and outstanding questions
Since the discovery of the noncanonical NF-κB pathway a decade ago, significant progress has been made in understanding its regulation and biological functions. In particular, the finding that NIK is regulated by the TRAF-cIAP E3-mediated degradation and activated via signal-induced NIK stabilization provides critical insights into the signaling mechanism of the noncanonical NF-κB pathway. These findings have yielded valuable information for drug design in the treatment of NF-κB-related diseases, such as autoimmunity, bone disorders, and cancer. However, there are a number of outstanding issues that need to be addressed in future studies.
A major question is about the molecular mechanism that regulates the dynamic function of cIAP1/2. Since cIAP1/2 is responsible for both constitutive degradation of NIK and inducible degradation of TRAF3, it is important to understand how the substrate selectivity of this E3 is regulated in resting and signal-induced cells. Recent evidence suggests that upon activation by receptor signals, TRAF2 mediates K63 ubiquitination of cIAP1/2 and, thereby, stimulates the K48 ubiquitin ligase activity of cIAP1/2 towards TRAF3 (30). However, it remains to be examined why cIAP1/2 is constitutively active towards NIK in resting cells and has to be activated by TRAF2 to ubiquitinate TRAF3 in signal-induced cells. It is also intriguing to understand how the K63 ubiquitination of cIAP1/2 stimulates its K48-specific ubiquitin ligase function. For example, does the K63 ubiquitin chain facilitate the association of an E2 to cIAP1/2?
Another question is on the functional selectivity of the noncanonical NF-κB pathway. Although this pathway is known to regulate specific genes, such as the lymphoid homing chemokines, it is still uncertain whether this involves the selective binding of RelB/p52 to a specific type of κB enhancer in these target gene promoters. One study suggests that RelB/p52 indeed selectively binds to the promoters of CXCL13, CCL21, and CCL19 genes in LTβR-stimulated cells via a novel type of κB enhancer (106), although such a binding preference could not be detected in an in vitro selection approach (127). In support of the promoter-selectivity model, our recent work suggests that the noncanonical NF-κB pathway is critical for regulating ICOSL expression in B cells (118). Stimulation of primary B cells and a B-cell line with BAFF induces the predominant binding of noncanonical NF-κB members to the ICOSL promoter via a κB-like element. However, it appears that this role of noncanonical NF-κB is dependent on the cell types and stimuli. Clearly, more studies are required to further explore the underlying molecular mechanisms.
Finally, although the biological roles of the noncanonical NF-κB pathway have been extensively studied, how this pathway functions in specific cell types is obscure. Some of the biological phenotypes detected with conventional KO mice are complex and may be contributed by impaired function of noncanonical NF-κB in multiple cell types. It is anticipated that important information will be obtained through the study of conditional knockout mice harboring cell-specific deficiencies in the noncanonical NF-κB signaling components.
Acknowledgments
Work in the author’s laboratory is supported by grants from the US National Institutes of Health (AI057555, AI064639, GM84459, and GM84459-S1).
Footnotes
The author has no conflicts of interest to declare.
