Abstract
In 2005, draft sequences of the genomes of Trypanosoma brucei, Trypanosoma cruzi and Leishmania major, also known as the Tri-Tryp genomes, were published. These protozoan parasites are the causative agents of three distinct insect-borne diseases, namely sleeping sickness, Chagas disease and leishmaniasis, all with a worldwide distribution. Despite the large estimated evolutionary distance among them, a conserved core of ~6,200 trypanosomatid genes was found among the Tri-Tryp genomes. Extensive analysis of these genomic sequences has greatly increased our understanding of the biology of these parasites and their host-parasite interactions. In this article, we review the recent advances in the comparative genomics of these three species. This analysis also includes data on additional sequences derived from other trypanosmatid species, as well as recent data on gene expression and functional genomics. In addition to facilitating the identification of key parasite molecules that may provide a better understanding of these complex diseases, genome studies offer a rich source of new information that can be used to define potential new drug targets and vaccine candidates for controlling these parasitic infections.
Keywords: Trypanosoma brucei, Trypanosoma cruzi, Leishmania major, genome, RNAseq
Tri-Tryp Diseases and The Tri-Tryp Genomes
Trypanosoma brucei, Trypanosoma cruzi and Leishmania major are unicellular protozoa of considerable medical importance since they are the etiologic agents of sleeping sickness (African trypanosomiasis), Chagas disease (American trypanosomiasis) and leishmaniasis, respectively. The geographic range of these parasites is determined by their insect vectors: in the case of sleeping sickness, a blood sucking fly of the genus Glossina, also known as the tsetse fly, for Chagas disease, a reduviid bug known as the “kissing bug” and for leishmaniasis, a phlebotomine sandfly. While sleeping sickness occurs in sub-Saharan Africa, Chagas disease is prevalent in Latin America. Leishmaniasis is considered to be endemic in 88 countries, 72 of which are developing countries in Asia, South America and Africa. Together, these three parasitic diseases represent a huge burden since approximately 0.5 million people are infected with T. brucei, 10 million with T. cruzi and an estimated 12 million with different species of Leishmania. In addition to the two human-infective subspecies, T. brucei gambiense and T. b. rhodesiense, other species and subspecies of African trypanosomes cause the disease known as nagana in domestic animals, imposing a further economic burden on several African countries. Different forms of leishmaniasis are caused by at least 20 leishmanial species: cutaneous leishmaniasis, with an estimated 1.5 million cases, and visceral leishmaniasis, with about 500,000 new cases annually, are the most common. Although control of the arthropod vectors of these diseases is an achievable goal and has been successful against the T. cruzi vector in parts of Latin America, the alarming resurgence of sleeping sickness in Africa and of leishmaniasis in parts of Asia and Latin America is a constant reminder of the need for better forms of chemotherapy and prevention of these diseases. More detailed information on African and American trypanosomiasis and the different forms of leishmaniasis, including vector distribution, disease control and treatment protocols can be found at http://apps.who.int/tdr/.
Trypanosoma brucei, T. cruzi and Leishmania spp. are hemoflagellates of the family Trypanosomatidae (order Kinetoplastida) that is characterized by the presence of a single flagellum and one mitochondrion containing a unique organelle known as the kinetoplast which contains the mitochondrial DNA (Simpson et al., 2006). Each parasite has a complex life cycle that involves humans as one of their various hosts (Figure 1). As some of the earliest divergent members of the Eukaryotae (Haag et al., 1998), these parasites have peculiar aspects of gene expression, including polycistronic transcription of most of their genomes (Martínez-Calvillo et al., 2010), RNA polymerase I-mediated transcription of protein-coding genes (Gunzl et al., 2003), RNA trans-splicing to generate mature, capped mRNAs (LeBowitz et al., 1993) and extensive RNA editing to generate functional mRNAs transcribed from mito-chondrial genes (Hajduk et al., 1993). Apart from their medical relevance, these peculiar characteristics make these parasites very interesting models for studying genome evolution and other aspects of genome function. On the other hand, the early evolutionary divergence of these organisms has resulted in biochemical characteristics that are not common in higher eukaryotes, such as enzymes related to antioxidant metabolism (Olin-Sandoval et al., 2010) as well as sterol and glycosylphosphatidylinositol (GPI) biosynthesis (Lepesheva et al., 2011; Koeller and Heise, 2011) that have been exploited as promising drug targets.
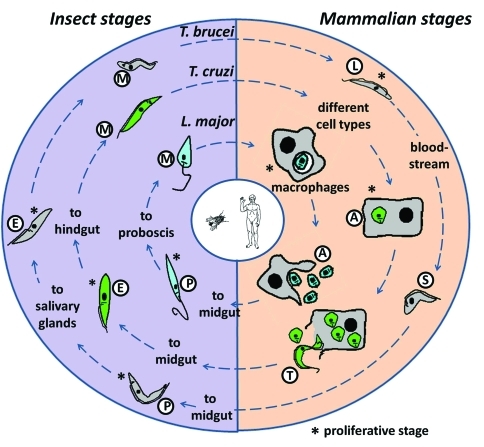
Figure 1.
The Tri-Tryp life cycles. Representation of the life cycles of Leishmania major, Trypanosoma cruzi and T. brucei, the etiological agents of leishmaniasis, Chagas disease and sleeping sickness, respectively, are shown, with the parasitic forms that are present in the insect vectors and the mammalian hosts. Leishmania major proliferates as promastigotes (P) in the sand fly midgut. The parasite is transmitted during bites by this fly and invades mammalian macrophages in the metacyclic promastigote (M) form. Inside the cell, the M form is converted into amastigotes (A) and divides before been released during cell lysis. Trypanosoma cruzi replicates as epimastigotes (E) in the reduviid bug midgut and develops into infective metacyclic trypomastigotes that are excreted in the feces (M) and invade different cell types when in contact with the mammalian host. After differentiation into proliferative amastigotes (A), these are transformed into bloodstream trypomastigotes (T) that cause cell lysis and invade new cells. Trypanosoma brucei differentiates from procyclic (P) to epimastigote (E) proliferative forms in the tsetse fly before being transformed into infective, metacyclic forms (M) in the salivary glands. After being injected into the host during a blood meal, M forms differentiate into long slender forms (L) that proliferate in the bloodstream and can reach the central nervous system. After increase of parasite numbers these last forms are replaced by non-proliferative stumpy forms (S).
Genome sequencing of Tri-Tryp parasites began in the early 90s with the analyses of 518 expressed sequence tags (ESTs) generated from mRNA isolated from bloodstream forms of T. b. rhodesiense (El-Sayed et al., 1995). Shortly thereafter, a comparison between EST and genomic sequences showed that sequencing random DNA fragments was as efficient as EST analyses for discovering new genes in the African trypanosome (El-Sayed and Donelson, 1997). In 1996, an EST analysis of cDNA libraries constructed with mRNA from L. major promastigotes was published (Levick et al., 1996), and the first EST analysis of T. cruzi epimastigote forms was published in 1997 (Brandão et al., 1997). During this period, pulsed-field gel electrophoretic analysis of chromosomes and the sequencing of large DNA fragments from cosmid, bacterial artificial chromosome and yeast artificial chromosome libraries were also undertaken to generate physical maps of Tri-Tryp genomes (Blackwell and Melville, 1999). In 1999, the sequence of a 257-kilobase region spanning almost the entire chromosome 1 of L. major revealed the unusual distribution of protein-coding genes that was later found to be characteristic of all Tri-Tryp genomes. The complete sequence of L. major chromosome 1 revealed 79 protein-coding genes, with the first 29 genes all encoded on one DNA strand and the remaining 50 genes encoded on the opposite strand (Myler et al., 1999).
The Tri-Tryp gene organization is reminiscent of bacterial operons, with protein coding genes densely packed within directional clusters in one strand separated by strand switch regions (i.e., changes in the coding strand) (Figure 2). Experimental evidence suggests that transcription initiates bi-directionally between two divergent gene clusters (Martínez-Calvillo et al., 2003, 2004) to produce polycistronic pre-mRNAs that are subsequently processed. Remarkably, with the exception of the spliced leader (SL) promoter, no promoter is recognized by RNA polymerase II and only a few transcription factors have been identified (Cribb and Serra, 2009; Cribb et al., 2010). Even more surprisingly, although orthologs of all conserved components of the RNA polymerase II complex were identified in the Tri-Tryp genome (Ivens et al., 2005), the transcription of some trypanosomatid genes such as VSG (Variant Surface Glycoprotein) and the procyclin genes of T. brucei, as well as several exogenous genes transfected into T. cruzi, are mediated by RNA polymerase I (Gunzl et al., 2003). Once the polycistronic pre-mRNA is produced, two coupled reactions (trans-splicing and poly-adenylation) result in mature monocistronic transcripts.
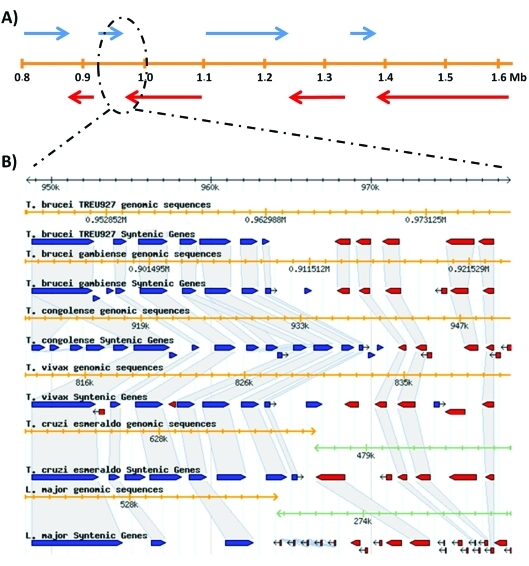
Figure 2.
Gene organization in the Tri-Tryp genome. Panel A shows the gene distribution in a 0.8 Mb region of T. brucei chromosome V with eight large polycistronic transcription units (blue arrows: plus strand encoded open reading frames or ORFs; red arrows: minus strand encoded ORFs). In panel B, a genomic region at around 960 kb is magnified to show the gene synteny in the genomes of various trypanosomatids (blue and red boxes correspond to + and − strand-encoded ORFs, respectively). The orange line in both panels corresponds to the chromosome position. Sequence information used to draw panel A and the graphic representation in panel B were obtained from the Tri-Tryp database (Aslett et al., 2010).
Trans-splicing means that every mature mRNA has an identical capped sequence of 39 nucleotides, known at the spliced leader (SL), at the 5′ end (Liang et al., 2003). Whilst no sequence consensus for polyadenylation or SL addition has been found, several studies have demonstrated that polypyrimidine-rich tracts located within intergenic regions guide SL addition and poly-adenylation, resulting in mature mRNAs (LeBowitz et al., 1993) (Figure 3). Intergenic sequences involved in the processing of T. cruzi, T. brucei and Leishmania mRNA have been thoroughly investigated by comparing mRNA with genomic sequences, initially using EST databases (Benz et al., 2005; Campos et al., 2008; Smith et al., 2008) and, more recently, using high-throughput RNA-sequencing (RNAseq) (Siegel et al., 2010; Kolev et al., 2010; Nilsson et al., 2010). In addition to providing valuable information on the mechanisms of gene expression in these organisms, these analyses also yielded data that allowed the optimization of transfection vectors used to express foreign genes and genetic manipulation in trypanosomatids.
