Abstract
Chronic overnutrition and physical inactivity are major risk factors for insulin resistance and type 2 diabetes. Recent research indicates that overnutrition generates an increase in hydrogen peroxide (H2O2) emission from mitochondria, serving as a release valve to relieve the reducing pressure created by fuel overload, as well as a primary signal to ultimately decrease insulin sensitivity. H2O2 is a major input to cellular redox circuits that link to cysteine residues throughout the entire proteome to regulate cell function. Here we review the principles of mitochondrial bioenergetics and redox systems biology and offer new insight as to how H2O2 emission may be linked via redox biology to the etiology of insulin resistance.
Nutrient overload and mitochondrial function/dysfunction
Decreased insulin sensitivity in skeletal muscle is a primary factor in the etiology of type 2 diabetes and, as such, identifying the underlying cause is critical to devising appropriate prevention and treatment strategies. Two of the leading theories that have emerged that explain insulin resistance, center on mitochondrial function/dysfunction, although interestingly with opposite views. In one theory, inherited or acquired mitochondrial dysfunction is thought to cause accumulation of intramyocellular lipids that leads to insulin resistance, and implies that strategies to accelerate flux through β-oxidation should improve insulin sensitivity [1]. In the second theory, the impact of cellular metabolic imbalance is viewed in the context of cellular and mitochondrial bioenergetics, positing that excess fuel relative to demand increases mitochondrial oxidant production and emission, ultimately leading to the development of insulin resistance. In this case, elevated flux through β-oxidation in the absence of added demand is viewed as an underlying cause of the disease [2]. The present paper focuses on the second theory, reviews the underlying principles and supporting data and provides a perspective on the role redox biology is likely to play in deciphering the link between nutrient overload and insulin resistance.
A primer on mitochondrial bioenergetics
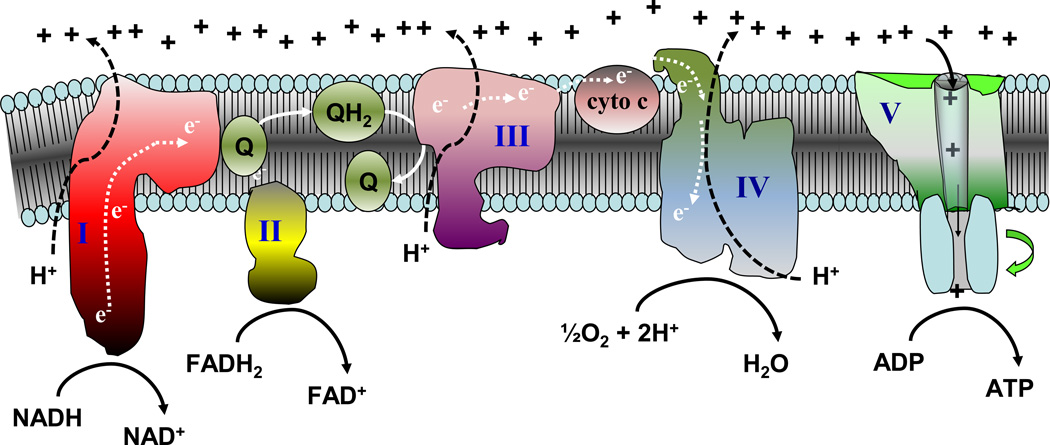
In 1961, Peter Mitchell published a unique hypothesis regarding cellular bioenergetic conservation [3], for which he was awarded the Nobel Prize in chemistry in 1978. Termed the “chemiosmotic theory of oxidative phosphorylation” (also known as chemiosmosis, see glossary), at its core is the notion of coupling hydrogen and electron transfer through an energy-conserving membrane to the phosphorylation of ADP to form ATP. As depicted in Figure 1, the mitochondrial electron transport system consists of several multi-polypeptide protein complexes (I–V) embedded in the inner mitochondrial membrane that receive electrons from reducing equivalents (i.e., NADH, FADH2) generated by dehydrogenases (e.g., pyruvate dehydrogenase, α-ketoglutarate dehydrogenase, acyl-CoA dehydrogenase, etc). These electrons are transferred through a series of electron carriers in the respiratory chain with O2 serving as the final electron acceptor, ultimately reducing ½O2 to H2O. Each of the electron carriers represents a redox couple (i.e., species capable of existing in a reduced or oxidized state) with a characteristic reduction potential – a measure of the tendency of the oxidized species to accept an electron(s)(see glossary). A negative reduction potential indicates the reduced species has a high tendency to donate (lose) an electron(s) (e.g., NADH) and a positive reduction potential indicates the oxidized species has a high tendency to accept (gain) an electron(s) (e.g., O2). The electron carriers in the respiratory chain are ordered in such a way that the reduction potentials progressively increase (i.e., become more positive) from one redox couple to another. In three of these complexes (I, III and IV), the difference in reduction potential (i.e., release of energy) across successive redox couples is sufficient to drive the translocation of protons from the matrix to the inner membrane space. This creates a proton gradient across the inner membrane that is derived from both the concentration (ΔpH) and the electrical potential (ΔũH+) difference across the membrane. By bioenergetic convention, ΔũH+ is converted to units of electrical potential (i.e., mV), and commonly referred to as the membrane potential (ΔΨ). Although ΔpH and ΔΨ together comprise the total proton motive force (Δp), ΔΨ is by far the dominant component and often used synonymously with Δp. The essence of the chemiosmotic theory is that the electrical-chemical potential energy created by the generation of Δp is sufficient to drive the synthesis of ATP as protons flow back through the ATP synthase complex into the matrix.
Figure 1. Schematic depiction of the mitochondrial electron transport system.
Reducing equivalents (NADH, FADH2) provide electrons that flow through complex I, the ubiquinone cycle (Q/QH2), complex III, cytochrome c, complex IV, and to the final acceptor O2 to form water. Electron flow through complexes I, III, and IV results in pumping of protons to the outer surface of the inner membrane, establishing a membrane potential that is used by the ATP synthase to drive the rephosphorylation of ADP. Animated versions depicting the bioenergetics governing the system are provided in the online version of the figure.
Several features of the respiratory system, some counterintuitive, are essential to understanding how cellular energy balance is normally governed and therefore how cellular energy surplus may affect the system. First, similar to an electrical circuit, the transport of electrons through the respiratory chain is an inherent property of the system – it occurs automatically. Second, for the most part, electron flow and proton pumping are tightly coupled; that is, one does not occur without the other. The third feature requires visualizing the boundaries that govern the system in real time and is depicted in the animated version of Figure 1 (see link to online video in legend to Figure 1). Consider starting with a de-energized system. To activate respiration, fuel (i.e., NADH, FADH2) is added which automatically initiates electron flow, proton pumping, and O2 consumption. However, as ΔΨ begins to build on the outer surface of the inner membrane, a “back pressure” is created which begins to oppose the pumping of protons, thereby gradually slowing electron flow and O2 consumption. Eventually the point is reached at which the force driving the pumping of protons out of the matrix (i.e., the reduction potential (see glossary) span from the NAD+/NADH couple to the O2/2H2O couple) is completely counter-balanced by the high ΔΨ. This creates “static head” equilibrium. Fortunately, mitochondria never reach this state (i.e., we never stop consuming O2) because the mitochondrial inner membrane is inherently leaky to protons. This basal rate of proton conductance ensures that ΔΨ is at some value less than maximum, thereby “allowing” electron flow, proton pumping, and O2 consumption to operate at an “idling” rate (known as state 2 respiration). If another avenue for protons to flow back into the matrix is created (e.g., via ATP synthase catalyzing re-phosphorylation of ADP), then the back pressure (ΔΨ) is further reduced and electron flow, proton pumping, and oxygen consumption increase accordingly (known as state 3 respiration). Once all of the ATP is resynthesized, the system slows back to the idling rate (known as state 4 respiration, see glossary) The critical point is that the mitochondrial respiratory system is a “primed engine”, automatically adjusting to each change in the rate of proton re-entry into the matrix with a corresponding change in electron flow and O2 consumption. The fourth and final feature should be apparent from the first three; the rate at which the respiratory system operates is governed by energy demand (“pull”), not energy supply (“push”). Proton conductance (via leak and ATP synthase) dictates the rate of electron flow and therefore the demand for reducing equivalents (e.g., NADH and FADH2) which, in turn, regulates the rate of substrate uptake and flux through catabolic pathways.
How then may an excess of metabolic fuel affect the system? To address this question requires consideration of redox state, which is defined as the product of the reduction potential and the reducing capacity (i.e., the concentration of the reduced species) of a redox couple (see glossary). The higher the concentration of the reduced species, the more reduced or “charged” the redox state. Thus, a provision of excess reducing equivalents due to over nutrition will increase the “pressure head” at the entry point of electrons into complex I (↑ NADH/NAD+) and/or the Q pool (↑ FADH2/FAD+), in either case elevating the redox state and membrane potential of the respiratory system. Several of the redox couples within the electron transport chain transfer single rather than two electrons and are therefore susceptible to leaking electrons directly to surrounding O2 to form the free-radical superoxide (O2•−). Under state 3 conditions, the redox state of the system is below the threshold at which electrons will leak to O2. However, at or near state 4 respiration, the respiratory system is in its most reduced state such that the rate at which O2•− is produced is extremely sensitive to the redox state of the system, increasing exponentially with even small increases in membrane potential [4–6]. Fortunately, O2•− is rapidly converted by manganese superoxide dismutase (MnSOD) to the two-electron non-radical hydrogen peroxide (H2O2). H2O2 in turn can be further reduced to H2O in the mitochondrial matrix by glutathione (GSH) or the thioredoxin/peroxiredoxin systems, or can freely diffuses out of the mitochondria where it again is buffered by GSH. Thus in a resting cell, the mitochondrial respiratory system functions as a redox pressure gauge that senses and reflects cellular metabolic balance. When in positive balance, electron leak serves as a release valve, accelerating mitochondrial O2•− production and H2O2 emission. The question becomes: Is mitochondrial O2•− and/or H2O2 casually linked to the etiology of high fat diet-induced insulin resistance?