References
- 1.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. NF-κB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Beinke S, Ley SC. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun SC. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin L, DeMartino GN, Greene WC. Cotranslational biogenesis of NF-kappaB p50 by the 26S proteasome. Cell. 1998;92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- 7.Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 8.Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV. RelB cellular regulation and transcriptional activity are regulated by p100. J Biol Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- 9.Sun S-C, Faye I. Cecropia immunoresponsive factor, an insect immunoresponsive factor with DNA-binding properties similar to nuclear factor kB. Eur J Biochem. 1992;204:885–892. doi: 10.1111/j.1432-1033.1992.tb16708.x. [DOI] [PubMed] [Google Scholar]
- 10.Sun S-C, Ganchi PA, Ballard DW, Greene WC. NF-kB controls expression of inhibitor IkBa: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 11.Sun S-C, Ganchi PA, Beraud C, Ballard DW, Greene WC. Autoregulation of the NF-kB transactivator Rel A (p65) by multiple cytoplasmic inhibitors containing ankyrin motifs. Proc Natl Acad Sci USA. 1994;91:1346–1350. doi: 10.1073/pnas.91.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malinin NL, Boldin MP, Kovalenko AV, Wallach D. MAP3K-related kinase involved in NF-kB induction by TNF, CD95 and IL1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 13.Shinkura R, et al. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat Genet. 1999;22:74–77. doi: 10.1038/8780. [DOI] [PubMed] [Google Scholar]
- 14.Yin L, et al. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Science. 2001;291:2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- 15.Fong A, Sun S-C. Genetic evidence for the essential role of beta-transducin repeat-containing protein in the inducible processing of NF-kB2/p100. J Biol Chem. 2002;277:22111–22114. doi: 10.1074/jbc.C200151200. [DOI] [PubMed] [Google Scholar]
- 16.Liang C, Zhang M, Sun SC. beta-TrCP binding and processing of NF-kappaB2/p100 involve its phosphorylation at serines 866 and 870. Cell Signal. 2006;18:1309–1317. doi: 10.1016/j.cellsig.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Senftleben U, et al. Activation of IKKa of a second, evolutionary conserved, NF-kB signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 18.Xiao G, et al. Retroviral oncoprotein Tax induces processing of NF-kappaB2/p100 in T cells: evidence for the involvement of IKKalpha. EMBO J. 2001;20:6805–6815. doi: 10.1093/emboj/20.23.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dejardin E, et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 20.Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappaB2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 21.Coope HJ, et al. CD40 regulates the processing of NF-kappaB2 p100 to p52. EMBO J. 2002;15:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayagaki N, et al. BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2. Immunity. 2002;17:515–524. doi: 10.1016/s1074-7613(02)00425-9. [DOI] [PubMed] [Google Scholar]
- 23.Novack DV, et al. The IkappaB function of NF-kappaB2 p100 controls stimulated osteoclastogenesis. J Exp Med. 2003;198:771–781. doi: 10.1084/jem.20030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munroe ME, Bishop GA. Role of tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) in distinct and overlapping CD40 and TNF receptor 2/CD120b-mediated B lymphocyte activation. J Biol Chem. 2004;279:53222–53231. doi: 10.1074/jbc.M410539200. [DOI] [PubMed] [Google Scholar]
- 25.Ramakrishnan P, Wang W, Wallach D. Receptor-specific signaling for both the alternative and the canonical NF-kappaB activation pathways by NF-kappaB-inducing kinase. Immunity. 2004;21:477–489. doi: 10.1016/j.immuni.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Rauert H, et al. Membrane tumor necrosis factor (TNF) induces p100 processing via TNF receptor-2 (TNFR2) J Biol Chem. 2010;285:7394–7404. doi: 10.1074/jbc.M109.037341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitoh T, Nakayama M, Nakano H, Yagita H, Yamamoto N, Yamaoka S. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J Biol Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- 28.Wicovsky A, et al. TNF-like weak inducer of apoptosis inhibits proinflammatory TNF receptor-1 signaling. Cell Death Dffer. 2009;16:1445–1459. doi: 10.1038/cdd.2009.80. [DOI] [PubMed] [Google Scholar]
