Abstract
Analysis of the 2.4-Å resolution crystal structure of the large ribosomal subunit from Haloarcula marismortui reveals the existence of an abundant and ubiquitous structural motif that stabilizes RNA tertiary and quaternary structures. This motif is termed the A-minor motif, because it involves the insertion of the smooth, minor groove edges of adenines into the minor groove of neighboring helices, preferentially at C-G base pairs, where they form hydrogen bonds with one or both of the 2′ OHs of those pairs. A-minor motifs stabilize contacts between RNA helices, interactions between loops and helices, and the conformations of junctions and tight turns. The interactions between the 3′ terminal adenine of tRNAs bound in either the A site or the P site with 23S rRNA are examples of functionally significant A-minor interactions. The A-minor motif is by far the most abundant tertiary structure interaction in the large ribosomal subunit; 186 adenines in 23S and 5S rRNA participate, 68 of which are conserved. It may prove to be the universally most important long-range interaction in large RNA structures.
It is well known that single-stranded RNAs fold back on themselves to form short, double-stranded helices that are stabilized primarily by Watson–Crick and GU wobble base pairs. In recent years, as increasing numbers of RNA structures have been determined, additional, rarer elements of RNA secondary structure (1, 2) have been identified such as tetraloops (3, 4), bulged-G motifs (5–7), and cross-stand purine stacks (5, 7, 8). Less is known about the ways RNAs with complex secondary structures fold to form RNA tertiary structure because few of the RNA structures known previously were large enough to have sufficient tertiary structure to analyze that problem. In contrast, the recently determined structures of the large ribosomal subunit from Haloarcula marismortui (9, 10) and the small ribosomal subunit from Thermus thermophilus (11, 12) contain a large number of long-range interactions between regions of RNA that are distant in the secondary structure. The ≈3,000 nt of the two RNAs of the large ribosomal subunit form a compact structure stabilized by tertiary interactions between secondary structure elements that include about 100 double helical stems. The structure of this large polyanion is stabilized, in part, by interactions with metal ions and proteins, which will be discussed elsewhere. Here we address the interactions occurring between and among RNA helices and single strands that stabilize RNA tertiary and quaternary structure.
Methods
For our study, each adenosine residue in the structure of the H. marismortui 23S rRNA (Protein DataBank entry 1FFK) was assessed for occurrences of A-minor interactions by using the graphics program o (13). A-minor interactions were selected based on the following predetermined geometric criteria. The C2 atom of the adenosine had to be within 3.7 Å of one of its neighboring atoms. The interacting atom had to lie within 45° of the adenine plane. Finally, the C2 face of the adenosine had to pack against the minor groove side of the receptor RNA. The surface accessibilities of the bases were measured by a 1.7-Å radius probe sphere according to Lee and Richards (14) by using cns (15) and were further averaged and normalized against the surface accessibilities of individual bases in a regular A-form helix (Protein DataBank entry 1SDR, excluding terminal residues and interhelical crystal packing). Figures were generated by using the graphics programs bobscript (16), ribbons (17), and spock (18).
Results and Discussion
Base-Pairing Stabilizes 23S rRNA Tertiary Structure.
Base pairs between remote nucleotides contribute significantly to the stabilization of RNA tertiary structure in the large ribosomal subunit, as was anticipated from phylogenetic analyses of rRNA sequences (19, 20). The long-range base-pairings in the 23S rRNA of H. marismortui have been catalogued (9), and they include base pairs and base triples of many different kinds. The number of hydrogen bonding interactions between bases that are remote in the secondary structure is about 100. The exact number depends on the criteria used to distinguish secondary from tertiary interactions.
A-Minor Motifs Are More Abundant than Tertiary Base Pairs.
The interaction we term the A-minor motif, because it involves adenines inserted into the minor grooves of RNA helices, is more abundant and may be of even greater significance than base-pairing in stabilizing RNA tertiary structure. In RNA, as in proteins, residues are highly conserved either because they are critical for function or because they are intimately involved in specifying tertiary structure. Examination of the RNA in the 50S subunit reveals that A residues are by far the most abundant of the conserved nucleotides involved in tertiary structure interactions. It had been realized earlier, when the first 23S rRNAs were sequenced, that A residues are more abundant than other bases in 23S rRNA sequences not involved in regular helix formation and that many of these nonbase-paired As are conserved (20, 21). It is these conserved As in irregular regions that engage in A-minor motif interactions. Indeed, 186 of the 721 adenine bases in H. marismortui 23S rRNA and 68 of its 106 As that are >95% conserved interact with RNA minor grooves via their N1-C2-N3 edges, which are smooth because they lack the exocyclic atoms of other bases (Table 1; Fig. 1 a and b).