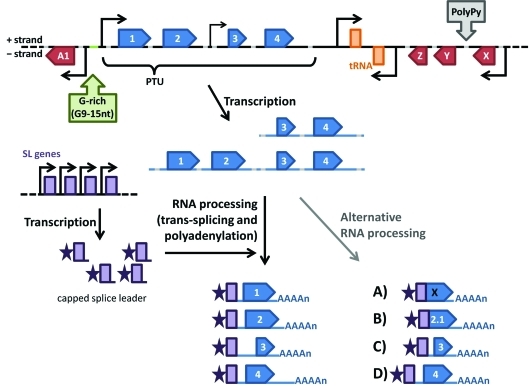
Figure 3.
Gene expression in trypanosomatids. Large clusters of unrelated genes (arrow boxes) are organized as polycistronic transcription units (PTUs) that are separated by divergent or convergent strand-switch regions. RNA Pol II transcription start sites (TSS) are usually located upstream of the first gene of the PTU (Martínez-Cavillo et al., 2004) or can be located as an internal TSS (Kolev et al., 2010). At the TSS (large bent arrow), the histone variants H2AZ and H2BV (Siegel et al., 2009), modified histones [K9/K14 acetylated and K4 tri-methylated histone (Respuela et al., 2008; Thomas et al., 2009; Wright et al., 2010) and K10 acetylated histone H4 (Siegel et al., 2009)], bromodomain factor BDF3 (Siegel et al., 2009) and transcription factors TRF4 and SNAP50 (Thomas et al., 2099) are frequently associated, with a few of these chromatin modifications also detected at internal TSS (small bent arrow) (Siegel et al., 2009; Wright et al., 2010). The polycistronic RNAs (pre-mRNAs) are individualized in monocistronic mRNAs after the addition of a capped splice leader RNA through a trans-splicing reaction coupled to polyadenylation. These processing reactions are guided by polypyrimidine tracts (PolyPy) that are present in every intergenic region. Mature mRNAs are exported to the cytoplasm where their stability and translation efficiencies are largely dependent on cis-acting elements present in their untranslated region (UTR) (Araujo et al., 2011). Transcriptomic analyses also showed that polycistronic pre-mRNAs can suffer alternative RNA processing that may result in changes in the initiator AUG, thereby altering protein translation (A), targeting and/or function (B). Alternative splicing and polyadenylation can also result in the inclusion/exclusion of regulatory elements present in the 5′ UTRs (C) or 3′ UTRs (D), thereby altering gene expression (Kolev et al., 2010; Nilsson et al., 2010; Siegel et al., 2010).
Comparative genomic analyses using the Tri-Tryp sequences have already provided interesting insights into the genetic and evolutionary bases of the distinct and shared lifestyles of these parasites. Probably the most striking finding is that the three genomes display high levels of synteny and share a conserved set of ~6,200 genes, 94% of which are arranged in syntenic directional gene clusters (El-Sayed et al., 2005a). Alignment of the deduced protein sequences of the majority of the clusters of orthologous genes across the three organisms reveals an average 57% identity between T. cruzi and T. brucei and 44% identity between T. cruzi and L. major that reflected the expected phylogenetic relationships (Lukes et al., 1997; Haag et al., 1998; Stevens et al., 1999; Wright et al., 1999). The majority of species-specific genes occurs on non-syntenic chromosomes and consists of members of large surface antigen families. Structural RNAs, retroelements and gene family expansion are also often associated with breaks in the conservation of gene synteny (El-Sayed et al., 2005a). Multi-gene family expansions are generally species-specific and most pronounced in the T. cruzi genome. As discussed below, a number of T. cruzi multi-gene families encode surface proteins, such as trans-sialidases, mucin-associated surface proteins (MASP) and mucins TcMUC and GP63 that likely play important roles in host-parasite interactions (Di Noia et al., 1995; Vargas et al., 2004; Baida et al., 2006; Bartholomeu et al., 2009). Based on their location in regions of synteny breaks these arrays may be subject to extensive rearrangements during the parasite’s evolution and are thus directly associated with the specificities of each of the three parasitic diseases.
The Genetic Diversity of T. Cruzi and the Genomes of Different Parasite Strains
Chagas disease, caused by T. cruzi, is endemic in more than 20 Latin American countries, where an estimated 10 million people are infected and the “domiciliation” of the triatomines exposes at least 90 million individuals to the risk of infection. With no vaccine or effective drug treatment available, the main strategy for control must rely on the prevention of transmission by the insect vectors and blood transfusions. The parasite proliferates in the midgut of several species of a triatomid hematophagous vector. After reaching the insect’s hindgut, epimastigote forms differentiate into non-dividing, infective metacyclic trypomastigotes that are excreted in the insect’s feces. Trypomastigotes can infect a mammalian host by passing through mucous membranes or skin lesions during feeding by the insect. Once inside the mammalian host, trypomastigotes invade different types of cells where they transform into proliferative intracellular amastigotes. After a number of cell divisions in the host cell cytoplasm, amastigotes differentiate into trypomastigotes that are released into the bloodstream after host cell rupture and, after being taken up by an insect during a blood meal, they start a new cycle (Brener, 1973) (Figure 1). The highly heterogenous T. cruzi population consists of a large number of strains with distinct characteristics related to morphology, growth rate, parasitemia curves, virulence, pathogenicity, drug sensitivity, antigenic profile, metacyclogenesis and tissue tropism (Buscaglia and Di Noia, 2003).
Despite the broad genetic diversity observed among different strains and isolates, early studies based on different genotyping strategies identified two major lineages in the parasite population, named T. cruzi I and T. cruzi II (Souto et al., 1996; Momen 1999). These divergent lineages occupy distinct ecological environments, namely, the sylvatic cycle (T. cruzi I) and the domestic cycle (T. cruzi II) of Chagas disease (Zingales et al., 1998), as well as distinct sylvatic host associations (Buscaglia and Di Noia, 2003). Further analyses led some authors to propose the sub-division of T. cruzi II into five sub-groups: T. cruzi IIa, IIb, IIc, IId and IIe (Brisse et al., 2000). Phylogenetic analyses of the T. cruzi strains became more confusing when additional data indicated the existence of not just two, but three major groups in the T. cruzi population, in addition to hybrid strains (Miles et al., 1978; Augusto-Pinto et al., 2003; de Freitas et al., 2006). After intense debate, in 2009 an international consensus recognized the existence of six major strains, also known as discrete typing units (DTUs) I–VI (Zingales et al., 2009) (Table 1). Since Chagas disease spawns a variety of clinical forms, these studies are highly relevant: understanding the genetic variation among strains can potentially explain differences in disease pathogenesis, host preferences and, most importantly, provides essential information for the identification of new drug targets and good antigenic candidates for better diagnosis and vaccine development. For instance, T. cruzi II strains and the hybrid strains belonging to T. cruzi V and VI are the predominant causes of human disease in South America (Zingales et al., 2009), whereas T. cruzi I strains are more abundant among wild hosts and vectors. Although detailed analysis of the biological and molecular factors underlying T. cruzi population structure and the epidemiology of Chagas disease are beyond the scope of this review, one must keep in mind that the genetic variability found in the T. cruzi population is an essential aspect to be considered when analyzing this parasites genome.
Table 1.
Classification of T. cruzi strains.
| Current designationa | Equivalence to former classificationsb | Examples of representative strains |
|---|---|---|
| T. cruzi I | T. cruzi I/DTU I | Sylvio X-10, Dm28c |
| T. cruzi II | T. cruzi II/DTU IIb | Esmeraldo, Y |
| T. cruzi III | T. cruzi III/DTU IIc | CM17 |
| T. cruzi IV | DTU IIa | CanIII |
| T. cruzi Vc | DTU IId | SO3 |
| T. cruzi VIc | DTU IIe | CL Brener |
DTU = discrete typing unit.
Momem (1999) (T. cruzi I and II classification), Brisse et al. (2000) (DTU I, IIa-e), de Freitas et al. (2006) (T. cruzi I, II and III),
Hybrid strains.
CL Brener, a clone derived from a hybrid T. cruzi strain belonging to T. cruzi VI, was chosen as a reference strain for the initial T. cruzi genome project. The hybrid nature of the CL Brener clone became clear only after the genome sequencing had begun, when analyses of nuclear and mitochondrial sequences showed that this strain resulted from a fusion event that had occurred between ancient genotypes corresponding to strains belonging to T. cruzi II and III groups (El-Sayed et al., 2005a; de Freitas et al., 2006). Prior to this knowledge, the choice of the clone CL Brener, initially classified as a member of sub-group IIe, was based on five characteristics: (1) it was isolated from the domiciliary vector Triatoma infestans, (2) its pattern of infectivity in mice was very well known, (3) it had preferential tropism for heart and muscle cells, (4) it showed a clear acute phase in accidentally infected humans, and (5) it was susceptible to drugs used to treat Chagas disease (Zingales et al., 1997). In addition, several genomic studies had previously used this strain for karyotype analyses (Branche et al., 2006) and the generation of physical maps and ESTs from all three stages of the parasite life cycle (Cano et al., 1995; Henriksson et al., 1995; Brandão et al., 1997; Verdun et al., 1998; Porcel et al., 2000; Cerqueira et al., 2005).
The T. cruzi CL Brener haploid genome, estimated to be 55 Mb, was sequenced using the WGS (whole genome shotgun) strategy. Because of its hybrid nature and the high level of allelic polymorphism, a 14X coverage, much higher than the usual 8–10X coverage, was required to distinguish the ambiguities derived from allelic variations from those produced by sequencing errors. In contrast to the other two Tri-Tryp genomes, the T. cruzi draft sequence (El-Sayed et al., 2005b) was published as an assembly of 5,489 scaffolds built by 8,740 contigs. Four years later, based on synteny maps for the T. brucei chromosomes, Weatherly et al. (2009) assembled the T. cruzi contigs and scaffolds initially in 11 pairs of homologous “T. brucei-like” chromosomes and, ultimately, in 41 T. cruzi chromosomes. Since trypanosomatid chromosomes do not condensate during mitosis and are therefore not visualized in metaphasic cells the predicted number of T. cruzi chromosomes was based on studies of pulsed-field gel electrophoresis (PFGE) analyses (Branche et al., 2006), which turned out to be similar to the number of assembled chromosomes. As mentioned above, the genome organization in T. cruzi is largely syntenic with the other Tri-Tryp (T. brucei and L. major) genomes, with most species-specific genes, such as surface protein gene families, occurring in internal and subtelomeric regions of non-syntenic chromosome (El-Sayed et al., 2005a).
Because of its hybrid nature, the CL Brener genome is represented by a redundant dataset since homologous regions displaying a high level of polymorphism were assembled separately, generating two set of contigs, each corresponding to one haplotype. To identify the two haplotypes, reads from the genome of the cloned Esmeraldo strain, a member of T. cruzi II, and representing one of the CL Brener parental strain (de Freitas et al., 2006), were generated. Thus, in the annotation data of the CL Brener genome, the two haplotypes are referred to as “Esmeraldo-like” or “non-Esmeraldo-like” sequences (Aslett et al., 2010).