Elevated mitochondrial H2O2 emission is a primary cause of insulin resistance
The establishment of a “causal” mechanism within any process requires adherence to the following general criteria: 1) the causal stimulus must precede the effect, and 2) removal/inhibition of the causal stimulus must attenuate/prevent the effect. Mitochondrial-derived oxidative stress is fairly well established as an underlying mechanism responsible for the pathological complications associated with diabetes [7]. However, its role as a primary factor in the development of insulin resistance (and subsequent overt diabetes), although long suspected, has been based largely on indirect evidence [8, 9]. The first direct evidence indicating a causal role was reported by Houstis et al. [10] who utilized two distinct means of inducing insulin resistance in 3T3-L1 adipocytes; TNFα and dexamethasone treatment. In both models, induction of insulin resistance resulted in elevated reactive oxygen species (ROS) production prior to any detectable decline in insulin sensitivity, fulfilling the first criteria. Further, using several different mitochondrial- and non-mitochondrial- targeted approaches, scavenging either O2•− or H2O2 was found to partially restore insulin sensitivity in all cases, fulfilling the second criteria [10]. Following this initial publication, strong experimental evidence from various animal models utilizing mitochondrial-targeted approaches has since established a link between mitochondrial-derived ROS and insulin resistance in vivo [11–14].
Which ROS species mediates the development of insulin resistance? O2•− is the parent molecule of all ROS and thus O2•− or any downstream free radical or non-radical oxidant product could serve as the initiating signal. Although there are several oxidases representing potential non-mitochondrial sources of O2•−, mitochondria are regarded as the primary source under conditions leading to insulin resistance [15]. Mitochondrial derived O2•− has been suggested to be the primary signal based on the ability of a variety of MnSOD mimetics to limit the development of insulin resistance in cultured myocytes and adipocytes and to improve glucose tolerance when administered to ob/ob or high fat fed mice [10, 13]. However, the SOD activity of commercial manganese(III) tetrakis (4-benzoic acid) porphyrin (MnTBAP), the principle MnSOD mimetic used in these studies, has recently been shown to be due to Mn-containing impurities; pure MnTBAP does not dismutate O2•− but more likely functions as a broad antioxidant scavenging peroxynitrites and/or carbonate radicals [16]. MnSOD has also been the target of genetic manipulation in mice under the assumption that overexpression will decrease, and partial knockout will increase, mitochondrial O2•− production. However, the high abundance (~10 µM) and kinetic efficiency of endogenous MnSOD suggests that O2•− is extremely short lived under most circumstances [17, 18]. Moreover, O2•− can also react with iron containing proteins to produce one to two molecules of H2O2 and/or with other molecules (e.g., nitric oxide, ubiquinone, cytochrome c3+) without producing H2O2 [17, 19], making it difficult to predict or interpret outcomes from the MnSOD models. More to the point however is that the rate constant at which O2•− oxidizes thiols is quite slow, on the order of 103M−1s−1 [20], which is insignificant in comparison to the rate constant at which O2•− is reduced to H2O2 by MnSOD (>109M−1s−1) [21]. Thus, in terms of a signaling role, O2•− is more likely to function simply as a precursor of H2O2 rather than as a primary second messenger [22].
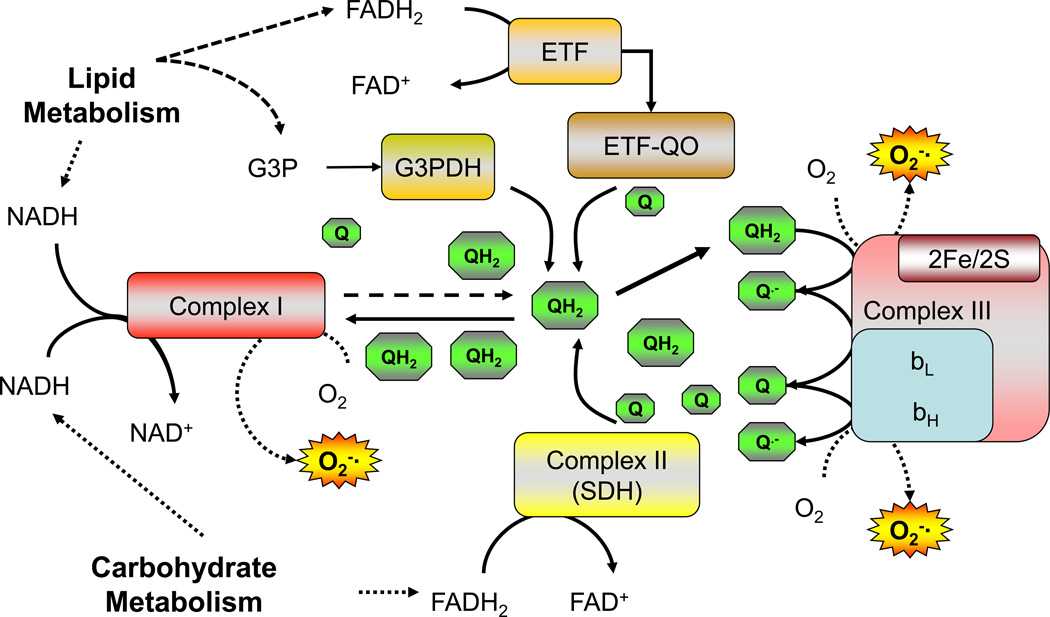
In contrast to O2•−, H2O2 is widely considered to fulfill the spacial, thermodynamic, and kinetic requirements of a second messenger [22, 23]. Mitochondria are regarded as the principle source of H2O2 under conditions of metabolic imbalance [24] and its emission rate from the mitochondria establishes the intracellular redox environment in favor of greater reducing (↓ H2O2) or oxidizing (↑ H2O2) conditions [25]. As stated earlier, when the rate of reducing equivalent supply outpaces demand, the increase in reduction potential within the complexes increases the probability of electrons leaking to form O2•−. Interestingly, not only the supply but also the relative proportion of electrons feeding into the system at complex I verses the Q pool may influence the reducing potential of the system. Oxidation of palmitate, which is known to generate high rates of mitochondrial H2O2 emission [26–28], yields ~2.5 times more electrons feeding directly into the Q pool (via β-oxidation and the electron-transferring flavoprotein dehydrogenase) which likely increases the ratio of reduced to oxidized Q (QH2/Q, Figure 2). In addition, unlike complex I, entry of electrons into the Q pool is not constrained by the membrane potential (i.e., back pressure) which likely favors oxidation of FADH2 and further reduction of the Q pool. A high QH2/Q ratio could increase O2•− production three ways; by limiting the availability of oxidized Q as an electron acceptor for complex I, by generating reverse electron transport back into complex I, and/or by increasing the pressure head at complex III (Figure 2). Any of all of these mechanisms could account for the elevated mitochondrial H2O2 emission associated with fat verses carbohydrate-based substrates and, in the broader context, have obvious implications regarding the potential impact of metabolic imbalance from high fat diets.
Figure 2. Schematic showing electron flow into the ubiquinone (Q) pool.
High rates of electron flux into the Q pool from lipid metabolism are proposed to increase O2•− generation at complex I due to decreased availability of oxidized Q and/or reverse electron flux from reduced Q (QH2) back into complex I. ETF, electron737 transferring flavoprotein; ETF-QO, electron-transferring flavoprotein dehydrogenase; G3PDH, glycerol-3-phosphate dehydrogenase.
To determine whether mitochondrial-derived H2O2 emission contributes to the etiology of high fat diet-induced insulin resistance, Anderson et al. [11] employed the use of a recently developed small antioxidant peptide called SS31 [29]. SS31 contains an electron-scavenging dimethyltyrosine as one of its four amino acids, is membrane permeable, and appears to localize to the mitochondrial inner membrane where it reduces H2O2 emission by 50–60% [11, 30]. Administration of SS31 to rats consuming a high fat diet for 6 wks normalized the mitochondrial H2O2 emitting potential, preserved the intracellular redox environment in muscle, and prevented the development of insulin resistance [11]. The exact mechanism of action of SS31 however is unclear, as it may scavenge electrons directly prior to O2•− formation or dismutate O2•− and/or H2O2 once formed. Thus, to determine whether H2O2 emission is the primary signaling mediator, transgenic mice expressing the human catalase gene targeted specifically to the mitochondrial matrix (MCAT) [31] were subjected to hyperinsulinemic-euglycemic clamps. Catalase converts H2O2 to H2O but is normally not expressed in mitochondria [32, 33]. In response to a high-fat diet, MCAT mice maintained whole-body and muscle-specific insulin sensitivity similar to wild-type mice on a chow diet [11]. Thus, these findings [11], coupled with other recent studies utilizing mitochondrial-targeted antioxidant approaches [12, 14], have established mitochondrial-derived H2O2 as a primary second messenger linking metabolic imbalance to the etiology of insulin resistance.
