Abstract
The genus Methylobacterium comprises pink-pigmented facultative methylotrophic (PPFM) bacteria, known to be an important plant-associated bacterial group. Species of this group, described as plant-nodulating, have the dual capacity of producing cytokinin and enzymes, such as pectinase and cellulase, involved in systemic resistance induction and nitrogen fixation under specific plant environmental conditions. The aim hereby was to evaluate the phylogenetic distribution of Methylobacterium spp. isolates from different host plants. Thus, a comparative analysis between sequences from structural (16S rRNA) and functional mxaF (which codifies for a subunit of the enzyme methanol dehydrogenase) ubiquitous genes, was undertaken. Notably, some Methylobacterium spp. isolates are generalists through colonizing more than one host plant, whereas others are exclusively found in certain specific plant-species. Congruency between phylogeny and specific host inhabitance was higher in the mxaF gene than in the 16S rRNA, a possible indication of function-based selection in this niche. Therefore, in a first stage, plant colonization by Methylobacterium spp. could represent generalist behavior, possibly related to microbial competition and adaptation to a plant environment. Otherwise, niche-specific colonization is apparently impelled by the host plant.
Keywords: phylogenetic diversity, methylotrofics, PPFM, plant-bacteria interaction
Introduction
The Methylobacterium genus, which belongs to the class Alphaproteobacteria, is described as pink-pigmented facultative methylotrophic (PPFM). Interestingly, this bacterial group presents the ability to metabolize one-carbon compounds as carbon sources (Toyama et al., 1998; Skovran et al., 2010).
A wide variety of Methylobacterium species have been isolated from plants (Pirttilä et al., 2000; Sy et al., 2001; Araújo et al., 2002; Yates et al., 2007; Ferreira et al., 2008; Andreote et al., 2009; Madhaiyan et al., 2011), the soil (Cao et al., 2011), cold lands, such as Antarctica (Moosvi et al., 2005), and the bottom of the Kuroshima Knoll sea in Japan (Inagaki et al., 2004). On considering bacteria-plant association, it has been shown that this genus can establish a beneficial interaction with the hosts, by fixing nitrogen (Sy et al., 2001; Yates et al., 2007), producing cellulase (Jayashree et al., 2011), or interacting with other plant pathogens (Araújo et al., 2002, Lacava et al., 2004, Madhaiyan et al., 2006a, 2006b). Curiously, in spite of the specific capacity for synthesizing hydrolytic enzymes (i.e. pectinase and cellulose), as yet, PPFMs have not been described as plant-pathogens, thereby indicating their additional capacity of offering host plant protection by inducing systemic resistance during the colonization process (Madhaiyan et al., 2006a, b). Additionally, a high level of PPFM inoculation can modulate the composition of the bacterial community associated with the host plant (Andreote et al., 2006), thereby implying that some competition may occur during this phase.
According to the most recent analysis, 34 species of the genus have been described to date (Kato et al., 2008; Weon et al., 2008; Madhaiyan et al., 2009), half of which (17) within the last five years, a clear indication that only a minor part of the diversity of this genushas been described so far. Thus, further studies of plant-associated members of the Methylobacterium genus will furnish additional knowledge on their distribution and ecology, thereby leading to research towards developing strains capable of enhancing plant fitness.
Since methylotrophic metabolism conferred by the mxaF gene is advantageous for Methylobacterium extorquens during plant colonization (Sy et al., 2005), it is plausible that the evolution of Methylobacterium-plant interaction has led to the selection of methylotrophic species/genotypes. Thus, in the present study, the genetic diversity of 60 Methylobacterium spp. strains obtained from eight different host plants was assessed by sequence analysis of 16S rRNA and mxaF genes, to so facilitate comprehension of the distribution of the Methylobacterium species in various host plants.
Material and Methods
Strains of Methylobacterium spp. and plant-species origins
Endophytic bacterial isolates (Table 1), obtained from the culture collection of the Laboratory of Microbial Genetics (ESALQ/USP, Piracicaba, Brazil), were isolated from previous studies of surface-disinfested Citrus spp. (18 isolates) (Araújo et al., 2002), eucalyptus (Eucalyptus grandis x Eucalyptus urophyla) (7 isolates) (Ferreira et al., 2008), Saccharum spp. (8 isolates) (Rossetto, 2008, Doctoral thesis, Universidade de São Paulo, Piracicaba), Coffea arabica (8 isolates), Borreria verticillata (12 isolates) and Capsicum annuum (7 isolates).
Table 1.