The haploid CL Brener genome has an estimated 12,000 genes. As with the other Tri-Tryps, the T. cruzi genes are organized in long polycistronic clusters that are transcribed by RNA polymerase II and processed into monocistronic mRNAs that accumulate differentially during the various stages of the parasite life cycle. As indicated before, one of the main characteristics revealed by the complete sequence of the T. cruzi genome was the dramatic expansion of families encoding surface proteins (El-Sayed et al., 2005a). Compared to T. brucei and L. major, T. cruzi has the largest set of multi-gene families, perhaps because of its unique capacity to invade and multiply within different types of host cells. Long terminal repeat (LTR) and non-LTR retroelements and other sub-telomeric also contribute to the large proportion of repetitive sequences (50% of the genome) in this genome. The largest protein gene family encodes a group of surface proteins known as trans-sialidases (TS), with 1,430 members. TSs are surface molecules identified as virulent factors of T. cruzi that are responsible for transferring sialic acid from host sialogly-coconjugates to the terminal ß-galactose on T. cruzi mucins. Mucin-associated surface proteins (MASP) are the second largest T. cruzi gene family, with a total of 1,377 members. Although MASP sequences correspond to ~6% of the parasite diploid genome, they were only identified during annotation of the T. cruzi genome. MASPs are glycosylphosphatidylinositol (GPI)-anchored surface proteins that are preferentially expressed in trypomastigotes; these proteins are characterized by highly conserved N- and C-terminal domains and a strikingly variable and repetitive central region (Bartholomeu et al., 2009). Together with the mucin and GP63 gene families, these four gene families account for ~17% of all protein-coding genes and are organized as dispersed clusters of tandem and interspersed repeats.
Other large families consist of the previously described RHS and DGF-1 genes whose functions are unknown and which, like the TS genes, occur mostly at subtelomeric locations. Examples of other gene families with more than 10 members also present in the T. cruzi genome include glycosyltransferases, protein kinases and phosphatases, kinesins, amino acid transporters and helicases, in addition to several gene families encoding hypothetical proteins (El-Sayed et al., 2005a). The collapse of nearly identical repeats in some gene families, such as the gene cluster encoding α- and β-tubulins, meant that not all copies of the family were included in the original genome assembly.
Arner et al. (2007) described an analysis of the total genomic repetitive content of protein coding sequences and concluded that 18% of all protein coding sequences existed in 14 or more copies. In addition to the need to evade the host immune system, the existence of highly repetitive gene families in the T. cruzi genome in which a large number of gene copies can lead to the enhanced expression of various proteins may help to overcome a major problem in this genome, namely, the lack of strong promoters capable of generating high levels of mRNA from single copy genes. It is also likely that many of the striking polymorphisms among T. cruzi isolates that are reflected in several epidemiological and pathological aspects of Chagas disease are partly attributable to variability within regions containing gene families. Whole genome comparisons of distinct T. cruzi lineages are beginning to improve our understanding of this question.
Soon after the CL Brener genome was completed several groups began sequencing the genome of representative strains of other major T. cruzi lineages. As indicated above, the hybrid nature of the CL Brener genome provided data for two genomes, with “Esmeraldo-like” and “non-Esmeraldo” contigs making it possible to distinguish information from T. cruzi II and III groups, respectively (see Table 1). Recently, Franzén et al. (2011) published a draft genome sequence of Sylvio X10, a strain belonging to T. cruzi I group, which is the predominant agent of Chagas disease in Central America and in the Amazon. Although rarely isolated from humans in endemic areas in southern countries of Latin America where most cases of Chagas disease with mega-syndromes occur, T. cruzi I strains are highly abundant among wild hosts and vectors (Zingales et al., 1998; Buscaglia and Di Noia, 2003). Thus, the distinct ecological niches occupied by T. cruzi I and II strains, together with the fact these strains are highly divergent in terms of phylogenetic analysis, prompted Franzén et al. (2011) to sequence the genome of a representative of T. cruzi I group and to undertake a comparative analysis with the CL Brener genome.
In agreement with previous analyses, the Sylvio X10 genome was estimated to be ~44 Mb in size, i.e., smaller than the CL Brener genome. Indeed, smaller genomes seems to be a general feature of T. cruzi I strains (Branche et al., 2006; Franzén et al., 2011). As expected, the architectures of the two genomes were very similar, with highly conserved syntenic regions corresponding to the gene-dense “core” of the coding regions organized for long polycistronic transcription. As with the CL Brener genome, the presence of repetitive sequences meant that the Sylvio X10 genome was represented as fragmented contigs. The technical difficulties associated with the assembly of repetitive sequences meant that only about 49% of the generated Sylvio X10 sequence data was incorporated into contigs, leaving 710,109 reads that were not included in the assembly. Consequently, the draft genome of Sylvio X10 was assembled into 7,092 contigs, which is slightly less than the number of contigs reported for the draft genome of CL Brener. The alignment of these contigs to both CL Brener haplotypes showed that the mean nucleotide identity was greater between Sylvio X10 and non-Esmeraldo (98.2%) than between Sylvio X10 and Esmeraldo (97.5%). This finding agrees with previous phylogenetic analyses indicating that sequences from T. cruzi I strains are more closely related to T. cruzi III (represented by the non-Esmeraldo CL Brener haplotype) than to T. cruzi II (represented by the Esmeraldo-like haplotype) (Cerqueira et al., 2008; Ruvalcaba-Trejo and Sturm, 2011).
In contrast to the hybrid CL Brener genome, for which the amount of heterozygosity in the core genome was estimated to be 5.5% (El-Sayed et al., 2005a), the diploid Sylvio X10 genome was homozygous (< 0.08% heterozygosity). Most importantly, analysis of the core gene content of CL Brener and Sylvio X10 revealed six open reading frames that were missing in the Sylvio X10 genome. Besides these six genes, estimations based on total sequence reads indicated that several multicopy gene families, including DGF, mucin, MASP and GP63 contained substantially fewer genes in Sylvio X10 than in CL Brener. A 5.9 Mb size difference between the Sylvio X10/1 and CL Brener genomes largely reflected the expansion of these gene families. However, the extent to which these genomic variations are related to strain differences in host preference and the ability to cause Chagas disease remains to be determined.
The advent of next-generation sequencing technologies has ushered in a new era in comparative sequencing by allowing the exploration of a wide range of evolutionary and pathological questions within the T. cruzi lineage. Several groups have initiated sequencing analyses of additional T. cruzi isolates. A consortium of laboratories funded by the National Institutes of Health/National Institutes of Allergy and Infectious Diseases (NIAID) and the National Human Genome Research Institute (NHGRI) is sequencing T. cruzi strains representative of each one of the six main groups, such as Esmeraldo (T. cruzi II), 3869 (T. cruzi III), Can III (T. cruzi IV), NRcl3 (T. cruzi V) and Tula cl2 (T. cruzi VI) (N. El-Sayed, personal communication). Our laboratory has been involved in the sequencing of another T. cruzi I strain (Dm28c) and CL-14, a non-virulent strain that belongs to the T. cruzi VI group (S. Teixeira, unpublished). In contrast to CL Brener, BALB/c mice injected with CL-14 trypomastigotes showed no parasitemia but developed high resistance against a lethal challenge with virulent trypomastigotes from the CL Brener or Y strains (Lima et al., 1995). Our goal in this work is to use comparative analyses of the CL Brener and CL-14 genomes to identify potential sequences that can restore the virulence of CL-14 and then test these in transfection protocols.
In addition to investigations of the nuclear genome, several studies have examined the mitochondrial genome of kinetoplastids which contains a mass of concatenated DNA known as kinetoplast DNA (kDNA) that is easily identified near the insertion of the flagellum (Brener, 1973). In T. cruzi, kDNA consists of a highly structured disk-shaped network of thousands of concatenated mini-circles 0.5–10 kb in size and dozens of concatenated maxicircles 20–40 kb in size. Whereas minicircle sequences are present exclusively in kinetoplastids, maxicircles are the homologues of mtDNA molecules found in other eukaryotes (Lukes et al., 1997). Following publication of the T. cruzi genome, Westenberger et al. (2006) described the complete sequences of maxicircle DNAs corresponding to groups T. cruzi II (from sequences of the Esmeraldo strain) and III (from CL Brener sequences). As with other trypanosomatid mitochondrial genes, sequence analyses showed that T. cruzi maxicircle DNA contained frameshift errors in most of its genes that were corrected at the RNA level by a complex U-insertion/deletion process known as RNA editing (Hajduk et al., 1993). Key elements of this repair process include gRNAs (guide RNAs) which are encoded mainly by minicircles, although a few gRNA sequences are also present in maxicircles. The gRNAs hybridize to the 3′ end of a target message and undertake direct U insertion and deletion by the so-called editosome machinery (Stuart and Panigrahi, 2002).
The complete sequences of the 25 kb T. cruzi maxi-circles revealed 18 tightly clustered mitochondrial protein-coding genes and two rRNA genes that were syntenic with previously sequenced maxicircles of T. brucei and Leishmania tarentolae. Fifteen of the 18 protein-coding genes were edited. Outside the coding region, strain-specific repetitive regions and a variable region that was unique for each strain were identified (Westenberger et al., 2006). More recently, comparative analyses of the mitochondrial genomes of T. cruzi I, II and III were reported after Ruvalcaba-Trejo and Sturm (2011) generated the sequence of the coding region of the maxicircle from Sylvio X10. In agreement with the nuclear genomic analysis, phylogenetic analysis of the maxicircle coding regions supported a close evolutionary relationship between T. cruzi I and III. Based on their mitochondrial DNA analyses, these authors proposed a model in which an ancestral strain belonging to T. cruzi I provided the maxicircle for the progeny of a TcI-TcII hybridization event that resulted in the generation of T. cruzi III and T. cruzi IV strains. A subsequent ‘back-cross’ hybridization between T. cruzi II and T. cruzi III strains resulted in the T. cruzi V and VI strains, such as CL Brener, that carry the maxicircle from their T. cruzi III ancestor.
Comparative Genomics of Leishmania Species That Cause Distinct Forms of Leishmaniasis
Leishmania spp. are parasitic protozoa transmitted by the bites of phlebotomine sand flies that are endemic in tropical and subtropical regions worldwide. More than 20 species are responsible for a wide spectrum of diseases, known as leishmaniasis (Murray et al., 2005). Parasites in this genus are classified into two subgenera according to the part of the sandfly gut where colonization and development occur: the subgenus Leishmania (Leishmania) consists of parasites with mid and foregut development, whereas the subgenus Leishmania (Viannia) consists of parasites that undergo hindgut development (Lainson et al., 1977; Bates, 2007). Depending on the species of Leishmania, infection of humans may result in diverse clinical forms of leishmaniasis with symptoms ranging from self-healing cutaneous lesions (L. major/L. tropica/L. mexicana) to fatal visceral leishmaniasis (L. donovani/L. infantum/L. chagasi). Infection by Leishmania can also result in mucosal leishmaniasis (mainly caused by L. braziliensis) and diffuse cutaneous leishmaniasis (mainly caused by L. amazonensis/L. guyanensis/L. aethiopica) (Desjeux, 1996). In addition to the species of Leishmania, other factors such as the genetic variability of the human host may determine the disease tropism and clinical manifestations in leishmaniasis (Blackwell et al., 2009; Sakthianandeswaren et al., 2009).