An introduction to redox systems biology
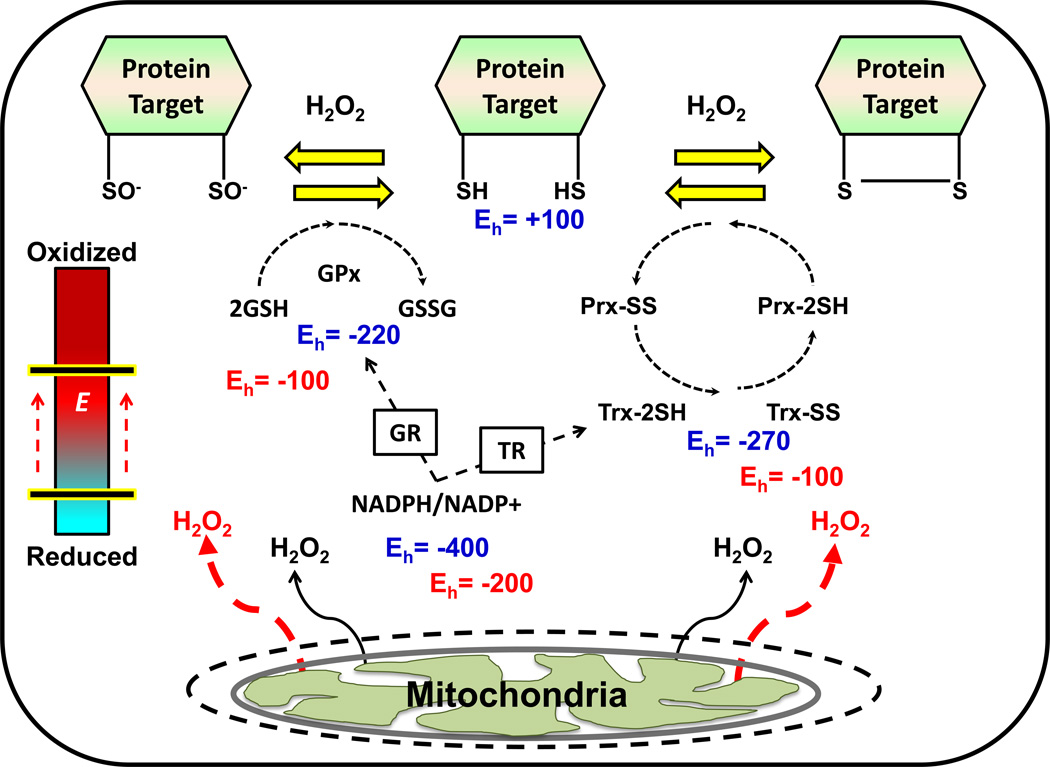
How then may elevations in mitochondrial H2O2 emission lead to disruptions in insulin signaling? To address this question requires consideration of several key aspects of redox systems biology, and more specifically, redox signaling. As stated above, H2O2 is widely accepted as a critical signaling molecule in biological systems [22, 23]. In fact, the continuous generation of H2O2, at concentrations far below those indicative of “oxidative stress”, is believed to be involved in the regulation of various redox signaling pathways, referred to as redox circuits (Figure 3, glossary) [23]. The glutathione (2GSH/GSSG) and thioredoxin (TrxRed/TrxOx) redox couples, both of which are expressed at high levels and derive their reducing power from NADPH/NADP+, serve as the common control nodes of the circuit (i.e., the wiring) and the direct link to cysteine (Cys) residues within specific redox-sensitive proteins (see glossary)1. Electron flux through these couples is thought to be rapid and continuous reflecting the rates of reducing (NADPH/NADP+, the “charge” within the circuit) and oxidizing (H2O2 production, the “dissipation of charge”) inputs. Under normal circumstances, the balance strongly favors reduction of protein Cys and thus an overall reduced cellular redox environment (see glossary). Of the ~214,000 Cys residues within the proteome, it is estimated >20,000 are redox sensitive (i.e., capable of oscillating between a reduced and oxidized state as a redox couple) [23]. The activity/function of these redox sensitive proteins is dependent upon the redox state of the sulfur atoms within these Cys moieties, thus giving rise to the term “sulfur switches” [23]. The sulfur atom within Cys can exist as a reduced thiol (−SH) or in a variety of oxidation states, including as a thiolate anion (−S−), sulfenate (−SO−), disulfide (−S-S) sulfinate (−SO2−), or sulfonate (−SO3−) [23]. Each of these redox-sensitive switches has a characteristic reduction potential, or sensitivity to the redox circuit, that reflects in large part the degree to which each redox sensitive Cys within the protein is spatially and kinetically insulated from the circuit. Alterations in the redox state of these “sulfur switches” induces a variety of effects, including changes in protein conformation, enzyme activity, transporter activity, ligand-receptor binding, protein-protein interaction, protein trafficking, protein degradation, and transcriptional binding to DNA [23]. While the sheer number of potential redox-regulated changes in protein function appears overwhelming, certain major themes in redox signaling networks have begun to emerge.
Figure 3. Simplified model of redox circuitry.
Theoretical Eh values are shown in blue for the 2GSH/GSSG, Trx2SH/TrxSS, and NADPH/NADP+ redox couples, as well as their protein target. Electrons flow through the circuit from the NADPH/NADP+ couple to thiol (SH) moieties within exposed Cys. The overall driving force for reduction is determined by the mV differences in Eh between NADPH/NADP+ and the redox sensitive Cys- containing proteins (e.g., ΔEh = −500 mV). The red text indicates the effects of a transient increase in mitochondrial H2O2 emission in the context of a redox mediated signaling event. Under these conditions, an oxidative shift in the redox environment (depicted as a positive shift in each couple’s Eh), would reduce the overall driving force for the reduction of exposed Cys residues within the target protein (ΔEh = −300 mV), thereby altering the redox state of the protein in favor of its oxidized form (e.g., either 2SO− or SS) and leading to a cellular response. These responses are thought to function as a means of rheostat regulation rather than as a simple “on/off” mechanism. The blue/red bar on the left labeled “E” represents the intracellular redox environment. GR, glutathione reductase; TR, thioredoxin reductase; Gpx, glutathione peroxidase; Prx, peroxiredoxin.
Redox regulation of phosphorylation/dephosphorylation
Mounting evidence indicates that the intracellular redox circuit is a master regulator of phosphorylation/dephosphorylation events due to the presence of redox-sensitive Cys residues within nearly all classes of protein phosphatase enzymes [34]. In general, phosphatase activity is depressed in response to an oxidative shift in the redox environment, thus leading to a concomitant increase in kinase activity either via direct oxidant-induced activation or secondary to phosphatase inactivation [34]. Deactivation of protein tyrosine phosphatases (PTPs) are mediated via the oxidation of a conserved redox sensitive Cys residue within their catalytic site, which must be in the reduced state (−SH) in order for the formation of a cysteinyl-phosphate intermediate during hydrolysis [35]. Both the magnesium-independent (PPP) and magnesium-dependent (PPM) phosphoprotein phosphatase family of Ser/Thr phosphatases do not utilize this Cys-dependent mechanism of hydrolysis; however both appear to be susceptible to oxidative deactivation [36, 37]. With respect to the PPP family, protein phosphatase 2A (PP2A), which together with protein phosphatase 1 (PP1) account for the large majority of all Ser/Thr phosphatase activity [38], have recently been shown to be susceptible to oxidative inactivation due to the presence of a conserved –CXXC- motif within their catalytic domains [36, 39].
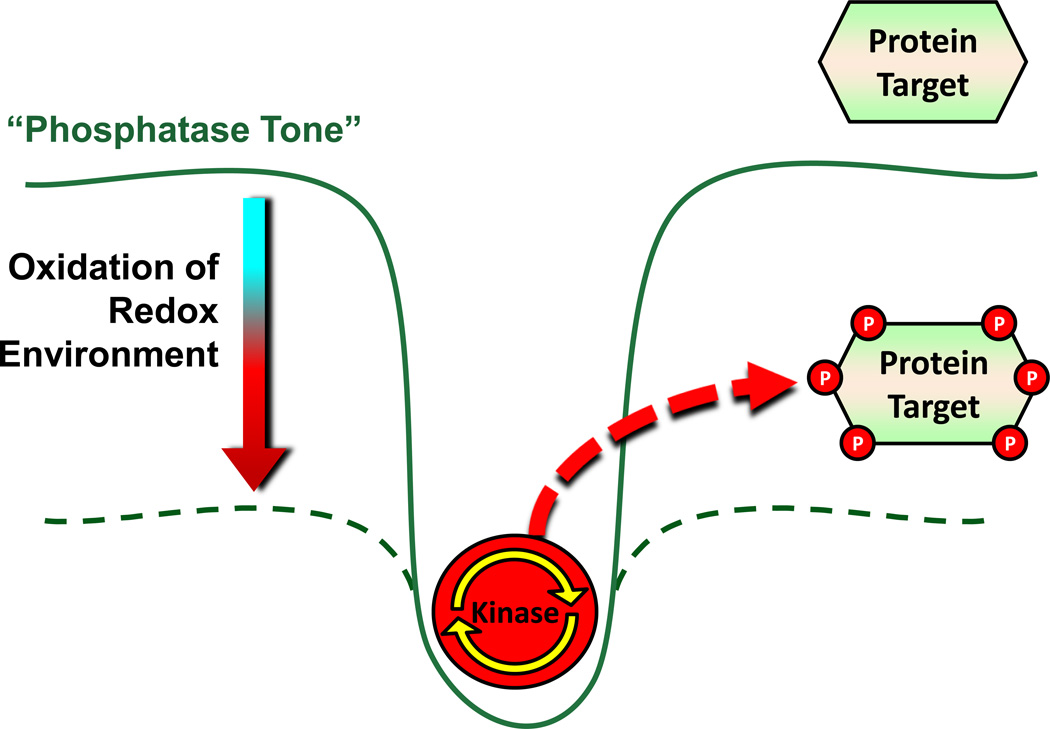
Under resting cellular conditions, the intracellular redox environment is in a relatively reduced state and, in general, phosphatase activity exceeds kinase activity by ~10-fold [40]. This creates what is referred to as an intracellular “phosphatase tone” [37]. This basal phosphatase tone places kinases at a disadvantage, setting a kinetic barrier that prevents random, ectopic phosphorylation events in vivo [37]. In this context, the intracellular redox environment can be viewed as a “potentiometer” in the “on” position with increasing H2O2 production serving as the ”dimming signal” to reduce the normally dominant phosphatase activity, via oxidation, and allow kinase-mediated signaling to occur (Figure 4). These effects are likely compartmentalized and transient, due to the extensive capabilities of the antioxidant redox couples operating in vivo, which rapidly restore the normal steady-state redox environment and overall phosphatase tone following disruption. Such a redox mechanism allows for the propagation of kinase signaling pathways within a tightly regulated range and, more importantly, ensures kinase signaling does not proceed uncontrollably. The implication is that regulation of the local and/or global intracellular redox environment may be a universal mechanism by which the activity, sensitivity, and/or responsiveness of specific kinase signaling pathways are regulated. Moreover, because Ser/Thr phosphorylation sites represent ~98% of the phosphoproteome of mammalian systems [38], recent evidence illustrating widespread susceptibility of Ser/Thr phosphatase enzymes to oxidative inhibition provides an interesting potential mechanism whereby alterations in the redox environment under certain pathological conditions could elicit far reaching affects within multiple cellular pathways. This concept is an emerging area of research interest within a variety of fields. For example, the oxidative inactivation of critical phosphatase enzymes, such as Map kinase phosphatase (MKP-1), phosphatase and tensin homologue (PTEN), mitochondrial matrix targeted PP2C (PP2Cm), and calcineurin have been linked to cellular senescence/aging (MKP-1 [41]), cancer (PTEN [42]), apoptosis (PP2Cm [43]), and inflammation (calcineurin [44]).