Table 1.
A-minor interactions in H. marismortui 23S rRNA
| Type | Number | Conserved >90% across all kingdoms | Conserved >90% in Archaea | Receptors
|
|||
|---|---|---|---|---|---|---|---|
| C-G | G-C | U-A | A-U | ||||
| I | 83 | 50 (60.2%) | 65 (78.3%) | 51 (61.4%) | 15 (18.1%) | 6 (7.2%) | 4 (4.8%) |
| II | 54 | 31 (57.4%) | 40 (74.1%) | 26 (48.1%) | 8 (14.8%) | 12 (22.2%) | 2 (3.7%) |
| III | 23 | 12 (52.2%) | 13 (56.5%) | ||||
| 0 | 10 | 6 (60%) | 8 (80%) | ||||
Figure 1.
(a) The smooth minor groove face of the adenosine nucleotide allows the base to pack tightly into the minor groove of an RNA helix. Its N1, N3, and 2′-OH atoms are available for hydrogen-bonding interactions. (b) Ribbon drawing of the overall structure of the 50S ribosomal subunit from H. marismortui highlighting the 186 adenosines (shown in red spheres) that make A-minor interactions based on distance and geometric criteria (see text). (c) Examples of the four major types of A-minor interactions found in H. marismortui 50S shown in surface representation. Each type is defined by the position of the 2′-OH group of the interacting adenosine relative to the positions of the two 2′-OH groups of the receptor base pair. Whereas type I and type II interactions are A-specific, type 0 and type III also are observed for other bases even though As are still preferred when the base packs against the ribose backbone.
The four RNA residues differ significantly in their solvent accessibility, in the large subunit, and adenine bases are once again unusual, as are uracils, reflecting their roles in the formation of RNA tertiary structure (Table 2). When the average solvent-accessible surface area of the individual bases in the large ribosomal subunit is calculated by using a 1.7-Å probe sphere and compared with their accessibility as free nucleotides or when a part of regular duplex structures, it is found that U residues are much more likely to be solvent-exposed than are A, G, and C residues, and 21% of all uracil bases have at least one unstacked face. Further, the uracil bases of the RNA structure of the large ribosomal subunit exhibit an average potential solvent accessibility that is 40% larger than their accessibility in a regular RNA helix. These statistics and their frequent location at the ends of helices suggest that Us may serve as helix breakers in large RNAs. A, G, and C residues are approximately equally well buried, and on average the solvent accessibility of these bases in the protein-free RNA structure is close to what it is in an A-form helix. Adenine bases are the most buried bases of all, on average, in the fully assembled subunit, which might be considered surprising given their overrepresentation in single-stranded regions. The reason As are so well protected from solvent, on average, is that many of them that occur in irregularly structured sequences are involved in the A-minor tertiary interactions and in protein–RNA contacts.
Table 2.
Average solvent accessibility of bases in the H. marismortui 50S ribosome relative to an A-form RNA helix
| Relative surface accessibility | A (720) | G (872) | C (722) | U (513) |
|---|---|---|---|---|
| 23S + 5S RNA* | 1.04 | 1.01 | 1.06 | 1.20 |
| 50S† | 0.73 | 0.81 | 0.89 | 1.02 |
Numbers in parentheses refer to the occurrences in the 50S structure.
Relative solvent accessibility of bases in the folded 23S and 5S RNA, excluding proteins and solvent molecules.
Relative solvent accessibility of bases in the assembled 50S particle, excluding solvent molecules.
There Are Four Variants of the A-Minor Motif.