The World Health Organization (WHO) estimates that there are over two million new cases of leishmaniasis each year, with more than 360 million people at risk of contracting this disease in 88 countries on five continents (Asia, Africa, Europe, North America and South America) (www.who.int/tdrdiseases/leish). As part of their life cycle, Leishmania spp. alternate between the alimentary tract of the sandfly vector, where they grow as extracellular flagellated promastigotes and differentiate into infective non-dividing metacyclic forms, and the phagolysosome of the vertebrate host macrophages, where they differentiate into aflagellated, replicative amastigotes (Figure 1). There is no effective vaccine against Leishmania and the available therapeutic arsenal is extremely limited (Mauel, 2002). Thus, completion of the genome sequences of several Leishmania species (Ivens et al., 2005; Peacock et al., 2007) represents a long awaited aspiration for groups involved in the discovery and development of new drugs and vaccine targets.
Leishmania major Friedlin was chosen as the Leishmania reference strain for the Tri-Tryp genome project. The L. major haploid genome (~32.8 Mb) is distributed among 36 relatively small chromosomes ranging from 0.28 to 2.8 Mb in size (Wincker, 1996) and was sequenced after shotgun cloning of large DNA fragments derived from chromosomal bands separated in agarose gels. Prior to publication of the Tri-Tryp genome sequence, the complete sequences of chromosomes 1 and 3 from L. major were published (Myler et al., 1999; Worthey et al., 2003) and an optical map of the entire genome was generated (Zhou et al., 2004). In 2007, the complete genomes of two other Leishmania species, L. infantum and L. braziliensis, were also described (Peacock et al., 2007). Leishmania infantum, also known as L. chagasi in Latin America, was chosen as the second Leishmania species to have its genome sequenced on the basis of its virulence in animals, transmissibility in sandflies and adaptability to laboratory experimentation (Denise et al., 2006). This species is the causative agent of visceral leishmaniasis, the most serious form of the disease and frequently fatal if left untreated. The New World species L. braziliensis, within the subgenus L. (Viannia), is the third and most divergent species sequenced. The L. infantum and L. braziliensis genome sequences were obtained by the whole-genome shotgun approach with five- and six-fold coverage, respectively (Peacock et al., 2007). Importantly, the three complete genomes are from strains that cause distinct types of leishmanial diseases, are adapted for maintenance and manipulation in the laboratory and are also frequently used in studies in vitro and in animal models of infection (Laurentino et al., 2004; Ivens et al., 2005; Denise et al., 2006). The complete sequences of all three Leishmania genomes can be accessed in the Tri-Tryp database; sequencing of the genome from a fourth species (L. mexicana) is in progress.
Surprisingly, comparative genomic studies of the three evolutionarily and geographically distinct species, L. major and L. infantum (Old World species) and L. braziliensis (New World species), showed very little divergence among the species in terms of genomic sequence and organization (Peacock et al., 2007; Lynn and McMaster, 2008), despite a divergence of 20–100 million years within the Leishmania genus (Lukes et al., 1997). The haploid genome of the three species has an estimated 8,300 genes, with more than 99% of them maintaining synteny in the three Leishmania genomes (Peacock et al., 2007). As with the other Tri-Tryps, the majority of Leishmania genes are annotated as genes of unknown function. Of the 8,300 genes, only 200 were identified as being differentially distributed between the three genomes. The L. braziliensis genome possesses 47 genes that are absent from the other two species, while 27 and 5 genes are specific for the L. infantum and L. major genomes, respectively. Such high conservation in overall genome sequences and genome synteny indicates that the Leishmania genome is highly stable and has not undergone large genomic rearrangements during speciation. This finding also suggests that only a few species-specific parasite genes may contribute to differential pathogenesis and tissue tropism (Peacock et al., 2007; Smith et al., 2007).
Thus far, the only gene for which there is experimental evidence indicating direct involvement in the differential tropism among Leishmania diseases is the A2 locus. Initially identified as an amastigote-specific gene family in L. donovani, A2 has been shown to play a major role in parasite virulence and visceralization (Zhang et al., 2003). Multiple copies of the A2 gene alternating with a distinct gene termed the A2rel gene are found in the genome of several species of the L. donovani group that is responsible for visceral diseases. Although its precise function is still unknown, the product of the A2 gene, an endoplasmic reticulum protein with a large repetitive domain, may be related to the parasite stress response (McCall and Matlashewski, 2010). In L. major, which does not cause visceral leishmaniasis, the A2 gene is a pseudogene and introduction of the L. donovani A2 gene into L. major enhanced the ability of L. major to survive in visceral organs of susceptible BALB/c mice (Zhang et al., 2003). More recently, the expression of A2 in L. tarentolae, a lizard parasite that is not pathogenic in mammals, significantly increased the infectivity of this species and enhanced its ability to survive in the liver of BALB/c mice (Mizbani et al., 2011). In addition to experiments suggesting a possible role for the A2 gene in the differential tropism of cutaneous and visceral leishmania parasites, the A2 gene has been used as a promising vaccine candidate and diagnostic antigen (Fernandes et al., 2008).
Another locus possibly involved in macrophage invasion and that varies considerably among the three Leishmania genomes is the GP63 locus. Also known as the major surface protease (MSP), GP63 constitutes a family of surface metalloproteases expressed in all trypanosomatids examined so far (Yao et al., 2003). GP63 sequences identified in the three Leishmania genomes and in the T. cruzi and T. brucei genomes vary in their gene copy number, and this may have implications for differences in the disease phenotype (Voth et al., 1998). A similar conclusion may apply to the amastin multi-gene family that encodes a family of amastigote-specific, highly glycosylated hydrophobic surface proteins also present in T. cruzi but which has been greatly expanded in the genus Leishmania (Teixeira et al., 1995; Jackson, 2010). Interestingly, in the T. brucei genome, which does not have an intracellular stage, only two copies of a highly divergent amastin sequence are present (Jackson, 2010). The identification of these species-specific genes represents an initial step towards the characterization of parasite factors that may determine the specificities of each type of parasitic infection. On the other hand, antigens common to all Leishmania species could be used as potential vaccine candidates (Peacock et al., 2007). However, apart from differences in gene sequences, it is possible that the distinct clinical manifestations observed in the different parasitic diseases may be a consequence of differential gene expression that occurs throughout the various stages of life cycle in each parasite species.
As discussed above, Leishmania genes are arranged in the genome as directional gene clusters that resemble prokaryotic polycistronic transcription units (Martínez-Calvillo et al., 2004). This type of gene organization and polycistronic transcription have profound implications on the regulation of gene expression, which must rely on post-transcriptional mechanisms (Boucher et al., 2002; Myung et al., 2002; Holzer et al., 2006; Leifso et al., 2007). Since the dependency on promoter-based transcription initiation mechanisms for the control of mRNA levels is greatly reduced, greater emphasis is placed on post-transcriptional regulatory mechanisms controlling mRNA stability and translation, as well as protein turnover (Clayton, 2002). Despite significant differences in life stage morphology, biochemical properties and disease phenotypes, comparative gene expression studies have revealed surprisingly few differences in gene expression when mRNA levels in different life-cycle stages or in the same stage but in different Leishmania species were compared (Peacock et al., 2007). Global interspecies analyses in L. major and L. infantum have shown that only 10%–12% of differentially expressed genes are unique to each species (Rochette et al., 2008; Depledge et al., 2009). A more careful examination of the protein expression patterns and the elucidation of regulatory mechanisms will provide unique insights into this important aspect of the parasite’s biology. This information will also improve our understanding of the host-parasite interaction and lead to the development of new strategies for contolling leishmaniasis.
Eukaryotic genomes contain an abundance of repeated DNA and some of these repeated sequences are mobile elements. Transposable elements (TEs) are defined as DNA sequences that are able to move from one location to another in the genome and have been identified in all organisms (prokaryotic and eukaryotic) examined so far. TEs can occupy a high proportion of a species’ genome. Retroposons, also known as non-long-terminal-repeat (LTR) retrotransposons, are ubiquitous elements that transpose through an RNA intermediate and are found in the genomes of most eukaryotes (Ivens et al., 2005; Peacock et al., 2007). Trypanosoma brucei and T. cruzi contain long autonomous retroposons of the ingi clade (Tbingi and L1Tc, respectively) and short nonautonomous truncated versions (TbRIME and NARTc, respectively), as well as degenerate ingi-related retroposons devoid of coding capacity (DIREs) that represent the most abundant transposable elements in these genomes (< 3% of the nuclear genome).
In contrast, L. major contains only remnants of extinct retroposons (LmDIREs) and short nonautonomous heterogenous elements (LmSIDERs). Recently, small degenerate retroposons (< 0.55 kb) containing the “79-bp signature” known as LmSIDERs (for short interspersed degenerate retroposons) have also been identified in the genomes of L. major (Ivens et al., 2005), L. infantum and L. braziliensis (Peacock et al., 2007). Unexpectedly, in L. braziliensis, the site-specific non-LTR retroposon SLACS/CZAR, which is associated with tandemly repeated spliced leader sequences in an arrangement similar to that of the SLACS or CZAR element in T. brucei or T. cruzi, respectively, is fully active (Aksoy et al., 1987; Villanueva et al., 1991). However, in contrast to the African trypanosome genomes and similar to L. major (Ivens et al., 2005) and L. infantum, no potentially active ingi-related retroposons were detected in the L. braziliensis genome (Bringaud et al., 2009).
Mobile elements are involved in creating mutations and genomic rearrangements and, in many eukaryotes, these effects can be regulated through an RNA silencing mechanism such as RNA interference (RNAi) (Shi et al., 2004b; Girard and Hannon, 2008). Since first reported in 1998 (Fire et al., 1998), RNAi has swept through all fields of eukaryotic biology and has proven to be a very useful tool for analyzing gene function in a variety of organisms in which the introduction or expression of short double-stranded RNAs leads to the rapid destruction of cognate mRNAs (Figure 4A) (Ngo et al., 1998). This approach has a number of advantages: it is fast, requires very little sequence information, and reduces the expression of multiple gene copies, an action that is especially advantageous in asexual diploid organisms (LaCount et al., 2000; Shi et al., 2000; Wang et al., 2000).
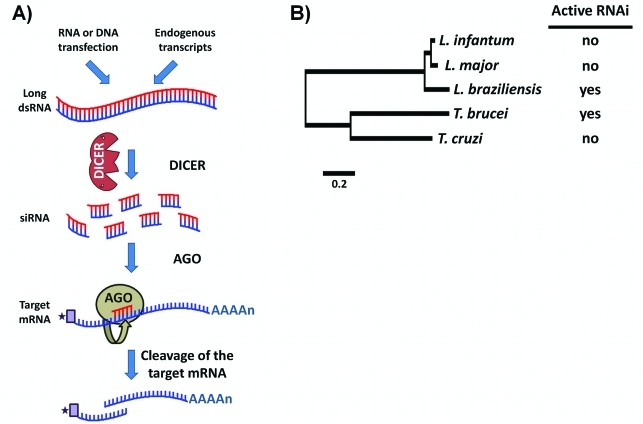
Figure 4.