Figure 4. Role of the redox environment in the establishment of “phosphatase tone”.
Under the normal reducing conditions of the intracellular redox environment, phosphatase tone is elevated, ensuring that net kinase activity is suppressed and specific protein targets are dephosphorylated. In response to an oxidative shift in the redox environment, phosphatase tone is lowered to a level which allows for kinase activity to dominate and thus leads to phosphorylation of target proteins. Figure adapted from Wright et al. [37].
Alterations in the intracellular redox environment during normal insulin signaling
As eluded to earlier, there is compelling evidence linking mitochondrial H2O2 emission to high fat diet-induced insulin resistance, consistent with the idea that an oxidative shift in intracellular redox environment disrupts the insulin signaling pathway. However, there is also compelling evidence that insulin stimulates H2O2 production which is required to activate insulin signaling [45–47]. These seemingly contradictory findings are an obvious source of confusion. However, careful consideration of the likely temporal and compartmental factors regulating redox changes and insulin signaling reveals a potential mechanism that is entirely consistent with the principles governing phosphorylation/dephosphorylation control in relation to redox environment and thus can account for both findings. The role of redox environment during normal insulin signaling will be considered first.
Insulin signaling begins with insulin binding to the insulin receptor, activating the tyrosine kinase activity intrinsic to the β-subunit of the receptor. Subsequent tyrosine phosphorylation of the insulin receptor substrate (IRS) docking proteins leads to the recruitment and activation of phosphoinositide 3-kinase (PI3K). The major substrate for PI3K is the membrane lipid phosphatidylinositol-4,5-bisphosphate [PI(4,5,)P2] which is phosphorylated to produce phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5,)P3]. The rise in PI (3,4,5,)P3 provides a lipid-based platform that attracts downstream pleckstrin homology domain-containing signaling proteins, including the Ser/Thr kinases phosphoinositide-dependent protein kinase-1 (PDK1) and Akt. PDK1 phosphorylates Akt as well as atypical protein kinase C isoforms λ and ζ (aPKC-λ/ζ). Both Akt and aPKC-λ/ζ have been linked to insulin-stimulated GLUT4 translocation and glucose uptake (Figure 5)[48].
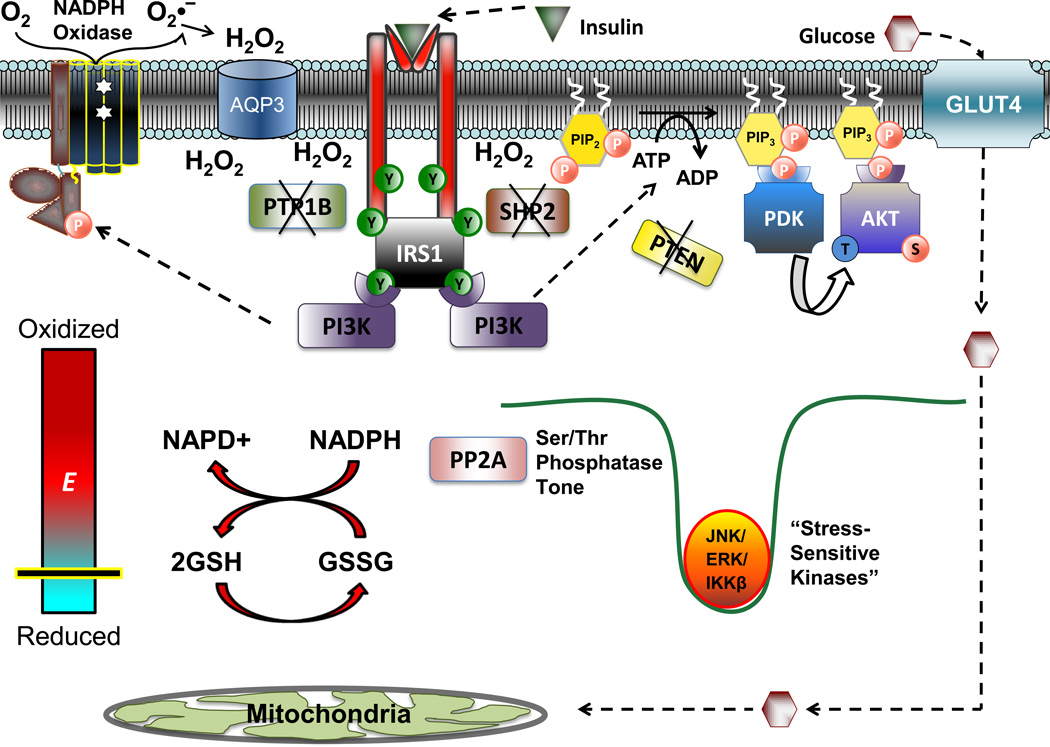
Figure 5. Proposed alterations in cell redox as a necessary component of insulin signaling.
In the absence of insulin, the principle nodes of regulation within the insulin signaling cascade are kept dephosphorylated via the membrane associated proteins PTP1B, SHP2, and PTEN. Following insulin binding and Tyr-phosphorylation of the insulin receptor and IRS1, the activation of membrane bound NADPH oxidase (potentially mediated by PI3K) results in the accumulation of H2O2 at the level of the plasma membrane to transiently inactivate PTP1B, SHP2 and PTEN, thus allowing propagation of kinase-mediated signaling, leading to GLUT4 translocation and glucose uptake. While the local redox environment at the plasma membrane, and redox sensitive proteins in direct proximity to the H2O2 source undergo an oxidative shift, the global redox state of the cell is maintained by the redox buffering systems(depicted by the thick red arrows illustrating the reaction between the 2GSH/GSSG and NADPH/NADP+couples). Maintenance of global redox ensures that Ser/Thr phosphatase activity, specifically PP2A (and potentially MKP and PP1, not shown), are maintained. The continued activation of these enzymes ensures that certain Ser/Thr kinases (JNK, ERK, IKKβ) remain inactive and that phosphomoieties are not allowed to accumulate within insulin signaling proteins.
To propagate the tyrosine kinase signal, as well as signaling through downstream protein and phospholipid kinases, insulin binding also initiates the deactivation of phosphatases that negatively regulate insulin signaling, including PTP1B, PTEN, and potentially Src homology 2 (SH2) domain-containing phosphatase (SHP2) [45, 49, 50]. Inactivation of these phosphatases is triggered by an oxidative shift in the redox environment localized at the level of the plasma membrane, effectively creating a redox environment conducive to kinase signaling (Figure 5)[45, 51]. Surprisingly, the source of the insulin-induced oxidative shift in muscle cells appears to be H2O2 generated extracellularly by the plasma membrane-bound multiprotein NADPH oxidase (NOX) complex [52, 53].
The NOX family of NADPH oxidase enzymes contains transmembrane electron carriers which utilize NADPH as an electron donor and molecular oxygen as an acceptor. NADPH oxidases localize to specific cellular membrane compartments via targeting proteins, facilitating H2O2 production within spatially confined areas to promote signaling via oxidation of redox-sensitive targets in proximity [54, 55]. In the well characterized phagocytic NADPH oxidase, the cytoplasmic/regulatory subunits (p40phox, p47phox, p67phox) in response to insulin migrate to the plasma membrane to combine with the catalytic subunits (p22phox, gp91phox) and a GTP-binding protein known as rac-1, forming the active enzyme complex [56]. The holoenzyme then catalyzes the electrogenic step-wise transfer of electrons from cytosolic NADPH through the plasma membrane (via the presence of coordinated heme molecules) into the extracellular space where they combine with molecular O2 to yield O2•−[57]. O2•− is then rapidly dismutated to H2O2 through the actions of extracellular Cu/Zn-SOD, thus creating a local concentration gradient across the plasma membrane that promotes simple or facilitated (via aquaporin 3,[58]) diffusion of H2O2 from the extracellular to the intracellular surface.
Although the mechanism by which insulin activates NADPH oxidase in skeletal muscle is not entirely clear, it has recently been shown that the NADPH oxidase regulatory subunit, p47phox, translocates to the plasma membrane in response to phosphorylation by PI3K and/or PKC [53]. Serine phosphorylation of p47phox is a necessary step, as inhibitors of either PI3K and/or PKC blunt p47phox translocation and H2O2 accumulation, as well as depress cellular signaling in response to insulin [53]. Because PI3K and PKC are both downstream of the insulin receptor/IRS1 axis, the initial insulin-mediated activation of NADPH oxidase may occur despite a dominance of PTP1B or SHP2 activity. In either case, insulin-induced H2O2 generation as well as kinase-mediated signal propagation throughout the cascade likely operates in a feed347 forward fashion to accelerate NADPH oxidase activity [53, 59]. In addition, receptor tyrosine kinase activation also leads to the transient phosphorylation and inactivation of membrane-associated peroxiredoxin I, a major H2O2 detoxifying enzyme [60]. Thus, insulin-stimulated tyrosine kinase and NADPH oxidase activation coordinate the generation of an oxidized redox environment localized along the plasma membrane to inhibit phosphatase activity, ensuring that kinase signaling proceeds through the various nodes of the insulin signaling cascade to affect the appropriate physiological responses.