- 29.Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 30.Vallabhapurapu S, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varfolomeev E, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 32.Vince JE, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 33.Zarnegar BJ, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betts JC, Nabel GJ. Differential regulation of NF-kappaB2(p100) processing and control by amino-terminal sequences. Mol Cell Biol. 1996;16:6363–6371. doi: 10.1128/mcb.16.11.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heusch M, Lin L, Geleziunas R, Greene WC. The generation of nfkb2 p52: mechanism and efficiency. Oncogene. 1999;18:6201–6208. doi: 10.1038/sj.onc.1203022. [DOI] [PubMed] [Google Scholar]
- 36.Liao G, Sun SC. Regulation of NF-kappaB2/p100 processing by its nuclear shuttling. Oncogene. 2003;22:4868–4874. doi: 10.1038/sj.onc.1206761. [DOI] [PubMed] [Google Scholar]
- 37.Blondel M, et al. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 2001;19:6085–6097. doi: 10.1093/emboj/19.22.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lassot I, et al. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(betaTrCP) ubiquitin ligase. Mol Cell Biol. 2001;21:2192–2202. doi: 10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadot E, Simcha I, Iwai K, Ciechanover A, Geiger B, Ben-Ze'ev A. Differential interaction of plakoglobin and beta-catenin with the ubiquitin-proteasome system. Oncogene. 2001;19:1992–2001. doi: 10.1038/sj.onc.1203519. [DOI] [PubMed] [Google Scholar]
- 40.Davis M, et al. Pseudosubstrate regulation of the SCF(b-TrCP) ubiquitin ligase by hnRNP-U. Genes Dev. 2002;16:439–451. doi: 10.1101/gad.218702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natoli G, Chiocca S. Nuclear ubiquitin ligases, NF-kappaB degradation, and the control of inflammation. Sci Sigal. 2008;1:pe1. doi: 10.1126/stke.11pe1. [DOI] [PubMed] [Google Scholar]
- 42.Qing G, Qu Z, Xiao G. Endoproteolytic processing of C-terminally truncated NF-kappaB2 precursors at kappaB-containing promoters. Proc Natl Acad Sci USA. 2007;104:5324–5329. doi: 10.1073/pnas.0609914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Zhang B, Yang L, Ding J, Ding HF. Constitutive production of NF-kappaB2 p52 is not tumorigenic but predisposes mice to inflammatory autoimmune disease by repressing Bim expression. J Biol Chem. 2008;283:10698–10706. doi: 10.1074/jbc.M800806200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IkappaB kinase alpha (IKKalpha) to p100 and IKKalpha-mediated phosphorylation. J Biol Chem. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 46.Amir RE, Haecker H, Karin M, Ciechanover A. Mechanism of processing of the NF-kappa B2 p100 precursor: identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCF(beta-TrCP) ubiquitin ligase. Oncogene. 2004;23:2540–2547. doi: 10.1038/sj.onc.1207366. [DOI] [PubMed] [Google Scholar]
- 47.Sun SC, Cesarman E. NF-κB as a Target for Oncogenic Viruses. Curr Top Microbiol Immunol. 2011;349:197–244. doi: 10.1007/82_2010_108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling L, Cao Z, Goeddel DV. NF-kB-inducing kinase activates IKK-a by phosphorylation of Ser-176. Proc Natl Acad Sci USA. 1998;95:3792–3797. doi: 10.1073/pnas.95.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun SC, Yamaoka S. Activation of NF-kB by HTLV-I and implications for cell transformation. Oncogene. 2005;24:5952–5964. doi: 10.1038/sj.onc.1208969. [DOI] [PubMed] [Google Scholar]