Retention and loss of RNAi genes in trypanosomatids. (A) Schematic representation of the mechanism of RNA interference (RNAi) present in some trypanosomes. Double-stranded RNA (dsRNA) derived endogenously or by transfection procedures is processed by an RNase III enzyme known as DICER into small interfering dsRNA molecules (siRNA) 19–23 nt long. siRNAs associate with Argonaute (AGO), the catalytic core of the RISC (RNA-induced silencing complex), with one siRNA strand being released and the guide siRNA strand then mediating the degradation or inhibiting the translation of the target mRNA. Genes encoding components of the RNAi machinery occur only in T. brucei and in a sub-group of Leishmania. (B) Phylogenetic analysis of trypanosomatid species based on the predicted protein sequences of the housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and ubiquitin. The neighbor-joining tree was generated using sequences found in the parasite genome databases (www.genedb.org) and MEGA4 software. The absence or presence of genes encoding components of active RNAi machinery (as identified by Lye et al., 2010) is indicated on the right.
Soon after it was described in C. elegans, RNAi was rapidly identified in T. brucei (Ngo et al., 1998). As discussed below, RNAi has proven to be a powerful new tool for functional genomic studies in T. brucei. Unexpectedly, experimental evidence as well as searches of genome databases quickly showed that RNAi is absent in L. major and T. cruzi (Robinson and Beverley, 2003; DaRocha et al., 2004). It came as a surprise when Peacock et al. (2007) revealed that the genome of L. braziliensis retained key genes involved with RNAi. These authors showed that L. braziliensis genome contains orthologs carrying domains characteristic of the Dicer protein as well as genes with the typical argonaute domains PAZ and PIWI, the latter containing conserved amino acid residues that are essential for functional TbAGO1 (Shi et al., 2004a). In addition, an N-terminal RGG domain, present in TbAGO1 and shown to be essential for its association with polyribosomes, is also present in the L. braziliensis Ago1 gene (Shi et al., 2004a). More recently, Lye et al. (2010) demonstrated that the RNAi pathway is functional in L. braziliensis and in other species within the Leishmania subgenus Viannia (L. guyanensis and L. panamensis) (Figure 4B). Thus, this divergent species of Leishmania appear to have retained not only the mechanisms for (RNAi)-mediated regulation but also potentially active retroposons (Peacock et al., 2007), which might have assisted to create the greater divergence within the L. braziliensis genome compared with the other Leishmania species. For molecular parasitologists, these findings came as very good news since, as shown in T. brucei, the efficacy of RNAi knockdown as a tool for systematic analysis of gene function is now also applicable in some species of Leishmania.
Functional Genomics of T. Brucei and Gene Expression Studies in a High Throughput Era
Infection by T. brucei occurs after metacyclic trypomastigotes are injected into the bloodstream by the bite of a tsetse fly. In the mammalian host, the parasite differentiates into long slender trypomastigotes (bloodstream form -BSF) that divide, colonize the body fluids and can transform into a nonproliferative bloodstream form, also known as the short stump form. After a blood meal, trypomastigotes acquired by the insect vector differentiate into procyclic trypomastigotes in the gut, replicate and then migrate to salivary glands where they transform into metacyclic trypomastigotes (Matthews, 2005) (Figure 1). The ability of the BSF to invade the central nervous system leads to the neurological manifestations associated with sleeping sickness.
In contrast to T. cruzi and Leishmania, T. brucei develops extracellularly throughout its entire life cycle. Its direct exposure to a strong antibody response in the bloodstream requires a sophisticated immune evasion protocol, known as variant surface protein (VSG) switching (Pays et al., 2007). VSGs are bloodstream-specific surface proteins with a hypervariable N-terminal domain that is exposed extracellularly and a more conserved C-terminal domain buried in the parasite’s surface coat (Van der Ploeg et al., 1982; Turner and Barry, 1989). Since they are tightly packed at the surface, VSGs are able to shield other invariant surface proteins from attack by the immune system. The tactic for evasion is based on antigenic variation in which the expression of a specific VSG is replaced by another gene from a supply of almost thousand copies at a rate of approximately one event per 100 cell divisions (Van der Ploeg et al., 1982; Turner and Barry, 1989). VSG genes are thus a family of T. brucei-specific genes whose monoallelic expression distinguishes African trypanosomes from the other two groups of pathogenic trypanosomatids; the lack of antigenic variation in T. cruzi and Leishmania spp. means that these species must evade the host’s immune system by entering and multiplying inside host cells.
After completion of the T. b. brucei genome, it was found that of the 9,068 predicted genes, including 904 pseudogenes, there were 1,700 T. brucei-specific genes, 806 of them (or 9% of the whole genome) corresponding to VSGs (El-Sayed et al., 2005b). Several VSG genes occur within hundreds of mini-chromosomes (50–150 kb in size) that are also part of the T. brucei genome. Surprisingly, analyses of all VSG sequences showed that only 57 are fully functional genes and have all the recognizable features of a typical VSG (Berriman et al., 2005). It has been long known that VSG variability can be generated in T. brucei by creating mosaic genes that include pseudogenes (Roth et al., 1989). Besides producing variability in VSG epitopes, these rearrangements may have the capacity to generate protein with new functions, as in the case of the SRA gene discussed below (Vanhamme et al., 2003).
As part of their “life in the bloodstream”, African trypanosomes that are pathogenic to humans have developed a mechanism to withstand other types of attack from the host immune system. In contrast to T. brucei brucei, which do not infect humans, T. b. gambiense and T. b. rhodesiense are resistant to trypanolytic factors (TLF) present in normal human serum (NHS). TLF activity has been attributed to apolipoprotein L1 (ApoL1) and haptoglobin (Hp)-related protein found in high density lipoprotein (for a review, see Wheeler, 2010). NHS resistance in T. b. rhodesiense is conferred by a truncated VSG known as serum resistance associated (SRA) protein which is located in endosomes and binds and neutralizes TLF (De Greef and Hamers, 1994; De Greef et al., 1992; Vanhamme et al., 2003; Pérez-Morga et al., 2005; Wheeler, 2010). Trypanosoma b. gambiense lacks the SRA gene but still infects humans. The killing of T. b. brucei requires the binding of TLF-1 and trafficking to the parasite acidic lysosome. It has been proposed that, in T. b. gambiense, changes in the receptor that binds TLF, as well as decreased expression of this gene, are responsible for the resistance of this T. brucei sub-species to attack by the human innate immune system (Kieft et al., 2010).
Since T. b. brucei is harmless to humans and T. b. gambiense is the most clinically relevant subspecies, Jackson et al. (2010) sequenced the genome of T. b. gambiense using the whole-genome shotgun approach combined with bacterial artificial chromosome large insert sequencing. Comparison of the sequences from a draft genome assembly of 281 contigs (~22.1 Mb) with the T. b. brucei genome data (Berriman et al., 2005) revealed a very similar composition in terms of gene content, gene synteny and sequence identity (86.4% of T. b. gambiense coding sequences vary by < 1% from their orthologs in T. b. brucei). Even though these subspecies behave differently in humans, there were very few differences in their genomes. One of these differences involved a gene encoding a putative iron-ascorbate oxidoreductase that is specific to a few T. b. brucei strains and is also absent in other trypanosomatids (L. major and T. cruzi). Thus, it seems that not only differences in gene content per se, but also individual single nucleotide polymorphisms (indels) and variations in gene expression, or a combination of these factors, may contribute to this phenotypic variation (Jackson et al., 2010).
Coordinated changes in gene expression are vital for trypanosomes since they must deal with rapid changes in their environment (nutrient availability, temperature, host defenses, the presence of drug, etc.) and be able to disseminate by alternating through different hosts. In addition, unique mechanisms for regulating gene expression are required since there is an almost complete absence of transcriptional control at the level of initiation. The lack of transcriptional regulatory elements, including a typical RNA polymerase II promoter, led to the conclusion that most factors involved in controlling gene expression act at the post-transcriptional level. By using approaches such as differential display, RNA fingerprinting, differential screening of cDNA libraries, random sequencing of cDNA clones and DNA microarrays, several groups have shown that significant changes in mRNA levels occur during the life cycle of all Tri-Tryps (Teixeira et al., 1995; El-Sayed et al., 1995; Mathieu-Daudé et al., 1996; Diehl et al., 2002; Cerqueira et al., 2005; Kabani et al., 2009). Experimental evidence also indicates that, in addition to mRNA stability, changes in polysomal mobilization constitute an important mechanism for regulating gene expression (Alves et al., 2010). With the advent of next generation sequencing technologies such as RNAseq, a global description of gene expression patterns in T. brucei has finally been achieved (Kolev et al., 2010; Nilsson et al., 2010; Siegel et al., 2010; Veitch et al., 2010). This approach has provided much more complete information about mRNA structure and expression levels, including the patterns of spliced leader (SL) and poly-A additions, as well as alternative pre-mRNA processing. Moreover, these global gene expression analyses have confirmed that only two T. brucei genes (poly-A polymerase and DNA/RNA helicase) contain introns.
Nilsson et al. (2010) showed that 2,500 alternative splicing events occur during processing of T. brucei genes, with a large number of these being regulated during the life cycle (a total of 600 genes have transcripts with more than one trans-spliced variant). The alternative splicing reactions can alter the message dramatically, as shown by Nilsson et al. (2010) and represented in Figure 3, with changes in the SL addition site leading to alterations in regulatory elements in the 5′UTR (resulting in the modification of gene expression) or the initiator AUG (generating a different N-terminus or even causing a complete change in the translated ORF). Aminoacyl tRNA synthetase transcripts are interesting examples of trans-spliced variants since differences in the enzyme N-terminus result in changes in the protein targeting signal: distinct mRNAs produce proteins that are directed to mitochondria or remain in the cytoplasm (Nilsson et al., 2010). Likewise, alternative splicing is a potential mechanism for dual localization of the T. cruzi LYT1 gene (Benabdellah et al., 2007). In addition to providing a mechanism for generating changes in the proteome, such flexibility in the selection of splicing sites is compatible with evidence that polycistronic transcription may also initiate at internal sites in gene clusters since correct processing of the 5′ end of mRNA can occur at various points in the primary transcript (Kolev et al., 2010).
The mechanisms involved in transcription initiation in trypanosomes have always been an intriguing question. Recently, chromatin immunoprecipitation in combination with conventional Sanger sequencing (Respuela et al., 2008) or next-generation sequencing technology (Siegel et al., 2009; Thomas et al., 2009; Wright et al., 2010) has provided a much awaited analysis of the distribution patterns of modified histones throughout the T. brucei genome. As summarized in Figure 3, the divergent strand-switch regions (SSR) have been found to contain an enrichment of histone variants (H2AZ and H2BV), acetylated histone 4 at lysine 10 (H4K10ac), histone 3 acetylated at residues K9 and K14 and trimethylated at K4, the bromodomain factor BDF3, and the transcription factors TRF4 and SNAP50 (Respuela et al., 2008; Siegel et al., 2009; Thomas et al., 2009; Wright et al., 2010). The histone variants H3V and H4V are present at transcription termination sites (Siegel et al., 2009). Similar patterns of chromatin modifications from transcription start sites (TSS) located at the beginning of polycistronic units were also found internally in the same polycistronic unit, suggesting the presence of internal TSS, in agreement with RNAseq data (Siegel et al., 2009; Kolev et al., 2010; Wright et al., 2010).