Under normal circumstances, propagation through the insulin signaling cascade, as well as the physiological processes under its control (e.g., glycogen synthesis), will continue as long as the local oxidized redox shift is maintained (i.e., as long as insulin-induced NADPH oxidase activity remains elevated). Once blood glucose is cleared and insulin levels decline, insulin-stimulated NADPH oxidase activity will decline and the local redox environment surrounding the plasma membrane will be restored to its reduced state via the redox buffering systems, restoring the normally dominant phosphatase activity and effectively resetting the insulin signaling pathway back to its basal/dephosphorylated state.
Insulin resistance as a disruption in the insulin signaling cascade
Research conducted over the past several years has established a role for the activation of stress sensitive Ser/Thr kinases, and their subsequent phosphorylation of inhibitory Ser/Thr residues within the insulin receptor and IRS1/2 as a potential mechanism of insulin resistance [61]. There are as many as 70 Ser/Thr residues in IRS proteins subject to phosphorylation and numerous Ser/Thr kinases have been implicated as potential “effectors” of insulin resistance including p70-S6 kinase 1 (S6K1), protein kinase C zeta/theta (PKCζ/θ), glycogen synthase kinase (GSK), C-Jun-N-Terminal kinase-1 (JNK1), extracellular signal regulated kinase-1 (ERK1), and inhibitory-κβ kinase β (IKKβ) [61]. In line with this notion, whole body knockouts for JNK1[62], ERK1[63], S6K1 [64], as well as IKKβ [65] have been shown to confer protection from high fat diet-induced insulin resistance. The majority of studies thus far have focused on the impact of phosphorylation at a single or a few Ser residues in IRS1 on insulin action, with Ser-307 (S-312 human) receiving much of the early attention [66]. However, it is now evident that Ser phosphorylation of a single site or even several sites within IRS1 cannot fully account for the impairment in insulin signaling seen during insulin resistance [67]. Moreover, recent evidence suggests that insulin resistance under some circumstances can occur in the absence of any change in insulin receptor/IRS1 activation (Tyr-phosphorylation) or IRS1 Ser-phosphorylation mediated inhibition, and may or may not be associated with aberrant phosphorylation of downstream targets [68, 69]. These findings are consistent with the growing sentiment that insulin resistance develops as a consequence of any number of combinations of aberant multiple Ser-phosphorylation events within multiple proximal and distal targets to differentially regulate the sensitivity of the insulin signaling pathway [45, 50, 67]. This concept implies that a more “global” mechanism of disruption capable of affecting the overall kinase/phosphatase activity within the insulin signaling cascade may underlie the etiology of the disease.
Insulin resistance induced by shift in global redox environment due to chronic nutrient overload
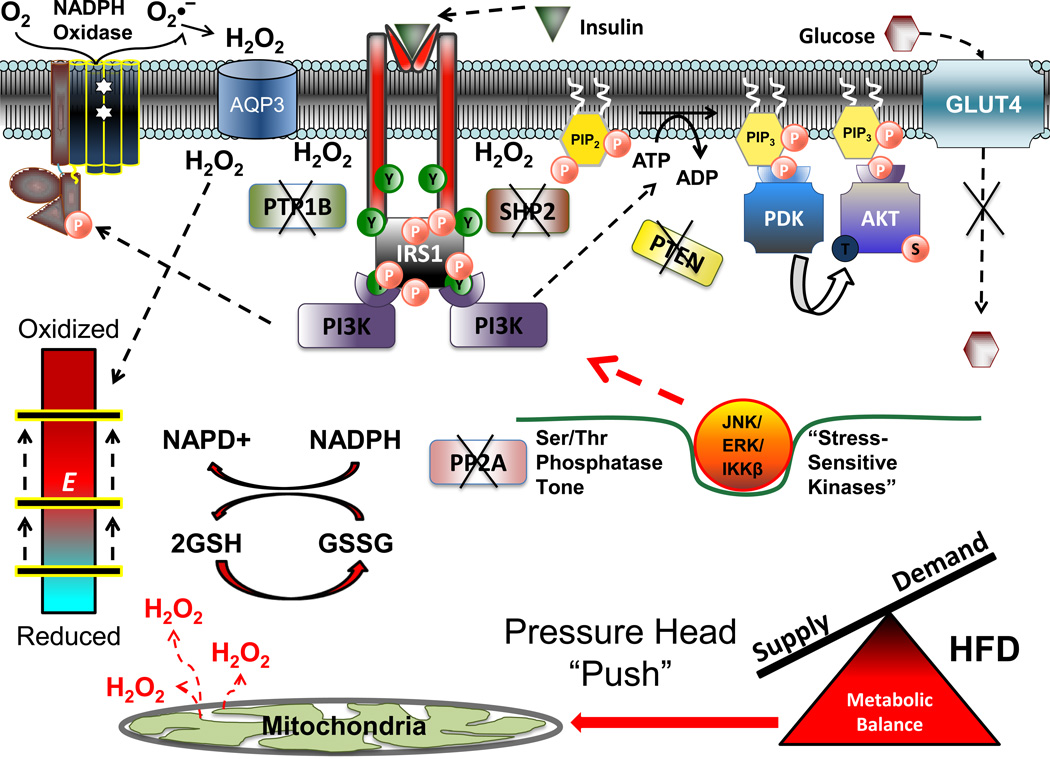
As discussed above, chronic nutrient overload generates an oversupply of reducing equivalents that is predicted to elevate the reducing pressure within the mitochondrial respiratory system. This pressure is at least partially relieved by the leaking of electrons, resulting in an acceleration of H2O2 production and emission as evidenced by reduced GSH:GSSG ratios in the skeletal muscle of both high fat-fed rats and humans [11]. Given the distribution of mitochondria throughout cells, this shift in the GSH buffering system reflects a global oxidative shift in the redox environment within the cell (i.e., a relative dissipation of charge); that is, a global shift in the reduction potential values (more positive) of the primary redox buffering couples and, by inference, in the redox state of regulatory Cys switches throughout the entire redox signaling network. As such, those Cys switches within cytosolic proteins particularly sensitive to the redox environment (i.e., those with more negative redox potentials) will become oxidized. Although redox sensitivity of Ser/Thr phosphatases is established, it has not been well studied particularly in the context of insulin resistance. In skeletal muscle however, acute exposure to different forms of oxidant stress, including H2O2, markedly reduces Ser/Thr phosphatase activity [37]. Moreover, oxidant-mediated inactivation of PP2A has recently been shown to be critical to the persistent hyperactivation of NF-κB following TNFα stimulation [70]. Taken together, these data are consistent with the notion that oxidative shifts in the global redox environment reduce Ser/Thr phosphatase tone, thereby lowering the kinetic barrier for cytosolic Ser/Thr kinases. The global shift in redox environment, in turn, is predicted to induce a global increase in the activity of stress-sensitive Ser/Thr kinases, many of whom are known effectors of insulin resistance (i.e., JNK1/ASK1, IKKβ/NF-κB, ERK, etc., reviewed in [61]), leading to the subsequent Ser/Thr phosphorylation of IRS1 and downstream signaling proteins to limit insulin sensitivity (Figure 6).
Figure 6. Proposed model of high fat diet-induced insulin resistance.
Chronic elevations in mitochondrial H2O2 emission as a result of a high fat diet serve to reduce the reserve capacity within the redox buffering systems and induce an oxidative shift in the cellular redox environment (bar shifted up to beginning of red zone on redox environment gauge). This global shift in cell redox may be sufficient to inactivate cellular Ser/Thr phosphatase (PP2A) activity, in turn promoting activation of stress-sensitive Ser/Thr kinases (JNK, ERK, IKKβ) and accumulation of inhibitory phosphomoieties within various insulin signaling proteins. Alternatively, the oxidative shift in global redox environment induced by the high fat diet may compromise the capacity of the redox buffering systems to buffer the H2O2 production from membrane bound NADPH oxidase in response to insulin, resulting in a further shift in global redox environment that is sufficient to inactivate both PTP and Ser/Thr phosphatases, all of which decreases signal propagation throughout the cascade and impairs glucose uptake.
Acute induction of insulin resistance in a healthy system
Skeletal muscle accounts for the vast majority of glucose and at least a portion of the lipid clearance after a meal, a process that often requires several hours [71, 72]. In light of the proposed importance of the global redox environment, this raises the obvious question as to how insulin sensitivity is maintained in the post-prandial state despite the persistent reducing pressure placed on the respiratory system. The concept of “auto-regulation” of the insulin receptor/IRS1 axis is fairly well established, owing to the identification of Ser sites within IRS1 that become phosphorylated and inhibit downstream signaling in response to persistent insulin exposure in healthy, insulin sensitive cells in culture [73–78]. However, the stoichiometric and temporal aspects of this process are not well established, and its relevance to regulation of the insulin receptor/IRS1 axis in vivo is unknown. In fact, insulin resistance does not develop during glucose infusion in rats (~10–18 mM [glucose]) for at least 3h and is independent of changes in phosphorylation of Akt or AS160 [79–81]. While the accumulation of intracellular metabolites and feedback regulation could account for hyperglycemia-induced insulin resistance, direct evidence to support this mechanism is not available [79]. Interestingly however, co-infusion of the antioxidants N-acetylcysteine or taurine prevents the development of insulin resistance induced by glucose infusion [82, 83], providing evidence of the importance of H2O2 production. These studies also highlight the remarkable capacity of the redox systems to buffer H2O2 and suggest in a healthy system there is sufficient reserve capacity in the redox signaling network to prevent any decline in the sensitivity to insulin for several hours after a meal.