In the A-minor motif, the ribose-phosphate backbone of the adenosine is closer to that of one strand of the receptor helix than the other, and the orientation of that near strand is necessarily antiparallel to that of the strand to which the A belongs. Four versions of the motif can be identified that differ with respect to the position of the O2′ and N3 atoms of the A residue relative to the O2′ atoms of the base pair in the receptor helix. In the type I version of the motif (Fig. 1c), both the O2′ and the N3 of the adenine residue are inside the minor groove of the receptor, which optimizes the fit of the adenine in the minor groove and maximizes the number of hydrogen bonds that can form. In the type II version of the motif, the O2′ of the A is outside the near strand O2′ whereas the N3 of the A is inside. The type III form of the motif is characterized by a placement of the A that puts both its O2′ and N3 outside the near strand O2′. A fourth, rarer, version of the motif is also encountered, type 0, in which N3 of the adenine is outside the O2′ of the far strand.
Type 0 and type III interactions are neither particularly A-specific nor selective with respect to receptor base pair. The type 0 motif is not base-specific because it is the ribose of the inserted residue that fills the minor groove of the receptor helix, not the base, and because the Watson–Crick faces of all bases include groups that can hydrogen-bond to 2′OH groups; the type III interaction is not A-specific either. Nevertheless, in both type 0 and type III motifs, inserted bases tend to pack against receptor riboses, and that contact is optimized when the base is an A. In contrast, both the type I and type II A-minor motifs are highly specific for adenine bases. Only As are able to fit snugly into minor grooves and hydrogen-bond with minor groove groups optimally. They have a strong preference for C-G receptor base pairs, which provide an optimal complementarity in shape and hydrogen bonds (Table 1; Fig. 1c).
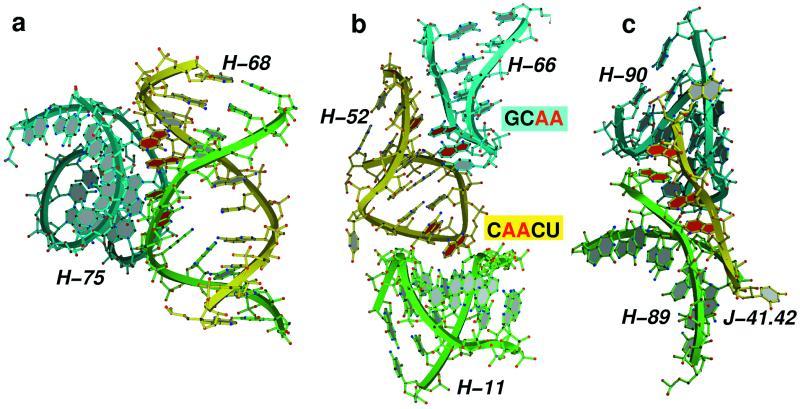
The packing orientations between helices stabilized by type I and II A-minor interactions show large variation in the interhelical angles (Fig. 2a) for two reasons. First, the adenine base and the base pair with which it interacts can depart from coplanarity by as much as 45o. Second, the minor groove edge of adenines can be presented to receptor helices in many different ways. They can be components of sheared GA or reverse Hoogsteen AU base pairs, for example, or be a bulged base or components of terminal loops. They are usually not forming Watson–Crick base pairs. For comparison, another reoccurring motif of helix–helix interaction, albeit much less frequent, leads to the packing of ordinary base-paired helices at a fixed interhelical angle of about 80o. This angle is required to allow the placement of the backbone from one helix in the minor groove of the other. Thus, this motif is sequence independent, but it requires the secondary structure of a base-paired helix instead. The base-paired nucleotide whose backbone atoms pack into the minor groove of the other helix in this specific arrangement are positioned in a way that corresponds to the type 0 interactions of adenosines in the A-minor motif. A-minor motifs are also common in loop–helix interactions (Fig. 2b), helix junctions, and places where sharp changes in backbone direction occur. There are even examples of single strands that stabilize the association of two distant helices through A-minor interactions (Fig. 2c).
Figure 2.
Examples of RNA tertiary structure stabilization by A-minor interactions in H. marismortui 23S rRNA. (a) A-minor interactions play an important role in stabilizing the interaction between helix 68 of domain IV (yellow and green), which forms the front rim of the active site cleft and helix 75 of domain V (blue). Four adenosines from two opposing strands of helix 68 (colored red) are stacked to allow packing within the minor groove of helix 75. (b) A-minor interactions (shown in red) are also critical in mediating loop–loop interactions, such as the one observed between the stem loops of helices 66 (blue) and 52 (yellow), and loop–helix interactions as the one observed between the helix 52 stem loop (yellow) and helix 11 (green). (c) The single-stranded junction between helices 41 and 42 (J41.42; yellow) donates adenosine residues for A-minor interactions that stabilize the interaction between helix 89 (green) and helix 90 (blue).