The increasing number of published genomes and transcriptomic data, partly as a consequence of the introduction of new sequencing platforms, has created enormous challenges for the field of functional genomics. One of the most useful techniques that has been used to identify gene function in T. brucei is a combination of the inducible system mediated by T7 RNA polymerase and the tetracy-cline repressor (Wirtz et al., 1999) in combination with RNAi gene knockdown. As indicated before, in contrast to T. cruzi and L. major, RNAi is functional in T. brucei (Ngo et al., 1998; Robinson and Beverley 2003; DaRocha et al., 2004; Lye et al., 2010). The first large scale functional genomics study using RNAi was reported by Morris et al. (2002), who transfected T. brucei procyclic forms with a random RNAi library. This library was generated by using a construct to inducibly express dsRNA via two head-to-head tetracycline-regulated T7 RNA polymerase promoters. By selecting parasites that expressed the dsRNA but were unable to bind lectin, these authors identified clones with a reduced glycosylation of surface proteins and showed that these clones had lower expression of hexokinase (Morris et al., 2002).
A few years later, the functions of 197 ORFs from T. b. brucei chromosome I were tested by RNAi. RNAi-induced parasites were tested for growth, nuclear and kinetoplast abnormalities by DAPI staining, and a pleio-tropic phenotype, morphology and motility. At least one of these phenotypes was found in 68 individual knockdowns (Subramaniam et al., 2006). In a similar approach, Portman et al. (2009) identified novel components of the para-flagellar rod (PFR) structure by comparing proteomic profiles before and after ablating the expression of individual PFR proteins. The silencing of PFR1 and PFR15 was evaluated by two-dimensional difference gel electrophoresis, and the spots with a two-fold change in volume were subjected to tandem MS protein identification. Proteomic analysis combined with RNAi knockdown led to the identification of 30 proteins as potential PFR components, 20 of which were novel proteins. High-throughput cloning systems also enhance functional analyses. By using the Gateway® technology to create yeast two-hybrid vectors, eight non-redundant protein-protein interactions were detected among proteins with a PFR structure (Lacomble et al., 2009). These authors were able to construct a map showing the complex interaction of PFR proteins.
The availability of efficient methods for genetic manipulation and for testing gene function through RNAi knockdown has led to major advances in genomic, transcriptomic and proteomic analyses of T. brucei, and has made this species a model organism for studying basic aspects of trypanosomatid biology. More recently, with the discovery of functional RNAi machinery in L. braziliensis and other Leishmania species, gene function studies can now be conducted in this group and will soon provide valuable new information about Leishmania-specific genes. However, as is becoming increasingly apparent from the data generated by comparative genomic analyses, each of the trypanosomatid species has its peculiarities. Researchers thus face the challenge of developing new protocols specific for studying each parasite and its corresponding diseases. With our current knowledge of each Tri-Tryp disease and the new research methods that are being developed we can expect many new studies to emerge from hypotheses based on the Tri-Tryp genomic data. As a consequence of these new findings, better methods of controlling and preventing these diseases will follow.
Acknowledgments
The work from SMT and RMCP was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), Instituto Nacional de Ciencia e Tecnologia de Vacinas (INCTV) and the Howard Hughes Medical Institute (HHMI). The work from WDR and MMKM was supported by FAPEMIG, Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Paraná (Fundação Araucária), CAPES/Reuni, PPSUS/MS and CNPq.
References
- Aksoy S, Lalor TM, Martin J, Van der Ploeg LH, Richards FF. Multiple copies of a retroposon interrupt spliced leader RNA genes in the African trypanosome, Trypanosoma gambiense. EMBO J. 1987;6:3819–3826. doi: 10.1002/j.1460-2075.1987.tb02718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves LR, Avila AR, Correa A, Holetz FB, Mansur FC, Manque PA, de Menezes JP, Buck GA, Krieger MA, Goldenberg S. Proteomic analysis reveals the dynamic association of proteins with translated mRNAs in Trypanosoma cruzi. Gene. 2010;452:72–78. doi: 10.1016/j.gene.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Araujo PR, Burle-Caldas GA, Silva-Pereira RA, Bartholomeu DC, DaRocha WD, Teixeira SM. Development of a dual reporter system to identify regulatory cis-acting elements in untranslated regions of Trypanosoma cruzi mRNAs. Parasitol Int. 2011;60:161–169. doi: 10.1016/j.parint.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Arner E, Kindlund E, Nilsson D, Farzana F, Ferella M, Tammi MT, Andersson B. Database of Trypanosoma cruzi repeated genes: 20,000 additional gene variants. BMC Genomics. 2007;8:e391. doi: 10.1186/1471-2164-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusto-Pinto L, Teixeira SM, Pena SD, Machado CR. Single-nucleotide polymorphisms of the Trypanosoma cruzi MSH2 gene support the existence of three phylogenetic lineages presenting differences in mismatch-repair efficiency. Genetics. 2003;164:117–126. doi: 10.1093/genetics/164.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baida RC, Santos MR, Carmo MS, Yoshida N, Ferreira D, Ferreira AT, El Sayed NM, Andersson B, da Silveira JF. Molecular characterization of serine-, alanine-, and proline-rich proteins of Trypanosoma cruzi and their possible role in host cell infection. Infect Immun. 2006;74:1537–1546. doi: 10.1128/IAI.74.3.1537-1546.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeu DC, Cerqueira GC, Leão AC, DaRocha WD, Pais FS, Macedo C, Djikeng A, Teixeira SM, El-Sayed NM. Genomic organization and expression profile of the mucin-associated surface protein (masp) family of the human pathogen Trypanosoma cruzi. Nucleic Acids Res. 2009;37:3407–3417. doi: 10.1093/nar/gkp172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PA. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int J Parasitol. 2007;37:1097–1106. doi: 10.1016/j.ijpara.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdellah K, González-Rey E, González A. Alternative trans-splicing of the Trypanosoma cruzi LYT1 gene transcript results in compartmental and functional switch for the encoded protein. Mol Microbiol. 2007;65:1559–1567. doi: 10.1111/j.1365-2958.2007.05892.x. [DOI] [PubMed] [Google Scholar]
- Benz C, Nilsson D, Andersson B, Clayton C, Guilbride DL. Messenger RNA processing sites in Trypanosoma brucei. Mol Biochem Parasitol. 2005;143:125–134. doi: 10.1016/j.molbiopara.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Blackwell JM, Melville SE. Status of protozoan genome analysis: Trypanosomatids. Parasitology. 1999;118(Suppl):S11–14. doi: 10.1017/s0031182099004163. [DOI] [PubMed] [Google Scholar]
- Blackwell JM, Fakiola M, Ibrahim ME, Jamieson SE, Jeronimo SB, Miller EN, Mishra A, Mohamed HS, Peacock CS, Raju M, et al. Genetics and visceral leishmaniasis: Of mice and man. Parasite Immunol. 2009;31:254–266. doi: 10.1111/j.1365-3024.2009.01102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher N, McNicoll F, Dumas C, Papadopoulou B. RNA polymerase I-mediated transcription of a reporter gene integrated into different loci of Leishmania. Mol Biochem Parasitol. 2002;119:153–158. doi: 10.1016/s0166-6851(01)00410-8. [DOI] [PubMed] [Google Scholar]
- Branche C, Ochaya S, Aslund L, Andersson B. Comparative karyotyping as a tool for genome structure analysis of Trypanosoma cruzi. Mol Biochem Parasitol. 2006;147:30–38. doi: 10.1016/j.molbiopara.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Brandão A, Urmenyi T, Rondinelli E, Gonzalez A, de Miranda AB, Degrave W. Identification of transcribed sequences (ESTs) in the Trypanosoma cruzi genome project. Mem Inst Oswaldo Cruz. 1997;92:863–866. doi: 10.1590/s0074-02761997000600024. [DOI] [PubMed] [Google Scholar]
- Brener Z. Biology of Trypanosoma cruzi. Annu Rev Microbiol. 1973;27:347–382. doi: 10.1146/annurev.mi.27.100173.002023. [DOI] [PubMed] [Google Scholar]
- Bringaud F, Berriman M, Hertz-Fowler C. Trypanosomatid genomes contain several subfamilies of ingi-related retroposons. Eukaryot Cell. 2009;8:1532–1542. doi: 10.1128/EC.00183-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse S, Dujardin JC, Tibayrenc M. Identification of six Trypanosoma cruzi lineages by sequence-characterised amplified region markers. Mol Biochem Parasitol. 2000;111:95–105. doi: 10.1016/s0166-6851(00)00302-9. [DOI] [PubMed] [Google Scholar]
- Buscaglia CA, Di Noia JM. Trypanosoma cruzi clonal diversity and the epidemiology of Chagas’ disease. Microbes Infect. 2003;5:419–427. doi: 10.1016/s1286-4579(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Campos PC, Bartholomeu DC, DaRocha WD, Cerqueira GC, Teixeira SM. Sequences involved in mRNA processing in Trypanosoma cruzi. Int J Parasitol. 2008;38:1383–1389. doi: 10.1016/j.ijpara.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Cano MI, Gruber A, Vazquez M, Cortés A, Levin MJ, González A, Degrave W, Rondinelli E, Zingales B, Ramirez JL, et al. Molecular karyotype of clone CL Brener chosen for the Trypanosoma cruzi genome project. Mol Biochem Parasitol. 1995;71:273–278. doi: 10.1016/0166-6851(95)00066-a. [DOI] [PubMed] [Google Scholar]
- Cerqueira GC, DaRocha WD, Campos PC, Zouain CS, Teixeira SM. Analysis of expressed sequence tags from Trypanosoma cruzi amastigotes. Mem Inst Oswaldo Cruz. 2005;100:385–389. doi: 10.1590/s0074-02762005000400008. [DOI] [PubMed] [Google Scholar]
- Cerqueira GC, Bartholomeu DC, DaRocha WD, Hou L, Freitas-Silva DM, Machado CR, El-Sayed NM, Teixeira SM. Sequence diversity and evolution of multigene families in Trypanosoma cruzi. Mol Biochem Parasitol. 2008;157:65–72. doi: 10.1016/j.molbiopara.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribb P, Serra E. One- and two-hybrid analysis of the interactions between components of the Trypanosoma cruzi spliced leader RNA gene promoter binding complex. Int J Parasitol. 2009;39:525–532. doi: 10.1016/j.ijpara.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Cribb P, Esteban L, Trochine A, Girardini J, Serra E. Trypanosoma cruzi TBP shows preference for C/G-rich DNA sequences in vitro. Exp Parasitol. 2010;124:346–349. doi: 10.1016/j.exppara.2009.11.003. [DOI] [PubMed] [Google Scholar]
- DaRocha WD, Otsu K, Teixeira SM, Donelson JE. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol Biochem Parasitol. 2004;133:175–186. doi: 10.1016/j.molbiopara.2003.10.005. [DOI] [PubMed] [Google Scholar]