The concept of redox reserve capacity
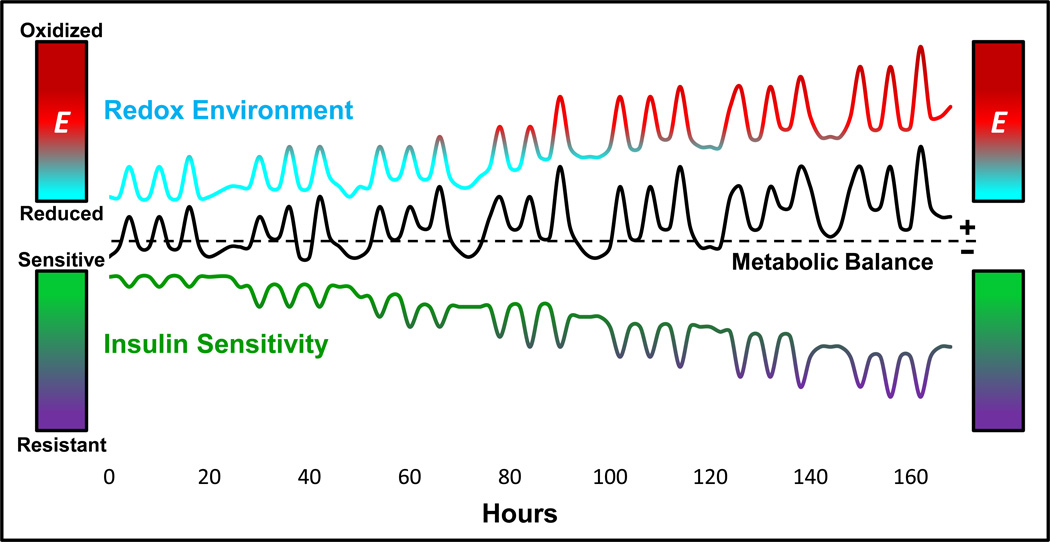
There are a number of intriguing implications raised by the concept of redox reserve capacity and its potential impact on global phosphatase tone and insulin sensitivity. First, as eluded to earlier, the sensitivity of redox-sensitive Cys(s) within any protein is a function of the intra- and inter-molecular environment in which it resides. Studies of protein chemistry indicate the reactivity of Cys within the entire redox-regulated proteome varies over seven orders of magnitude [84], meaning that the reduction potentials range from very negative (i.e., high potential to become oxidized) to very positive (i.e., low potential to become oxidized). This infers that the redox reserve capacity is a unique property of each redox couple, reflecting the “threshold” level of redox signaling or oxidant shift in redox environment that is necessary to trigger oxidation at that particular Cys. Second, proximity to the source of H2O2 production is undoubtedly a key determinant of a redox protein’s sensitivity to redox signaling. Third, given the dynamics of living animals, the redox environment is unlikely to ever be in “steady-state”. Each meal will induce a transient increase in mitochondrial H2O2 production and an oxidative shift in redox environment [11], temporarily decreasing the redox reserve capacity (Figure 7). Incoming substrates directed to replacing energy reserves (i.e., glycogen, triglycerides) will minimize H2O2 production, whereas large and frequent meals in sedentary individuals will accentuate and prolong the oxidative shift in redox environment. Fourth, under conditions of metabolic imbalance due to consistent nutrient overload and inactivity, an inability to completely restore the redox environment between meals will induce a progressive decline in redox reserve capacity, ultimately affecting global phosphatase tone and insulin sensitivity (Figure 7). Finally, it is intriguing to consider the possibility that the burst of H2O2 production and influx across the plasma membrane normally required to propagate insulin signaling may in fact be the oxidative input that pushes the global redox environment beyond the threshold needed to inhibit insulin signaling (Figure 6). This is consistent with the notion that insulin stimulation may be necessary to induce insulin resistance [85].
Figure 7. Potential dynamic alterations in insulin sensitivity and redox environment as a function of metabolic balance over time.
Frequent intake of energy dense meals in conjunction with a sedentary lifestyle promotes acute and persistent fluctuations, positive metabolic balance, as well as concomitant shifts in both the redox environment and insulin sensitivity. Summation of these acute fluctuations over an extended period of time (1 week) result in a gradual shift in baseline levels for redox environment and insulin sensitivity, as well as exacerbated shifts in response to meal challenges, ultimately leading to relative insulin resistant state.
Concluding remarks
In the present review, the etiology of diet-induced insulin resistance is viewed in the context of mitochondrial bioenergetics and cellular redox biology. Nutrient overload places reducing pressure on the mitochondria, generating a transient elevation in H2O2 emission and an oxidative shift in redox environment, representing a primary mechanism by which cells sense and respond to metabolic imbalance. In contrast to the local oxidation of redox environment at the plasma membrane necessary for normal insulin signaling, the concept is put forward that insulin resistance develops as a consequence of persistent transient periods of mitochondrial H2O2 emission that outpace restoration of the redox reserve capacity, resulting in a cumulative oxidative shift in the global redox environment. This is predicted to depress phosphatase tone throughout the cell, ultimately leading to a persistent dominance in stress-sensitive Ser/Thr kinase activity targeted to insulin signaling proteins and disruption in the normally well-coordinated pattern of insulin signaling. This concept is consistent with the principles of cellular energetics (i.e., energy supply versus energy demand) and suggests that in the absence of lifestyle (e.g., exercise) and/or dietary (e.g., caloric restriction) interventions designed to restore metabolic balance, pharmacological strategies must be devised that are directed at maintaining the integrity of the redox buffering systems operating in vivo, either through the development of mitochondrial targeted H2O2 scavenging compounds and/or mimetics of the primary redox buffering system.
Box 1. Commonly employed me thods for detecting mitochondrial H2O2.
The existence of H2O2 as a signaling molecule has been demonstrated for some time. However, elucidation of the precise mechanism by which H2O2 exerts signaling effects, as well as any potential role that this reactive metabolite may play in human disease has in many ways been limited by a lack of experimental methods available for probing H2O2 within living systems. At present, the most widely used methods for assessing H2O2 emission from isolated mitochondria or permeabilized cells are fluorescence-based detection systems (e.g., Amplex Red [86], p-hydroxyphenylacetate (PHPA [27]), dichlorofluorescein (DCF [10]). Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine; Invitrogen), and the newly developed Amplex Ultra Red Reagent (Invitrogen), react with H2O2 (1:1 stoichiometry) to produce the highly fluorescent resorufin, with a low-end detection limit of 50 nM. However, most redox-sensitive probes are at least partially reactive with lipid hydroperoxides and other redox metabolites, requiring some caution with interpretation of data [87]. The recent development of the genetically encoded biosensors for H2O2 including HyPer [88] and the redox-sensitive green fluorescent proteins (RoGFP) [89], the latter including a mitochondrial-targeted version, have for the first time made it possible to monitor changes in redox environment in intact cells. A particularly exciting advancement has come from the development of MitoB, a ratiometric mass spectrometry probe comprised of a triphenylphosphonium (TPP) cation (allows for accumulation within the mitochondrial matrix) conjugated to an arylboronic acid (reacts with H2O2 to form a phenol; MitoP) [90]. By quantifying the MitoP/MitoB ratio using liquid chromatography-tandem mass spectrometry, this probe was shown to report on the level of H2O2 within living Drosophila [90]. The continual development of compounds such as MitoB and subsequent application to other systems, as well as the creation and/or refinement of additional measures of the redox environment are critical to deciphering the complex role that H2O2 plays in human physiology.
Box 2. Outstanding issues.
These are but a few of the many.
How are cellular redox circuits organized and regulated in vivo?
The vast number of redox-sensitive Cys-thiols estimated to be present within the proteome (>20,000) represents a mind-numbing array of possible regulatory circuits. Compared with other signaling mechanisms (e.g., phosphorylation, acetylation), the field of redox biology is in its infancy. Elucidating the overall organization and regulation of such circuits in the context of health and disease undoubtedly represents one of the most exciting challenges for science in the 21st century.
How does insulin-induced H2O2 bypass the redox-buffering systems and elicit a “targeted” response?
The proximity of PTPs to the H2O2-emitting source activated by insulin (membrane bound NADPH oxidase), raises the possibility that direct oxidation of Cys by H2O2 may occur; however, these reactions would be in competition with the redox buffering systems. Another possibility is that the “H2O2 signal” could be transmitted to Cys through organization of PTP enzymes within a redox circuit, as illustrated in Figure 2. How reductions in antioxidant reserve capacity may disrupt the specificity of redox signaling cascades is unknown.
How is specificity of Ser/Thr phosphatase activity distributed within the insulin signaling cascade?
The specific Ser/Thr phosphatase enzymes exerting both positive and negative control over individual phosphorylation sites within the cascade are not known. Ser/Thr phosphatases are known to be directed to various cellular compartments through interactions with different targeting proteins. How these interactions and targeting directives are influenced acutely and chronically by changes in the redox environment and associated changes in “global” phosphatase activity remains to be determined.
What are the dynamic relationships between cellular redox environment and insulin sensitivity?
As depicted in Figure 7, cellular redox environment is likely in a constant state of flux depending on the acute and relatively chronic metabolic state of the cell. How redox biology dynamics impacts the overall redox environment of the cell and translates into regulatory control under conditions of both positive and negative metabolic balance is of obvious importance to understand.
Supplementary Material
Acknowledgements
P.D.N. is supported by grants from the National Institutes of Health (R01-DK073488 and RO1-DK074825).
Glossary
- Chemiosmosis
The movement of hydrogen ions across the mitochondrial inner membrane, down their electrochemical gradient, to drive the re-synthesis of ATP.
- Phosphatase Tone
Qualitative term reflecting the dominance of phosphatase over kinase activity in a resting cell.
- Redox Buffering Systems
Refers to the principle cellular thiol/disulfide redox couples glutathione (2GSH/GSSG) and thioredoxin (TrxRed/TrxOx) and their associated enzyme systems (glutathione peroxidase (GPx), glutathione reductase (GR), thioredoxin reductase (TR) peroxiredoxin (Prx), glutaredoxin (Grx). Both buffering systems derive reducing power from the NADPH/NADP+ redox couple.
- Redox Capacity
Refers to the concentration of the reduced species within a given redox couple (i.e., indicates how many electrons are available for transfer).
- Redox Circuit
Analogous to an electrical circuit, a set of linked oxidation/reduction reactions initiated by a difference in reduction potential between two redox couples (i.e., voltage) and propagated as electron flow (i.e., electrical current) among successive redox couples with higher reduction potentials.