A-Minor Motifs Often Cluster.
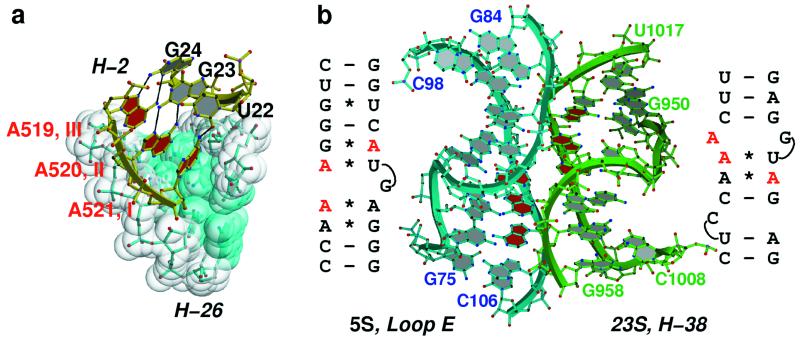
Adenines involved in A-minor interactions often stack on other As that are similarly engaged to form what we call A patches. The number of As in an A patch that belongs to the same RNA strand seldom exceeds three, and as a general rule, the A-minor interactions formed by such a run of As decrease in type order going from the 5′ to 3′ direction. Thus as we see in Fig. 3a, the first A in a three-adenosine patch makes a type III interaction with the receptor helix, the second makes a type II, and the third forms a type I interaction. A patches longer than three residues usually are generated by As emanating from the two strands of the same helical element that are stacked on each other (Fig. 2a). The type order of the interactions in such a stack is low in the middle and high at its ends; in the example shown in Fig. 2a the interactions are in succession: type II, I, I, II. The stacking interaction between adenosines in the center of such a patch is necessarily a cross-strand purine stack.
Figure 3.
(a) Interaction between the A patch on helix 2 (in yellow stick) with the minor groove of helix 26 (shown in surface representation). A519, A520, and A521 of helix 2 make type III, type II, and type I interactions, respectively, typical of A-patch packing geometry in the A-minor motif. (b) 5S rRNA solely interacts with 23S rRNA via a symmetric A-patch interaction between three stacked adenosines of the 5S rRNA loop E helix and three stacked adenosines in helix 38 of 23S rRNA.
Helical RNAs that include sheared GAs followed by reverse-Hoogsteen UA pairs include cross-strand A stacks (5, 7, 8). Commonly they interact with distant secondary structure elements by using A-minor motifs. The loop E region of H. marismortui 5S rRNA includes such a motif, and the A patch it generates makes A-minor interactions with helix 38 from domain II of 23S rRNA (Fig. 3b). As it happens, helix 38 contains an A-rich bulge close to the place where it interacts with the 5S A-patch, and the As in that bulge form an A patch that makes reciprocal interactions with 5S rRNA. The resulting helix–helix interaction involving six As (Fig. 3b) is the only 5S rRNA interaction with 23S rRNA of any consequence.
A-Minor Motifs Are Common in All Large RNAs.
A-minor motifs are not unique to 23S and 5S rRNAs. For example, the A platform-mediated GAAA tetraloop-tetraloop receptor interaction and the ribose zipper motifs reported in the P4-P6 domain of the Tetrahymena self-splicing ribozyme (22) include A-minor motifs. We note that there are only two A platforms in the large ribosomal subunit of H. marismortui, neither of which are conserved; however, there are several examples of GAAA tetraloop interactions with helices through A-minor interactions. A-minor motifs are also present in the hepatitis delta virus ribozyme (23) and in the RNA fragment that binds L11 (24).
A-Minor Motifs Are Functionally Significant.