- 50.Matta H, Chaudhary PM. Activation of alternative NF-kappa B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1 beta-converting enzyme inhibitory protein (vFLIP) Proc Natl Acad Sci USA. 2004;101:9399–9404. doi: 10.1073/pnas.0308016101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki Y, et al. NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF:BAFF-R-mediated survival signals in B cells. Proc Natl Acad Sci USA. 2008;105:10883–10888. doi: 10.1073/pnas.0805186105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang C, et al. NIK stabilization in osteoclasts results in osteoporosis and enhanced inflammatory osteolysis. PLoS One. 2010;5:e15383. doi: 10.1371/journal.pone.0015383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosebeck S, et al. Cleavage of NIK by the API2-MALT1 fusion oncoprotein leads to noncanonical NF-kappaB activation. Science. 2011;331:468–472. doi: 10.1126/science.1198946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He JQ, et al. Rescue of TRAF3-null mice by p100 NF-kappa B deficiency. J Exp Med. 2006;203:2413–2418. doi: 10.1084/jem.20061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grech AP, Amesbury M, Chan T, Gardam S, Basten A, Brink R. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-kappaB activation in mature B cells. Immunity. 2004;21:629–642. doi: 10.1016/j.immuni.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 56.Xie P, Stunz LL, Larison KD, Yang B, Bishop GA. Tumor necrosis factor receptor-associated factor 3 is a critical regulator of B cell homeostasis in secondary lymphoid organs. Immunity. 2007;27:253–267. doi: 10.1016/j.immuni.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardam S, Sierro F, Basten A, Mackay F, Brink R. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity. 2008;28:391–401. doi: 10.1016/j.immuni.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 58.Gardam S, et al. Deletion of cIAP1 and cIAP2 in murine B lymphocytes constitutively activates cell survival pathways and inactivates the germinal center response. Blood. 2011;117:4041–4051. doi: 10.1182/blood-2010-10-312793. [DOI] [PubMed] [Google Scholar]
- 59.Conze DB, Zhao Y, Ashwell JD. Non-canonical NF-κB activation and abnormal B cell accumulation in mice expressing ubiquitin protein ligase-inactive c-IAP2. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000518. e1000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Razani B, et al. Negative feedback in non-canonical NF-κB signaling modulates NIK stability through IKKα-mediated phosphorylation. Sci Sig. 2010;3:ra41. doi: 10.1126/scisignal.2000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun SC. Controlling the fate of NIK: a central stage in noncanonical NF-kappaB signaling. Sci Sigal. 2010;3:pe18. doi: 10.1126/scisignal.3123pe18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin X, Mu Y, Cunningham ETJ, Marcu KB, Geleziunas R, Greene WC. Molecular determinants of NF-kappaB-inducing kinase action. Mol Cell Biol. 1998;18:5899–5907. doi: 10.1128/mcb.18.10.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahoney DJ, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci USA. 2008;105:11778–11783. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 65.Hostager BS, Haxhinasto SA, Rowland SL, Bishop GA. Tumor necrosis factor receptor-associated factor 2 (TRAF2)-deficient B lymphocytes reveal novel roles for TRAF2 in CD40 signaling. J Biol Chem. 2003;278:45382–45390. doi: 10.1074/jbc.M306708200. [DOI] [PubMed] [Google Scholar]
- 66.Lavorgna A, De Filippi R, Formisano S, Leonardi A. TNF receptor-associated factor 1 is a positive regulator of the NF-kappaB alternative pathway. Mol Immunol. 2009;46:3278–3282. doi: 10.1016/j.molimm.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 67.Zheng C, Kabaleeswaran V, Wang Y, Cheng G, Wu H. Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation. Mol Cell. 2010;38:101–113. doi: 10.1016/j.molcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tusche MW, et al. Differential requirement of MALT1 for BAFF-induced outcomes in B cell subsets. J Exp Med. 2009;206:2671–2683. doi: 10.1084/jem.20091802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z, et al. Emu-BCL10 mice exhibit constitutive activation of both canonical and noncanonical NF-kappaB pathways generating marginal zone (MZ) B-cell expansion as a precursor to splenic MZ lymphoma. Blood. 