- Redox Couple
Species capable of oscillating between a reduced and oxidized form. Aside from NAD(P)H/NAD(P)+ and FAD-containing proteins, the majority of redox couples operating in vivo are cysteine-thiol (−SH) containing proteins/peptides.
- Redox Environment
Refers to the theoretical summation of the redox state for all redox couples in a system, thus reflecting the global tendency for electron transfer within that system.
- Redox Reserve Capacity
Refers to the magnitude of disequilibrium in reduction potential available between two redox couples prior to loss of electron flow, and thus the ability to maintain the downstream redox couple in a reduced state.
- Redox Sensitive Proteins
Proteins containing one or more cysteine-thiols sensitive to the redox environment and thus capable of oscillating between a reduced and oxidized form.
- Redox State
The product of the reduction potential (i.e., voltage) and redox capacity (i.e., total charge stored) of a given redox couple.
- Reduction Potential
A measure (in mV) of the inherent tendency of a given redox pair to accept an electron(s), typically defined under standard conditions (E°, pH 7.0, 25° C. From the concentrations of the reduced and oxidized species, the half-cell reduction potential (Eh) for a single redox couple or overall reduction potential between two redox couples (ΔE) can be calculated using the Nernst equation.
- State 2 respiration
The rate of O2 consumption required to support basal proton conductance (i.e., proton “leak” prior to addition of ADP).
- State 3 respiration
The rate of O2 consumption required to support any level of ADP-stimulated proton conductance (i.e., ATP synthesis).
- State 4 respiration
Experimentally, the rate of O2 consumption required to support respiration once all ADP is converted to ATP. Functionally, the same as state 2, with state 4 usually the term used for both states.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Although the thioether of methionine is also capable of undergoing reversible oxidation-reduction, it is unclear at present the extent to which these residues are reversibly oxidized 23. Jones, D.P. (2008) Radical-free biology of oxidative stress. Am J Physiol Cell Physiol 295, C849–868.
References
- 1.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 2.Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin resistance. Cell Metabolism. doi: 10.1016/j.cmet.2012.04.010. ((in press)) Invited Perspective. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 4.Korshunov SS, et al. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997;416:15–18. doi: 10.1016/s0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- 5.Liu SS. Generating, partitioning, targeting and functioning of superoxide in mitochondria. Biosci Rep. 1997;17:259–272. doi: 10.1023/a:1027328510931. [DOI] [PubMed] [Google Scholar]
- 6.Liu SS. Cooperation of a "reactive oxygen cycle" with the Q cycle and the proton cycle in the respiratory chain--superoxide generating and cycling mechanisms in mitochondria. J Bioenerg Biomembr. 1999;31:367–376. doi: 10.1023/a:1018650103259. [DOI] [PubMed] [Google Scholar]
- 7.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 8.Evans JL, et al. Oxidative Stress and Stress-Activated Signaling Pathways: A Unifying Hypothesis of Type 2 Diabetes. Endocr Rev. 2002;23:599–622. doi: 10.1210/er.2001-0039. [DOI] [PubMed] [Google Scholar]
- 9.Evans JL, et al. The molecular basis for oxidative stress-induced insulin resistances. Antioxid Redox Signal. 2005;7:1040–1052. doi: 10.1089/ars.2005.7.1040. [DOI] [PubMed] [Google Scholar]
- 10.Houstis N, et al. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 11.Anderson EJ, et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J Clin Invest. 2009;119:573–581. doi: 10.1172/JCI37048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, et al. Reduction of mitochondrial H2O2 by overexpressing peroxiredoxin 3 improves glucose tolerance in mice. Aging Cell. 2008;7:866–878. doi: 10.1111/j.1474-9726.2008.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoehn KL, et al. Insulin resistance is a cellular antioxidant defense mechanism. Proc Natl Acad Sci U S A. 2009;106:17787–17792. doi: 10.1073/pnas.0902380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H-Y, et al. Targeted Expression of Catalase to Mitochondria Prevents Age-Associated Reductions in Mitochondrial Function and Insulin Resistance. Cell Metabolism. 2010;12:668–674. doi: 10.1016/j.cmet.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura S, et al. Palmitate induces insulin resistance in H4IIEC3 hepatocytes through reactive oxygen species produced by mitochondria. J Biol Chem. 2009;284:14809–14818. doi: 10.1074/jbc.M901488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batinic-Haberle I, et al. Pure MnTBAP selectively scavenges peroxynitrite over superoxide: Comparison of pure and commercial MnTBAP samples to MnTE-2-PyP in two models of oxidative stress injury, an SOD-specific Escherichia coli model and carrageenan-induced pleurisy. Free Radical Biology and Medicine. 2009;46:192–201. doi: 10.1016/j.freeradbiomed.2008.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buettner GR, et al. A new paradigm: manganese superoxide dismutase influences the production of H2O2 in cells and thereby their biological state. Free Radic Biol Med. 2006;41:1338–1350. doi: 10.1016/j.freeradbiomed.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowald A, Klipp E. Alternative pathways might mediate toxicity of high concentrations of superoxide dismutase. Ann N Y Acad Sci. 2004;1019:370–374. doi: 10.1196/annals.1297.065. [DOI] [PubMed] [Google Scholar]
- 19.Gardner R, et al. Why does SOD overexpression sometimes enhance, sometimes decrease, hydrogen peroxide production? A minimalist explanation. Free Radic Biol Med. 2002;32:1351–1357. doi: 10.1016/s0891-5849(02)00861-4. [DOI] [PubMed] [Google Scholar]
- 20.Winterbourn CC, Metodiewa D. Reaction of superoxide with glutathione and other thiols. Methods Enzymol. 1995;251:81–86. doi: 10.1016/0076-6879(95)51112-1. [DOI] [PubMed] [Google Scholar]
- 21.Forman HJ, Fridovich I. Superoxide dismutase: a comparison of rate constants. Arch Biochem Biophys. 1973;158:396–400. doi: 10.1016/0003-9861(73)90636-x. [DOI] [PubMed] [Google Scholar]
- 22.Forman HJ, et al. Signaling Functions of Reactive Oxygen Species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson KA, Buse MG. Mechanisms of high-glucose/insulin-mediated desensitization of acute insulin-stimulated glucose transport and Akt activation. Am J Physiol Endocrinol Metab. 2008;294:E870–E881. doi: 10.1152/ajpendo.00644.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 26.Anderson EJ, et al. Induction of endogenous uncoupling protein 3 suppresses mitochondrial oxidant emission during fatty acid-supported respiration. J Biol Chem. 2007;282:31257–31266. doi: 10.1074/jbc.M706129200. [DOI] [PubMed] [Google Scholar]
- 27.Seifert EL, et al. Electron transport chain-dependent and -independent mechanisms of mitochondrial H2O2 emission during long-chain fatty acid oxidation. J Biol Chem. 2010;285:5748–5758. doi: 10.1074/jbc.M109.026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.St-Pierre J, et al. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 29.Szeto HH. Mitochondria-targeted cytoprotective peptides for ischemia-reperfusion injury. Antioxid Redox Signal. 2008;10:601–619. doi: 10.1089/ars.2007.1892. [DOI] [PubMed] [Google Scholar]
- 30.Zhao K, et al. Cell-permeable peptide antioxidants targeted to inner mitochondrial membrane inhibit mitochondrial swelling, oxidative cell death, and reperfusion injury. J Biol Chem. 2004;279:34682–34690. doi: 10.1074/jbc.M402999200. [DOI] [PubMed] [Google Scholar]
- 31.Schriner SE, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 32.Phung CD, et al. Hydrogen peroxide metabolism in skeletal muscle mitochondria. Arch Biochem Biophys. 1994;315:479–482. doi: 10.1006/abbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- 33.Kudin AP, et al. Characterization of superoxide-producing sites in isolated brain mitochondria. J Biol Chem. 2004;279:4127–4135. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
- 34.Chiarugi P. PTPs versus PTKs: the redox side of the coin. Free radical research. 2005;39:353–364. doi: 10.1080/10715760400027987. [DOI] [PubMed] [Google Scholar]
- 35.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Foley TD, et al. Oxidative inhibition of protein phosphatase 2A activity: role of catalytic subunit disulfides. Neurochem Res. 2007;32:1957–1964. doi: 10.1007/s11064-007-9394-x. [DOI] [PubMed] [Google Scholar]
- 37.Wright VP, et al. Redox modulation of global phosphatase activity and protein phosphorylation in intact skeletal muscle. J Physiol. 2009;587:5767–5781. doi: 10.1113/jphysiol.2009.178285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barford D, et al. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 39.Fetrow JS, et al. Structure-based functional motif identifies a potential disulfide oxidoreductase active site in the serine/threonine protein phosphatase-1 subfamily. Faseb J. 1999;13:1866–1874. doi: 10.1096/fasebj.13.13.1866. [DOI] [PubMed] [Google Scholar]
- 40.Maher P. Redox control of neural function: background, mechanisms, and significance. Antioxidants & redox signaling. 2006;8:1941–1970. doi: 10.1089/ars.2006.8.1941. [DOI] [PubMed] [Google Scholar]
- 41.Dasgupta J, et al. Reactive oxygen species control senescence-associated matrix metalloproteinase-1 through c-Jun-N-terminal kinase. J Cell Physiol. 2010;225:52–62. doi: 10.1002/jcp.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Covey TM, et al. Alkylation of the tumor suppressor PTEN activates Akt and beta-catenin signaling: a mechanism linking inflammation and oxidative stress with cancer. PLoS One. 2010:e13545. doi: 10.1371/journal.pone.0013545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu G, et al. A novel mitochondrial matrix serine/threonine protein phosphatase regulates the mitochondria permeability transition pore and is essential for cellular survival and development. Genes Dev. 2007;21:784–796. doi: 10.1101/gad.1499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JE, et al. Hydrogen peroxide triggers the proteolytic cleavage and the inactivation of calcineurin. J Neurochem. 2007;100:1703–1712. doi: 10.1111/j.1471-4159.2006.04340.x. [DOI] [PubMed] [Google Scholar]