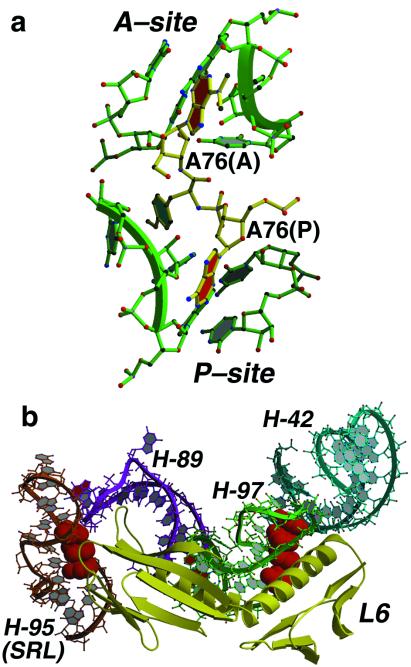
A-minor motifs appear in functionally important interactions and are structurally important. In our study of the interaction of peptidyl transferase substrate analogues with the ribosome (10) (Fig. 4a) we have observed that analogues of the 3′ terminal A of tRNA bound to the A site form a type I interaction with the G2618-U2541 base pair in 23S rRNA (U2506-G2583 in Escherichia coli). Similarly, analogues of the 3′ terminal A of tRNA bound to the P site make a type I interaction with A2485-C2536 (A2450-C2501 in E. coli). This may explain, at least in part, why the 3′ terminal residue of all tRNA molecules is always an A.
Figure 4.
(a) Structure of a peptidyl transferase transition state analogue (29) (yellow) bound to the active site of the 50S ribosomal subunit of H. marismortui (in green). Each of the adenosine-like groups on the inhibitor that are analogous to A76 residues of the P-site and A-site tRNAs (with base planes in red) make type I A-minor interactions with base pairs in domain V of 23S rRNA. (b) The interaction between ribosomal protein L6 (yellow) and 23S rRNA is mediated by conserved A patches (shown in red space filling) on helix 95 (orange) and helix 97 (green).
There are five A patches in other functionally interesting locations on the surface of the large ribosomal subunit, some of which may interact with RNAs or proteins. One is located in helix 89 (A2504, A2417; E. coli A2469, A2482) in the active site cleft. In the model we have proposed for tRNA interactions with the large subunit, this patch packs against the T stem of a tRNA bound to the A site in a way that allows A-motif formation. Interestingly, the same region of the T stem is recognized by EF-Tu in EF-Tu-tRNA complexes (25, 26). Two A patches, A806, A807 and A808 at the end of helix 34 (E. coli A715, A716, C717) and A1767, A1778, A1779 in helix 62 (E. coli A1689, A1700, A1701) are likely to contact the 30S subunit in the 70S ribosome (27). Interactions such as those shown in Fig. 2b might occur in these cases. The end of the sarcin-ricin loop, which is critical for factor binding to the ribosome, is the fourth such A patch, and the fifth is found inside the tunnel.
The A patches in the sarcin-ricin loop and in the tunnel are likely to interact with protein, not RNA, and there are numerous examples in the large ribosomal subunit of conserved A patches that do indeed form part of binding sites for proteins. The N-terminal domain of L6, for example, binds to the A patch formed by A2783, A2784, A2792, and A2793 in helix 97 (E. coli A2748, A2749, A2757, A2758) and its C-terminal domain interacts with an A patch consisting of A2694 and A2702 (E. coli A2657, A2665) in the sarcin-ricin loop at the end of helix 95 (Fig. 4b). In addition, L3, L15, L18, L22, L24, L37e, and L37ae all interact with conserved A patches. Perhaps unexpectedly, the interactions of proteins with A patches are all idiosyncratic; each protein does it a different way.
While this manuscript was in preparation, V. Ramakrishnan and coworkers (12) reported the existence of the adenosine interactions we call A-minor motifs in the 30S ribosomal subunit and noted their structural and functional importance. Their observations provide strong support for the universality of the A-minor motif and its importance in stabilizing both tertiary and quaternary structures of RNA.