2009;114:4158–4168. doi: 10.1182/blood-2008-12-192583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhattacharyya S, Borthakur A, Dudeja PK, Tobacman JK. Lipopolysaccharide-induced activation of NF-kappaB non-canonical pathway requires BCL10 serine 138 and NIK phosphorylations. Exp Cell Res. 2010;316:3317–3327. doi: 10.1016/j.yexcr.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Derudder E, Dejardin E, Pritchard LL, Green DR, Korner M, Baud V. RelB/p50 dimers are differentially regulated by tumor necrosis factor-α and lymphotoxin-β receptor activation: critical roles for p100. J Biol Chem. 2003;278:23278–23284. doi: 10.1074/jbc.M300106200. [DOI] [PubMed] [Google Scholar]
- 72.Yilmaz ZB, Weih DS, Sivakumar V, Weih F. RelB is required for Peyer's patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 2003;22:121–130. doi: 10.1093/emboj/cdg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madge LA, May MJ. Classical NF-kappaB activation negatively regulates noncanonical NF-kappaB-dependent CXCL12 expression. J Biol Chem. 2010;285:38069–38077. doi: 10.1074/jbc.M110.147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu J, et al. The tumor suppressor gene WWOX links the canonical and noncanonical NF-κB pathways in HTLV-I Tax-mediated tumorigenesis. Blood. 2011;117:1652–1661. doi: 10.1182/blood-2010-08-303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aqeilan RI, Croce CM. WWOX in biological control and tumorigenesis. J Cell Physiol. 2007;212:307–310. doi: 10.1002/jcp.21099. [DOI] [PubMed] [Google Scholar]
- 77.Salah Z, Aqeilan R, Huebner K. WWOX gene and gene product: tumor suppression through specific protein interactions. Future Oncol. 2010;6:249–259. doi: 10.2217/fon.09.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 79.Gerondakis S, Siebenlist U. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb Perspect Biol. 2010;2:1–29. doi: 10.1101/cshperspect.a000182. a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacque E, Tchenio T, Piton G, Romeo PH, Baud V. RelA repression of RelB activity induces selective gene activation downstream of TNF receptors. Proc Natl Acad Sci USA. 2005;102:14635–14640. doi: 10.1073/pnas.0507342102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gringhuis SI, et al. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-kappaB activation through Raf-1 and Syk. Nat Immunol. 2009;10:203–213. doi: 10.1038/ni.1692. [DOI] [PubMed] [Google Scholar]
- 82.Sasaki CY, Ghosh P, Longo DL. Recruitment of RelB to the Csf2 promoter enhances RelA-mediated transcription of granulocyte-macrophage colony-stimulating factor. J Biol Chem. 2011;286:1093–1120. doi: 10.1074/jbc.M110.119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suto H, Katakai T, Sugai M, Kinashi T, Shimizu A. CXCL13 production by an established lymph node stromal cell line via lymphotoxin-beta receptor engagement involves the cooperation of multiple signaling pathways. Int Immunol. 2009;21:467–476. doi: 10.1093/intimm/dxp014. [DOI] [PubMed] [Google Scholar]
- 84.Kim JY, et al. TNFα induced noncanonical NF-κB activation is attenuated by RIP1 through stabilization of TRAF2. J Cell Sci. 2011;124:647–656. doi: 10.1242/jcs.075770. [DOI] [PubMed] [Google Scholar]
- 85.Gentle IE, et al. In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8. J Biol Chem. 2011;286:13282–13291. doi: 10.1074/jbc.M110.216226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zarnegar B, Yamazaki S, He JQ, Cheng G. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc Natl Acad Sci USA. 2008;105:3503–3508. doi: 10.1073/pnas.0707959105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood. 2010;115:3541–3552. doi: 10.1182/blood-2009-09-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pham LV, et al. Constitutive BR3 receptor signaling in diffuse, large B-cell lymphomas stabilizes nuclear factor-κB-inducing kinase while activating both canonical and alternative nuclear factor-κB pathways. Blood. 2011;117:200–210. doi: 10.1182/blood-2010-06-290437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neely RJ, et al. The RET/PTC3 oncogene activates classical NF-κB by stabilizing NIK. Oncogene. 