- 45.Bashan N, et al. Positive and negative regulation of insulin signaling by reactive oxygen and nitrogen species. Physiol Rev. 2009;89:27–71. doi: 10.1152/physrev.00014.2008. [DOI] [PubMed] [Google Scholar]
- 46.Loh K, et al. Reactive oxygen species enhance insulin sensitivity. Cell Metab. 2009;10:260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiganis T. Reactive oxygen species and insulin resistance: the good, the bad and the ugly. Trends Pharmacol Sci. 2011;32:82–89. doi: 10.1016/j.tips.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Thong FS, et al. Turning signals on and off: GLUT4 traffic in the insulin-signaling highway. Physiology (Bethesda) 2005;20:271–284. doi: 10.1152/physiol.00017.2005. [DOI] [PubMed] [Google Scholar]
- 49.Goldstein B, et al. Role of insulin-induced reactive oxygen species in the insulin signaling pathway. Antioxid Redox Signal. 2005;7:1021–1031. doi: 10.1089/ars.2005.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szypowska AA, Burgering BM. The peroxide dilemma: opposing and mediating insulin action. Antioxid Redox Signal. 2011;15:219–232. doi: 10.1089/ars.2010.3794. [DOI] [PubMed] [Google Scholar]
- 51.Goldstein BJ, et al. Role of insulin-induced reactive oxygen species in the insulin signaling pathway. Antioxidants & redox signaling. 2005;7:1021–1031. doi: 10.1089/ars.2005.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahadev K, et al. The NAD(P)H oxidase homolog Nox4 modulates insulin-stimulated generation of H2O2 and plays an integral role in insulin signal transduction. Mol Cell Biol. 2004;24:1844–1854. doi: 10.1128/MCB.24.5.1844-1854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Espinosa A, et al. NADPH oxidase and hydrogen peroxide mediate insulin-induced calcium increase in skeletal muscle cells. J Biol Chem. 2009;284:2568–2575. doi: 10.1074/jbc.M804249200. [DOI] [PubMed] [Google Scholar]
- 54.Chen K, et al. Downstream targets and intracellular compartmentalization in Nox signaling. Antioxid Redox Signal. 2009;11:2467–2480. doi: 10.1089/ars.2009.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ushio-Fukai M. Compartmentalization of redox signaling through NADPH oxidase-derived ROS. Antioxid Redox Signal. 2009;11:1289–1299. doi: 10.1089/ars.2008.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bedard K, Krause K-H. The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiological Reviews. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 57.Leto TL, et al. Targeting and regulation of reactive oxygen species generation by Nox family NADPH oxidases. Antioxid Redox Signal. 2009;11:2607–2619. doi: 10.1089/ars.2009.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller EW, et al. Aquaporin-3 mediates hydrogen peroxide uptake to regulate downstream intracellular signaling. Proc Natl Acad Sci U S A. 2010;107:15681–15686. doi: 10.1073/pnas.1005776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Espinosa A, et al. Myotube depolarization generates reactive oxygen species through NAD(P)H oxidase; ROS-elicited Ca2+ stimulates ERK CREB, early genes. Journal of cellular physiology. 2006;209:379–388. doi: 10.1002/jcp.20745. [DOI] [PubMed] [Google Scholar]
- 60.Woo HA, et al. Inactivation of Peroxiredoxin I by Phosphorylation Allows Localized H2O2 Accumulation for Cell Signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 61.Boura-Halfon S, Zick Y. Phosphorylation of IRS proteins, insulin action, and insulin resistance. Am J Physiol Endocrinol Metab. 2009;296:E581–E591. doi: 10.1152/ajpendo.90437.2008. [DOI] [PubMed] [Google Scholar]
- 62.Hirosumi J, et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 63.Bost F, et al. The extracellular signal-regulated kinase isoform ERK1 is specifically required for in vitro and in vivo adipogenesis. Diabetes. 2005;54:402–411. doi: 10.2337/diabetes.54.2.402. [DOI] [PubMed] [Google Scholar]
- 64.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 65.Yuan M, et al. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkbeta. Science. 2001;293:1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
- 66.Danielsson A, et al. Attenuation of insulin-stimulated insulin receptor substrate-1 serine 307 phosphorylation in insulin resistance of type 2 diabetes. J Biol Chem. 2005;280:34389–34392. doi: 10.1074/jbc.C500230200. [DOI] [PubMed] [Google Scholar]
- 67.Tanti JF, Jager J. Cellular mechanisms of insulin resistance: role of stress-regulated serine kinases and insulin receptor substrates IRS serine phosphorylation. Curr Opin Pharmacol. 2009;9:753–762. doi: 10.1016/j.coph.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 68.Hoehn KL, et al. IRS1-independent defects define major nodes of insulin resistance. Cell Metab. 2008;7:421–433. doi: 10.1016/j.cmet.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouzakri K, et al. Malonyl CoenzymeA Decarboxylase Regulates Lipid and Glucose Metabolism in Human Skeletal Muscle. Diabetes. 2008;57:1508–1516. doi: 10.2337/db07-0583. [DOI] [PubMed] [Google Scholar]
- 70.Loukili N, et al. Oxidants Positively or Negatively Regulate Nuclear Factor ΰB in a Context-dependent Manner. Journal of Biological Chemistry. 2010;285:15746–15752. doi: 10.1074/jbc.M110.103259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeFronzo RA, et al. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes. 1981;30:1000–1007. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- 72.Bickerton AS, et al. Preferential uptake of dietary Fatty acids in adipose tissue and muscle in the postprandial period. Diabetes. 2007;56:168–176. doi: 10.2337/db06-0822. [DOI] [PubMed] [Google Scholar]
- 73.Mothe I, Van Obberghen E. Phosphorylation of insulin receptor substrate-1 on multiple serine residues, 612, 632, 662, and 731, modulates insulin action. J Biol Chem. 1996;271:11222–11227. doi: 10.1074/jbc.271.19.11222. [DOI] [PubMed] [Google Scholar]
- 74.Paz K, et al. A molecular basis for insulin resistance. Elevated serine/threonine phosphorylation of IRS-1 and IRS-2 inhibits their binding to the juxtamembrane region of the insulin receptor and impairs their ability to undergo insulin-induced tyrosine phosphorylation. J Biol Chem. 1997;272:29911–29918. doi: 10.1074/jbc.272.47.29911. [DOI] [PubMed] [Google Scholar]
- 75.Paz K, et al. Phosphorylation of insulin receptor substrate-1 (IRS-1) by protein kinase B positively regulates IRS-1 function. J Biol Chem. 1999;274:28816–28822. doi: 10.1074/jbc.274.40.28816. [DOI] [PubMed] [Google Scholar]
- 76.Weigert C, et al. The Phosphorylation of Ser318 of Insulin Receptor Substrate 1 Is Not per se Inhibitory in Skeletal Muscle Cells but Is Necessary to Trigger the Attenuation of the Insulin-stimulated Signal. Journal of Biological Chemistry. 2005;280:37393–37399. doi: 10.1074/jbc.M506134200. [DOI] [PubMed] [Google Scholar]
- 77.Weigert C, et al. Interplay and Effects of Temporal Changes in the Phosphorylation State of Serine-302, -307, and -318 of Insulin Receptor Substrate-1 on Insulin Action in Skeletal Muscle Cells. Mol Endocrinol. 2008;22:2729–2740. doi: 10.1210/me.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luo M, et al. Phosphorylation of Human Insulin Receptor Substrate-1 at Serine 629 Plays a Positive Role in Insulin Signaling. Endocrinology. 2007;148:4895–4905. doi: 10.1210/en.2007-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brandon AE, et al. The evolution of insulin resistance in muscle of the glucose infused rat. Arch Biochem Biophys. 2011;509:133–141. doi: 10.1016/j.abb.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoy AJ, et al. Lipid and insulin infusion-induced skeletal muscle insulin resistance is likely due to metabolic feedback and not changes in IRS-1, Akt, or AS160 phosphorylation. Am J Physiol Endocrinol Metab. 2009;297:E67–E75. doi: 10.1152/ajpendo.90945.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kraegen EW, et al. Increased malonyl-CoA and diacylglycerol content and reduced AMPK activity accompany insulin resistance induced by glucose infusion in muscle and liver of rats. American Journal of Physiology - Endocrinology And Metabolism. 2006;290:E471–E479. doi: 10.1152/ajpendo.00316.2005. [DOI] [PubMed] [Google Scholar]
- 82.Haber CA, et al. N-acetylcysteine and taurine prevent hyperglycemia-induced insulin resistance in vivo: possible role of oxidative stress. American Journal of Physiology - Endocrinology And Metabolism. 2003;285:E744–E753. doi: 10.1152/ajpendo.00355.2002. [DOI] [PubMed] [Google Scholar]
- 83.Lin Y, et al. The hyperglycemia-induced inflammatory response in adipocytes: the role of reactive oxygen species. J Biol Chem. 2005;280:4617–4626. doi: 10.1074/jbc.M411863200. [DOI] [PubMed] [Google Scholar]
- 84.Nagy P, Winterborun CC. Redox Chemistry of Biological Thiols. In: Fishbein JC, editor. Advances in Molecular Toxicology. Elsevier; 2010. pp. 183–222. [Google Scholar]
- 85.Cao W, et al. Excess exposure to insulin may be the primary cause of insulin resistance. Am J Physiol Endocrinol Metab. 2010;298:E372. doi: 10.1152/ajpendo.00677.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perry CG, et al. Inhibiting myosin-ATPase reveals a dynamic range of mitochondrial respiratory control in skeletal muscle. Biochem J. 2011;437:215–222. doi: 10.1042/BJ20110366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhattacharya A, et al. Denervation induces cytosolic phospholipase A2-mediated fatty acid hydroperoxide generation by muscle mitochondria. J Biol Chem. 2009;284:46–55. doi: 10.1074/jbc.M806311200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belousov VV, et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 89.Meyer AJ, Dick TP. Fluorescent protein-based redox probes. Antioxid Redox Signal. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 90.Cocheme HM, et al. Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011;13:340–350. doi: 10.1016/j.cmet.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.