- de Freitas JM, Augusto-Pinto L, Pimenta JR, Bastos-Rodrigues L, Gonçalves VF, Teixeira SM, Chiari E, Junqueira AC, Fernandes O, Macedo AM, et al. Ancestral genomes, sex, and the population structure of Trypanosoma cruzi. PLoS Pathog. 2006;2:e24. doi: 10.1371/journal.ppat.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Greef C, Hamers R. The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Mol Biochem Parasitol. 1994;68:277–284. doi: 10.1016/0166-6851(94)90172-4. [DOI] [PubMed] [Google Scholar]
- De Greef C, Chimfwembe E, Kihang’a Wabacha J, Bajyana Songa E, Hamers R. Only the serum-resistant bloodstream forms of Trypanosoma brucei rhodesiense express the serum resistance associated (SRA) protein. Ann Soc Belg Med Trop. 1992;72(Suppl 1):13–21. [PubMed] [Google Scholar]
- Denise H, Poot J, Jimenez M, Ambit A, Hermman DC, Vermeulen AN, Coombs GH, Mottram JC. Studies on the CPA cysteine peptidase in the Leishmania infantum genome strain JPCM5. BMC Mol Biol. 2006;7:42. doi: 10.1186/1471-2199-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge DP, Evans KJ, Ivens AC, Aziz N, Maroof A, Kaye PM, Smith DF. Comparative expression profiling of Leishmania: Modulation in gene expression between species and in different host genetic backgrounds. PLoS Negl Trop Dis. 2009;3:e476. doi: 10.1371/journal.pntd.0000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjeux P. Leishmaniasis. Public health aspects and control. Clin Dermatol. 1996;14:417–423. doi: 10.1016/0738-081x(96)00057-0. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Sánchez DO, Frasch AC. The protozoan Trypanosoma cruzi has a family of genes resembling the mucin genes of mammalian cells. J Biol Chem. 1995;270:24146–24149. doi: 10.1074/jbc.270.41.24146. [DOI] [PubMed] [Google Scholar]
- Diehl S, Diehl F, El-Sayed NM, Clayton C, Hoheisel JD. Analysis of stage-specific gene expression in the bloodstream and the procyclic form of Trypanosoma brucei using a genomic DNA-microarray. Mol Biochem Parasitol. 2002;123:115–123. doi: 10.1016/s0166-6851(02)00138-x. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Donelson JE. A survey of the Trypanosoma brucei rhodesiense genome using shotgun sequencing. Mol Biochem Parasitol. 1997;84:167–178. doi: 10.1016/s0166-6851(96)02792-2. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Alarcon CM, Beck JC, Sheffield VC, Donelson JE. cDNA expressed sequence tags of Trypanosoma brucei rhodesiense provide new insights into the biology of the parasite. Mol Biochem Parasitol. 1995;73:75–90. doi: 10.1016/0166-6851(95)00098-l. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005a;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005b;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- Fernandes AP, Costa MM, Coelho EA, Michalick MS, de Freitas E, Melo MN, Luiz Tafuri W, Resende DM, Hermont V, Abrantes CF, et al. Protective immunity against challenge with Leishmania (Leishmania) chagasi in beagle dogs vaccinated with recombinant A2 protein. Vaccine. 2008;26:5888–5895. doi: 10.1016/j.vaccine.2008.05.095. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Franzén O, Ochaya S, Sherwood E, Lewis MD, Llewellyn MS, Miles MA, Andersson B. Shotgun sequencing analysis of Trypanosoma cruzi I Sylvio X10/1 and comparison with T. cruzi VI CL Brener. PLoS Negl Trop Dis. 2011;5:e984. doi: 10.1371/journal.pntd.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, Lee PT, Lee MG. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell. 2003;2:542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J, O’hUigin C, Overath P. The molecular phylogeny of trypanosomes: Evidence for an early divergence of the Salivaria. Mol Biochem Parasitol. 1998;91:37–49. doi: 10.1016/s0166-6851(97)00185-0. [DOI] [PubMed] [Google Scholar]
- Hajduk SL, Harris ME, Pollard VW. RNA editing in kinetoplastid mitochondria. FASEB J. 1993;7:54–63. doi: 10.1096/fasebj.7.1.8422975. [DOI] [PubMed] [Google Scholar]
- Henriksson J, Porcel B, Rydåker M, Ruiz A, Sabaj V, Galanti N, Cazzulo JJ, Frasch AC, Pettersson U. Chromosome specific markers reveal conserved linkage groups in spite of extensive chromosomal size variation in Trypanosoma cruzi. Mol Biochem Parasitol. 1995;73:63–74. doi: 10.1016/0166-6851(95)00096-j. [DOI] [PubMed] [Google Scholar]
- Holzer TR, McMaster WR, Forney JD. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol Biochem Parasitol. 2006;146:198–218. doi: 10.1016/j.molbiopara.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP. The evolution of amastin surface glycoproteins in trypanosomatid parasites. Mol Biol Evol. 2010;27:33–45. doi: 10.1093/molbev/msp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, Quail MA, Chukualim B, Capewell P, MacLeod A, Melville SE, et al. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human African trypanosomiasis. PLoS Negl Trop Dis. 2010;4:e658. doi: 10.1371/journal.pntd.0000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani S, Fenn K, Ross A, Ivens A, Smith TK, Ghazal P, Matthews K. Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics. 2009;10:e427. doi: 10.1186/1471-2164-10-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft R, Capewell P, Turner CM, Veitch NJ, MacLeod A, Hajduk S. Mechanism of Trypanosoma brucei gambiense (group 1) resistance to human trypanosome lytic factor. Proc Natl Acad Sci USA. 2010;107:16137–16141. doi: 10.1073/pnas.1007074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeller CM, Heise N. The sphingolipid biosynthetic pathway is a potential target for chemotherapy against Chagas disease. Enzyme Res. 20112011:e648159. doi: 10.4061/2011/648159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomble S, Portman N, Gull K. A protein-protein interaction map of the Trypanosoma brucei paraflagellar rod. PLoS One. 2009;4:e7685. doi: 10.1371/journal.pone.0007685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount DJ, Bruse S, Hill KL, Donelson JE. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol Biochem Parasitol. 2000;111:67–76. doi: 10.1016/s0166-6851(00)00300-5. [DOI] [PubMed] [Google Scholar]
- Lainson R, Ward RD, Shaw JJ. Leishmania in phlebotomid sandflies: VI. Importance of hindgut development in distinguishing between parasites of the Leishmania mexicana and L. braziliensis complexes. Proc R Soc Lond B. 1977;199:309–320. doi: 10.1098/rspb.1977.0141. [DOI] [PubMed] [Google Scholar]
- Laurentino EC, Ruiz JC, Fazelini G, Myler PJ, Degrave W, Ferreira MA, Ribeiro LMC, Cruz AK. A survey of Leishmania braziliensis genome by shotgun sequencing. Mol Biochem Parasitol. 2004;13:81–86. doi: 10.1016/j.molbiopara.2004.05.001. [DOI] [PubMed] [Google Scholar]
- LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: The Leishmania genome is constitutively expressed. Mol Biochem Parasitol. 2007;152:35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Lepesheva GI, Villalta F, Waterman MR. Targeting Trypanosoma cruzi sterol 14a -demethylase (CYP51) Adv Parasitol. 2011;75:65–87. doi: 10.1016/B978-0-12-385863-4.00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levick MP, Blackwell JM, Connor V, Coulson RM, Miles A, Smith HE, Wan KL, Ajioka JW. An expressed sequence tag analysis of a full-length, spliced-leader cDNA library from Leishmania major promastigotes. Mol Biochem Parasitol. 1996;76:345–348. doi: 10.1016/0166-6851(95)02569-3. [DOI] [PubMed] [Google Scholar]
- Liang XH, Haritan A, Uliel S, Michaeli S. Trans and cis splicing in trypanosomatids: Mechanism, factors, and regulation. Eukaryot Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima MT, Lenzi HL, Gattass CR. Negative tissue parasitism in mice injected with a noninfective clone of Trypanosoma cruzi. Parasitol Res. 1995;81:6–12. doi: 10.1007/BF00932410. [DOI] [PubMed] [Google Scholar]
- Lukes J, Jirkû M, Dolezel D, Kral’ová I, Hollar L, Maslov DA. Analysis of ribosomal RNA genes suggests that trypanosomes are monophyletic. J Mol Evol. 1997;44:521–527. doi: 10.1007/pl00006176. [DOI] [PubMed] [Google Scholar]
- Lye LF, Owens K, Shi H, Murta SM, Vieira AC, Turco SJ, Tschudi C, Ullu E, Beverley SM. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6:e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn MA, McMaster RW. Leishmania: Conserved evolution – Diverse diseases. Trends Parasitol. 2008;24:103–105. doi: 10.1016/j.pt.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Martínez-Calvillo S, Yan S, Nguyen D, Fox M, Stuart K, Myler PJ. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol Cell. 2003;11:1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- Martínez-Calvillo S, Nguyen D, Stuart K, Myler PJ. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot Cell. 2004;3:506–517. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Calvillo S, Vizuet-de-Rueda JC, Florencio-Martínez LE, Manning-Cela RG, Figueroa-Angulo EE. Gene expression in trypanosomatid parasites. J Biomed Bio-technol. 20102010:525241. doi: 10.1155/2010/525241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu-Daudé F, Cheng R, Welsh J, McClelland M. Screening of differentially amplified cDNA products from RNA arbitrarily primed PCR fingerprints using single strand conformation polymorphism (SSCP) gels. Nucleic Acids Res. 1996;24:1504–1507. doi: 10.1093/nar/24.8.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KR. The developmental cell biology of Trypanosoma brucei. J Cell Sci. 2005;118:283–290. doi: 10.1242/jcs.01649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauel J. Vaccination against Leishmania infections. Curr Drug Targets Immune Endocr Metabol Disord. 2002;2:201–226. doi: 10.2174/1568008023340631. [DOI] [PubMed] [Google Scholar]
- McCall LI, Matlashewski G. Localization and induction of the A2 virulence factor in Leishmania: Evidence that A2 is a stress response protein. Mol Microbiol. 2010;77:518–530. doi: 10.1111/j.1365-2958.2010.07229.x. [DOI] [PubMed] [Google Scholar]
- Miles MA, Souza A, Povoa M, Shaw JJ, Lainson R, Toye PJ. Isozymic heterogeneity of Trypanosoma cruzi in the first autochthonous patients with Chagas’ disease in Amazonian Brazil. Nature. 1978;272:819–821. doi: 10.1038/272819a0. [DOI] [PubMed] [Google Scholar]
- Mizbani A, Taslimi Y, Zahedifard F, Taheri T, Rafati S. Effect of A2 gene on infectivity of the nonpathogenic parasite Leishmania tarentolae. Parasitol Res. 2011;109:793–799. doi: 10.1007/s00436-011-2325-4. [DOI] [PubMed] [Google Scholar]
- Momen H. Taxonomy of Trypanosoma cruzi: A commentary on characterization and nomenclature. Mem Inst Oswaldo Cruz. 1999;94(Suppl 1):181–184. doi: 10.1590/s0074-02761999000700025. [DOI] [PubMed] [Google Scholar]
- Morris JC, Wang Z, Drew ME, Englund PT. Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J. 2002;21:4429–4438. doi: 10.1093/emboj/cdf474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- Myler PJ, Audleman L, deVos T, Hixson G, Kiser P, Lemley C, Magness C, Rickel E, Sisk E, Sunkin S, et al. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc Natl Acad Sci USA. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung KS, Beetham JK, Wilson ME, Donelson JE. Comparison of the post-transcriptional regulation of the mRNAs for the surface proteins PSA (GP46) and MSP (GP63) of Leishmania chagasi. J Biol Chem. 2002;277:16489–16497. doi: 10.1074/jbc.M200174200. [DOI] [PubMed] [Google Scholar]
- Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, Roditi I, Ochsenreiter T. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 2010;6:e1001037. doi: 10.1371/journal.ppat.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olin-Sandoval V, Moreno-Sánchez R, Saavedra E. Targeting trypanothione metabolism in trypanosomatid human parasites. Curr Drug Targets. 2010;11:1614–1630. doi: 10.2174/1389450111009011614. [DOI] [PubMed] [Google Scholar]
- Pays E, Salmon D, Morrison LJ, Marcello L, Barry JD. Antigenic variation in Trypanosoma brucei. In: Barry JD, McCulloch R, Mottram J, Acosta-Serrano A, editors. Trypanosomes After the Genome. Horizon Bioscience; Wymondham: 2007. pp. 339–372. [Google Scholar]
- Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, Homblé F, Vanhamme L, Tebabi P, Pays A, Poelvoorde P, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309:469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- Porcel BM, Tran AN, Tammi M, Nyarady Z, Rydâker M, Urmenyi TP, Rondinelli E, Pettersson U, Andersson B, Aslund L. Gene survey of the pathogenic protozoan Trypanosoma cruzi. Genome Res. 2000;10:1103–1107. doi: 10.1101/gr.10.8.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman N, Lacomble S, Thomas B, McKean PG, Gull K. Combining RNA interference mutants and comparative proteomics to identify protein components and dependences in a eukaryotic flagellum. J Biol Chem. 2009;284:5610–5619. doi: 10.1074/jbc.M808859200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Respuela P, Ferella M, Rada-Iglesias A, Aslund L. Histone acetylation and methylation at sites initiating divergent polycistronic transcription in Trypanosoma cruzi. J Biol Chem. 2008;283:15884–15892. doi: 10.1074/jbc.M802081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2003;128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- Rochette A, Raymond F, Ubeda JM, Smith M, Messier N, Boisvert S, Rigault P, Corbeil J, Ouellette M, Papadopoulou B. Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics. 2008;9:255. doi: 10.1186/1471-2164-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Bringaud F, Layden RE, Baltz T, Eisen H. Active late-appearing variable surface antigen genes in Trypanosoma equiperdum are constructed entirely from pseudogenes. Proc Natl Acad Sci USA. 1989;86:9375–9379. doi: 10.1073/pnas.86.23.9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvalcaba-Trejo LI, Sturm NR. The Trypanosoma cruzi Sylvio X10 strain maxicircle sequence: The third musketeer. BMC Genomics. 2011;12:58. doi: 10.1186/1471-2164-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Foote SJ, Handman E. The role of host genetics in leishmaniasis. Trends Parasitol. 2009;25:383–391. doi: 10.1016/j.pt.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Shi H, Djikeng A, Tschudi C, Ullu E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: Control of small interfering RNA accumulation and retroposon transcript abundance. Mol Cell Biol. 2004;24:420–427. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Djikeng A, Mark T, Wirtz E, Tschudi C, Ullu E. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA. 2000;6:1069–1076. doi: 10.1017/s1355838200000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Chamond N, Tschudi C, Ullu E. Selection and characterization of RNA interference-deficient trypanosomes impaired in target mRNA degradation. Eukaryot Cell. 2004;3:1445–1453. doi: 10.1128/EC.3.6.1445-1453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AG, Stevens JR, Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Smith DF, Peacock CS, Cruz AK. Comparative genomics: From genotype to disease phenotype in the leishmaniases. Int J Parasitol. 2007;37:1173–1186. doi: 10.1016/j.ijpara.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Blanchette M, Papadopoulou B. Improving the prediction of mRNA extremities in the parasitic protozoan Leishmania. BMC Bioinformatics. 2008;9:158. doi: 10.1186/1471-2105-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol Biochem Parasitol. 1996;83:141–152. doi: 10.1016/s0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- Stevens JR, Noyes HA, Dover GA, Gibson WC. The ancient and divergent origins of the human pathogenic trypanosomes, Trypanosoma brucei and T. cruzi. Parasitology. 1999;118:107–116. doi: 10.1017/s0031182098003473. [DOI] [PubMed] [Google Scholar]
- Stuart K, Panigrahi AK. RNA editing: Complexity and complications. Mol Microbiol. 2002;45:591–596. doi: 10.1046/j.1365-2958.2002.03028.x. [DOI] [PubMed] [Google Scholar]
- Subramaniam C, Veazey P, Redmond S, Hayes-Sinclair J, Chambers E, Carrington M, Gull K, Matthews K, Horn D, Field MC. Chromosome-wide analysis of gene function by RNA interference in the African trypanosome. Eukaryot Cell. 2006;5:1539–1549. doi: 10.1128/EC.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira SM, Kirchhoff LV, Donelson JE. Post-transcriptional elements regulating expression of mRNAs from the amastin/tuzin gene cluster of Trypanosoma cruzi. J Biol Chem. 1995;270:22586–22594. doi: 10.1074/jbc.270.38.22586. [DOI] [PubMed] [Google Scholar]
- Thomas S, Green A, Sturm NR, Campbell DA, Myler PJ. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics. 2009;10:e152. doi: 10.1186/1471-2164-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CM, Barry JD. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology. 1989;99:67–75. doi: 10.1017/s0031182000061035. [DOI] [PubMed] [Google Scholar]
- Van der Ploeg LH, Valerio D, De Lange T, Bernards A, Borst P, Grosveld FG. An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res. 1982;10:5905–5923. doi: 10.1093/nar/10.19.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, Pays A, Tebabi P, Van Xong H, Jacquet A, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422:83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- Vargas N, Pedroso A, Zingales B. Chromosomal polymorphism, gene synteny and genome size in T. cruzi I and T. cruzi II groups. Mol Biochem Parasitol. 2004;138:131–141. doi: 10.1016/j.molbiopara.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Veitch NJ, Johnson PC, Trivedi U, Terry S, Wildridge D, MacLeod A. Digital gene expression analysis of two life cycle stages of the human-infective parasite, Trypanosoma brucei gambiense, reveals differentially expressed clusters of co-regulated genes. BMC Genomics. 2010;11:e124. doi: 10.1186/1471-2164-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Di Paolo N, Urmenyi TP, Rondinelli E, Frasch AC, Sanchez DO. Gene discovery through expressed sequence Tag sequencing in Trypanosoma cruzi. Infect Immun. 1998;66:5393–5398. doi: 10.1128/iai.66.11.5393-5398.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva MS, Williams SP, Beard CB, Richards FF, Aksoy S. A new member of a family of site-specific retrotransposons is present in the spliced leader RNA genes of Trypanosoma cruzi. Mol Cell Biol. 1991;11:6139–6148. doi: 10.1128/mcb.11.12.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth BR, Kelly BL, Joshi PB, Ivens AC, McMaster WR. Differentially expressed Leishmania major gp63 genes encode cell surface leishmanolysin with distinct signals for glycosylphosphatidylinositol attachment. Mol Biochem Parasitol. 1998;93:31–41. doi: 10.1016/s0166-6851(98)00013-9. [DOI] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Weatherly DB, Boehlke C, Tarleton RL. Chromosome level assembly of the hybrid Trypanosoma cruzi genome. BMC Genomics. 2009;10:e255. doi: 10.1186/1471-2164-10-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenberger SJ, Cerqueira GC, El-Sayed NM, Zingales B, Campbell DA, Sturm NR. Trypanosoma cruzi mitochondrial maxicircles display species- and strain-specific variation and a conserved element in the non-coding region. BMC Genomics. 2006;7:e60. doi: 10.1186/1471-2164-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RJ. The trypanolytic factor-mechanism, impacts and applications. Trends Parasitol. 2010;26:457–464. doi: 10.1016/j.pt.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Wincker P. The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 1996;24:1688–1694. doi: 10.1093/nar/24.9.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Worthey EA, Martinez-Calvillo S, Schnaufer A, Aggarwal G, Cawthra J, Fazelinia G, Fong C, Fu G, Hassebrock M, Hixson G, et al. Leishmania major chromosome 3 contains two long convergent polycistronic gene clusters separated by a tRNA gene. Nucleic Acids Res. 2003;31:4201–4210. doi: 10.1093/nar/gkg469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AD, Li S, Feng S, Martin DS, Lynn DH. Phylogenetic position of the kinetoplastids, Cryptobia bullocki, Cryptobia catostomi, and Cryptobia salmositica and mono-phyly of the genus Trypanosoma inferred from small subunit ribosomal RNA sequences. Mol Biochem Parasitol. 1999;99:69–76. doi: 10.1016/s0166-6851(98)00184-4. [DOI] [PubMed] [Google Scholar]
- Wright JR, Siegel TN, Cross GA. Histone H3 trime-thylated at lysine 4 is enriched at probable transcription start sites in Trypanosoma brucei. Mol Biochem Parasitol. 2010;172:141–144. doi: 10.1016/j.molbiopara.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Donelson JE, Wilson ME. The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol Biochem Parasitol. 2003;132:1–16. doi: 10.1016/s0166-6851(03)00211-1. [DOI] [PubMed] [Google Scholar]
- Zhang WW, Mendez S, Ghosh A, Myler P, Ivens A, Clos J, Sacks DL, Matlashewski G. Comparison of the A2 gene locus in Leishmania donovani and Leishmania major and its control over cutaneous infection. J Biol Chem. 2003;278:35508–35515. doi: 10.1074/jbc.M305030200. [DOI] [PubMed] [Google Scholar]
- Zhou S, Kile A, Kvikstad E, Bechner M, Severin J, Forrest D, Runnheim R, Churas C, Anantharaman TS, Myler P, et al. Shotgun optical mapping of the entire Leishmania major Friedlin genome. Mol Biochem Parasitol. 2004;138:97–106. doi: 10.1016/j.molbiopara.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Zingales B, Pereira ME, Almeida KA, Umezawa ES, Nehme NS, Oliveira RP, Macedo A, Souto RP. Biological parameters and molecular markers of clone CL Brener – The reference organism of the Trypanosoma cruzi genome project. Mem Inst Oswaldo Cruz. 1997;92:811–814. doi: 10.1590/s0074-02761997000600016. [DOI] [PubMed] [Google Scholar]
- Zingales B, Souto RP, Mangia RH, Lisboa CV, Campbell DA, Coura JR, Jansen A, Fernandes O. Molecular epidemiology of American trypanosomiasis in Brazil based on dimorphisms of rRNA and mini-exon gene sequences. Int J Parasitol. 1998;28:105–112. doi: 10.1016/s0020-7519(97)00178-1. [DOI] [PubMed] [Google Scholar]
- Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]