2011;6:87–96. doi: 10.1038/onc.2010.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dejardin E. The alternative NF-kappaB pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem Pharmacol. 2006;72:1161–1179. doi: 10.1016/j.bcp.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 91.Zhu M, Fu Y. The complicated role of NF-kappaB in T-cell selection. Cell Mol Immunol. 2010;7:89–93. doi: 10.1038/cmi.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Novack DV. Role of NF-κB in the skeleton. Cell Res. 2011;21:169–182. doi: 10.1038/cr.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mebius RE. Organogenesis of lymphoid tissues. Nat Rev Immunol. 2003;3:292–303. doi: 10.1038/nri1054. [DOI] [PubMed] [Google Scholar]
- 94.Turley SJ, Fletcher AL, Elpek KG. The stromal and haematopoietic antigen-presenting cells that reside in secondary lymphoid organs. Nat Rev Immunol. 2010;10:813–825. doi: 10.1038/nri2886. [DOI] [PubMed] [Google Scholar]
- 95.van de Pavert SA, Mebius RE. New insights into the development of lymphoid tissues. Nat Rev Immunol. 2010;10:664–674. doi: 10.1038/nri2832. [DOI] [PubMed] [Google Scholar]
- 96.Ruddle NH, Akirav EM. Secondary lymphoid organs: responding to genetic and environmental cues in ontogeny and the immune response. J Immunol. 2009;183:2205–2212. doi: 10.4049/jimmunol.0804324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De Togni P, et al. Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science. 1994;264:703–707. [PubMed] [Google Scholar]
- 98.Banks TA, et al. Lymphotoxin-alpha-deficient mice. Effects on secondary lymphoid organ development and humoral immune responsiveness. J Immunol. 1995;155:1685–1693. [PubMed] [Google Scholar]
- 99.Alimzhanov MB, et al. Abnormal development of secondary lymphoid tissues in lymphotoxin beta-deficient mice. Proc Natl Acad Sci USA. 1997;94:9302–9307. doi: 10.1073/pnas.94.17.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koni PA, Sacca R, Lawton P, Browning JL, Ruddle NH, Flavell RA. Distinct roles in lymphoid organogenesis for lymphotoxins a and b revealed in lymphotoxin b-deficient mice. Immunity. 1997;6:491–500. doi: 10.1016/s1074-7613(00)80292-7. [DOI] [PubMed] [Google Scholar]
- 101.Futterer A, Mink K, Luz A, Kosco-Vilbois MH, Pfeffer K. The lymphotoxin beta receptor controls organogenesis and affinity maturation in peripheral lymphoid tissues. Immunity. 1998;9:59–70. doi: 10.1016/s1074-7613(00)80588-9. [DOI] [PubMed] [Google Scholar]
- 102.Kuprash DV, Alimzhanov MB, Tumanov AV, Anderson AO, Pfeffer K, Nedospasov SA. TNF and lymphotoxin beta cooperate in the maintenance of secondary lymphoid tissue microarchitecture but not in the development of lymph nodes. J Immunol. 1999;163:6575–6580. [PubMed] [Google Scholar]
- 103.Weih F, Caamaño J. Regulation of secondary lymphoid organ development by the nuclear factor-kappaB signal transduction pathway. Immunol Rev. 2003;195:91–105. doi: 10.1034/j.1600-065x.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- 104.Miyawaki S, et al. A new mutation, aly, that induces a generalized lack of lymph nodes accompanied by immunodeficiency in mice. Eur J Immunol. 1994;24:429–434. doi: 10.1002/eji.1830240224. [DOI] [PubMed] [Google Scholar]
- 105.Koike R, et al. The splenic marginal zone is absent in alymphoplastic aly mutant mice. Eur J Immunol. 1996;26:669–675. doi: 10.1002/eji.1830260324. [DOI] [PubMed] [Google Scholar]
- 106.Bonizzi G, et al. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. EMBO J. 2004;23:4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matsushima A, et al. Essential role of nuclear factor (NF)-kappaB-inducing kinase and inhibitor of kappaB (IkappaB) kinase alpha in NF-kappaB activation through lymphotoxin beta receptor, but not through tumor necrosis factor receptor I. J Exp Med. 2001;193:631–636. doi: 10.1084/jem.193.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Paxian S, et al. Abnormal organogenesis of Peyer's patches in mice deficient for NF-kappaB1, NF-kappaB2, and Bcl-3. Gastroenterology. 2002;122:1853–1868. doi: 10.1053/gast.2002.33651. [DOI] [PubMed] [Google Scholar]