Conclusions
The abundance of A-minor motif interactions in the 50S ribosomal subunit as well as the extensive conservation of the nucleotides involved implies that these interactions are of paramount importance in stabilizing the tertiary structure of globular RNA folds. Thermodynamic studies of the contribution of adenosine interactions in the minor groove of the P4-P6 domain of the Tetrahymena group I ribozyme indicate that they do in fact contribute significantly to the free energy of stabilization (28). It is likely that the stability of A-minor interactions, particularly the type I interaction, arises from the tight packing of the A into the minor groove of a C-G base pair. The high degree of phylogenetic conservation of both the As and the G-C base pairs with which they interact as well as the significance of the A-minor interactions in specifying the globular fold of rRNA lead us to wonder whether their existence could be identified in other RNA sequences and thus predict the tertiary fold of those RNAs.
Acknowledgments
We thank D. Klein, J. Hansen, R. Batey, and E. Doherty for helpful discussions. This work was supported by grants from the National Institutes of Health to T.A.S (GM22778) and P.B.M (GM54216) and by a grant from the Agouron Institute (to T.A.S. and P.B.M.). N.B. was supported by a Burroughs Welcome Fund Career Award.
Note Added in Proof.
A-minor interactions between adenines in 16s rRNA and the helix formed when mRNA codons interact with tRNA anticodons have been implicated in the decoding mechanism (30).
References
- 1.Moore P B. Annu Rev Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- 2.Westhof E, Fritsch V. Struct Fold Des. 2000;8:R55–R65. doi: 10.1016/s0969-2126(00)00112-x. [DOI] [PubMed] [Google Scholar]
- 3.Tuerk C, Gauss P, Thermes C, Groebe D R, Gayle M, Guild N, Stormo G, d'Aubenton-Carafa Y, Uhlenbeck O C, Tinoco I., Jr Proc Natl Acad Sci USA. 1988;85:1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woese C R, Winkler S, Gutell R R. Proc Natl Acad Sci USA. 1990;87:8467–8468. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wimberly B, Varani G, Tinoco I., Jr Biochemistry. 1993;32:1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- 6.Szewczak A A, Moore P B, Chan Y-L, Wool I G. Proc Natl Acad Sci USA. 1993;90:9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correll C C, Freeborn B, Moore P B, Steitz T A. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 8.Dallas A, Moore P B. Structure (London) 1997;5:1639–1653. doi: 10.1016/s0969-2126(97)00311-0. [DOI] [PubMed] [Google Scholar]
- 9.Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 10.Nissen P, Hansen J, Ban N, Moore P B, Steitz T A. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 11.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 12.Wimberly B T, Brodersen D E, Clemons W M, Jr, Morgan-Warren R J, Carter A P, Vonrhein C, Hartsch T, Ramakrishnan V. Nature (London) 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 13.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 14.Lee B, Richards F M. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 15.Brünger A T, Adams P D, Clore G M, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, Read R J, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 17.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 18.Christopher J A. The Structural Properties Observation and Calculation Kit Program Manual. Texas A & M Univ., College Station: The Center for Macromolecular Design; 1998. [Google Scholar]
- 19.Gutell R. In: Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Biosynthesis. Dahlberg A, Zimmerman R, editors. Boca Raton, FL: CRC; 1996. pp. 111–128. [Google Scholar]
- 20.Gutell R R, Gray M W, Schnare M N. Nucleic Acids Res. 1993;21:3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ware V C, Tague B W, Clark C G, Gourse R L, Brand R C, Gerbi S A. Nucleic Acids Res. 1983;11:7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 23.Ferré-D'Amaré A R, Zhou K, Doudna J A. Nature (London) 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 24.Wimberly B T, Guymon R, McCutcheon J P, White S W, Ramakrishnan V. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 25.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B F, Nyborg J. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 26.Nissen P, Kjeldgaard M, Thirup S, Clark B F C, Nyborg J. Biochimie. 1996;78:921–933. doi: 10.1016/s0300-9084(97)86714-4. [DOI] [PubMed] [Google Scholar]
- 27.Cate J H, Yusupov M M, Yusupova G Z, Earnest T N, Noller H F. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 28.Doherty, E. A., Batey, R. T., Masquida, B. & Doudna, J. A. (2001) Nat. Struct. Biol., in press. [DOI] [PubMed]
- 29.Welch M, Chastang M, Yarus M. Biochemistry. 1995;34:385–390. doi: 10.1021/bi00002a001. [DOI] [PubMed] [Google Scholar]
- 30.Carter A P, Clemons W M, Broderson D E, Morgan-Warren R J, Wimberly B T, Ramakrishnan V. Nature (London) 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]