- 109.Carragher D, et al. A stroma-derived defect in NF-kappaB2−/ − mice causes impaired lymph node development and lymphocyte recruitment. J Immunol. 2004;173:2271–2279. doi: 10.4049/jimmunol.173.4.2271. [DOI] [PubMed] [Google Scholar]
- 110.Lo JC, et al. Coordination between NF-kappaB family members p50 and p52 is essential for mediating LTbetaR signals in the development and organization of secondary lymphoid tissues. Blood. 2006;107:1048–1055. doi: 10.1182/blood-2005-06-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tucker E, et al. A novel mutation in the Nfkb2 gene generates an NF-kappa B2 "super repressor". J Immunol. 2007;179:7514–7522. doi: 10.4049/jimmunol.179.11.7514. [DOI] [PubMed] [Google Scholar]
- 112.Alcamo E, Hacohen N, Schulte LC, Rennert PD, Hynes RO, Baltimore D. Requirement for the NF-kappaB family member RelA in the development of secondary lymphoid organs. J Exp Med. 2002;195:233–244. doi: 10.1084/jem.20011885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mebius RE, Kraal G. Structure and function of the spleen. Nat Rev Immunol. 2005;5:606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- 114.Matsumoto M, et al. Essential role of NF-kappa B-inducing kinase in T cell activation through the TCR/CD3 pathway. J Immunol. 2002;169:1151–1158. doi: 10.4049/jimmunol.169.3.1151. [DOI] [PubMed] [Google Scholar]
- 115.Ishimaru N, Kishimoto H, Hayashi Y, Sprent J. Regulation of naive T cell function by the NF-kappaB2 pathway. Nat Immunol. 2006;7:763–772. doi: 10.1038/ni1351. [DOI] [PubMed] [Google Scholar]
- 116.Jin W, Zhou XF, Yu J, Cheng X, Sun SC. Regulation of Th17 cell differentiation and EAE induction by the MAP3K NIK. Blood. 2009;113:6603–6610. doi: 10.1182/blood-2008-12-192914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 118.Hu H, Wu X, Jin W, Chang M, Cheng X, Sun SC. Noncanonical NF-κB regulates inducible costimulator (ICOS) ligand expression and T follicular helper cell development. Proc Natl Acad Sci USA. 2011;108:12827–12832. doi: 10.1073/pnas.1105774108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Simpson TR, Quezada SA, Allison JP. Regulation of CD4 T cell activation and effector function by inducible costimulator (ICOS) Curr Opin Immunol. 2010;22:326–332. doi: 10.1016/j.coi.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 120.King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu Rev Immunol. 2008;26:741–766. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 121.Linterman MA, Vinuesa CG. Signals that influence T follicular helper cell differentiation and function. Semin Immunopathol. 2010;32:183–196. doi: 10.1007/s00281-009-0194-z. [DOI] [PubMed] [Google Scholar]
- 122.Walters S, et al. Increased CD4+Foxp3+ T cells in BAFF-transgenic mice suppress T cell effector responses. J Immunol. 2009;182:793–801. doi: 10.4049/jimmunol.182.2.793. [DOI] [PubMed] [Google Scholar]
- 123.Tamura C, et al. Impaired function of dendritic cells in alymphoplasia (aly/aly) mice for expansion of CD25+CD4+ regulatory T cells. Autoimmunity. 2006;39:445–453. doi: 10.1080/08916930600833390. [DOI] [PubMed] [Google Scholar]
- 124.Tas SW, et al. Noncanonical NF-kappaB signaling in dendritic cells is required for indoleamine 2,3-dioxygenase (IDO) induction and immune regulation. Blood. 2007;110:1540–1549. doi: 10.1182/blood-2006-11-056010. [DOI] [PubMed] [Google Scholar]
- 125.Hofmann J, Mair F, Greter M, Schmidt-Supprian M, Becher B. NIK signaling in dendritic cells but not in T cells is required for the development of effector T cells and cell-mediated immune responses. J Exp Med. 2011;208:1917–1929. doi: 10.1084/jem.20110128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Powolny-Budnicka I, Riemann M, Tänzer S, Schmid RM, Hehlgans T, Weih F. RelA and RelB transcription factors in distinct thymocyte populations control lymphotoxin-dependent interleukin-17 production in γδ T cells. Immunity. 2011;34:364–374. doi: 10.1016/j.immuni.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 127.Britanova LV, Makeev VJ, Kuprash DV. In vitro selection of optimal RelB/p52 DNA-binding motifs. Biochem Biophys Res Commun. 2008;365:583–588. doi: 10.1016/j.bbrc.2007.10.200. [DOI] [PubMed] [Google Scholar]