Identification of Methylobacterium spp. isolated from different hosts by the partial sequence of the 16S rRNA and mxaF genes.
| Isolate | Host | Identification* | Phylogenetic groups | |
|---|---|---|---|---|
| 16 S rRNA | mxaF | |||
| TC3-5 | Coffee | M. populi | 4 | II |
| TC3-6 | Coffee | Methylobacterium sp. | 4 | II |
| TC3-7 | Coffee | Methylobacterium sp. | 5 | VII |
| TC3-10 | Coffee | Methylobacterium sp.. | 5 | VII |
| TC3-11 | Coffee | Methylobacterium sp. | 5 | VII |
| TC3-13 | Coffee | M. extorquens | 4 | II |
| TC3-14 | Coffee | Methylobacterium sp. | 5 | VII |
| MC3-1 | Coffee | Methylobacterium sp. | 7 | VI |
| F4 | Sugarcane | Methylobacterium sp. | 7 | VII |
| F5 | Sugarcane | M. fujisawaense | 3 | VII |
| F7 | Sugarcane | Methylobacterium sp. | 5 | VII |
| F8 | Sugarcane | Methylobacterium sp. | 5 | VII |
| F9 | Sugarcane | Methylobacterium sp. | 6 | VII |
| F10 | Sugarcane | Methylobacterium sp. | 7 | VII |
| F11 | Sugarcane | Methylobacterium sp. | 7 | VII |
| D5 | Sugarcane | Methylobacterium sp. | 5 | VII |
| AR1.6/1 | Citrus | Methylobacterium sp. | 6 | VII |
| AR1.6/2 | Citrus | Methylobacterium sp. | 4 | II |
| AR1.6/8 | Citrus | Methylobacterium sp. | 4 | II |
| AR5/1 | Citrus | Methylobacterium sp. | 5 | II |
| AR5.1/5 | Citrus | Methylobacterium sp. | 6 | VII |
| ER1/21 | Citrus | M. mesophilicum | 5 | III |
| ER1.6/2 | Citrus | Methylobacterium sp. | 4 | V |
| SR1.6/2 | Citrus | Methylobacterium sp. | 7 | V |
| SR1.6/4 | Citrus | M. radiotolerans | 7 | VI |
| SR1.6/6 | Citrus | Methylobacterium sp. | 5 | III |
| SR1.6/9 | Citrus | Methylobacterium sp. | 7 | VII |
| SR1.6/13 | Citrus | Methylobacterium sp. | 4 | II |
| SR3/27 | Citrus | Methylobacterium sp. | 3 | II |
| SR5/3 | Citrus | M. fujisawaense | 3 | IV |
| SR5/4 | Citrus | M. fujisawaense | 3 | II |
| PR1/3 | Citrus | M. mesophilicum | 5 | III |
| PR3/10 | Citrus | Methylobacterium sp. | 5 | III |
| PR3/11 | Citrus | Methylobacterium sp. | 5 | IV |
| TP2-1 | Sweet pepper | M. fujisawaense | 4 | VII |
| TP4-1 | Sweet pepper | Methylobacterium sp. | 4 | II |
| TP4-2 | Sweet pepper | M. hispanicum | 2 | I |
| TP5 | Sweet pepper | M. hispanicum | 2 | I |
| TP7 | Sweet pepper | Uncultured methylotrophic bacterium | 7 | VI |
| TP8 | Sweet pepper | M. hispanicum | 2 | V |
| MP2-3 | Sweet pepper | M. hispanicum | 2 | V |
| Aw04 | Borreria verticillata | Methylobacterium sp. | 2 | III |
| Aw05 | Borreria verticillata | M. radiotolerans | 7 | VI |
| Aw06 | Borreria verticillata | Methylobacterium sp. | 7 | VI |
| Aw08 | Borreria verticillata | M. radiotolerans | 7 | VI |
| Aw09 | Borreria verticillata | M. radiotolerans | 7 | VI |
| Aw10 | Borreria verticillata | M. radiotolerans | 7 | VI |
| Aw11 | Borreria verticillata | M. radiotolerans | 7 | VI |
| Aw12 | Borreria verticillata | M. radiotolerans | 7 | VI |
| Aw13 | Borreria verticillata | M. radiotolerans | 7 | VI |
| Aw15 | Borreria verticillata | M. radiotolerans | 7 | VI |
| Aw16 | Borreria verticillata | M. hispanicum | 2 | I |
| Aw18 | Borreria verticillata | M. radiotolerans | 7 | VI |
| R1E | Eucalyptus | Methylobacterium spp. | 3 | III |
| R2E | Eucalyptus | Methylobacterium spp. | 7 | III |
| R3E | Eucalyptus | Methylobacterium spp. | 1 | VII |
| R10E | Eucalyptus | Methylobacterium spp. | 7 | VII |
| R12E | Eucalyptus | Methylobacterium spp | 6 | VII |
| R14E | Eucalyptus | Methylobacterium spp. | 5 | VII |
| R16E | Eucalyptus | Methylobacterium spp. | 3 | VII |
Identification based on the RDP database (http://simo.marsci.uga.edu/public_db/rdp_query.htm) and phylogenetic analysis in this study (Figure 1).
DNA extraction and sequencing methodology
After cultivation, bacterial DNA was extracted according to previously described methodology (Araújo et al., 2002). A partial sequence of the 16S rRNA gene (27–1401, according to E. coli position) was amplified with the primers R1378 (Heuer et al., 1997) and P027F (Lane et al., 1985). PCRs were performed in 50 μL of a reaction containing 1 X enzyme buffer, 3.75 mM of MgCl2, 0.2 mM of each dNTP, 0.2 μM of each primer and 0.1U/μL of Taq DNA Polymerase (Invitrogen, Brazil). Initial denaturation was carried out at 94 °C for 4 min, followed by 35 thermal cycles of 30 s at 94 °C, 1 min at 62.5 °C and 1 min at 72 °C, with a final extension at 72 °C for 7 min. Partial amplification of the mxaF gene was obtained with mxa1003f and mxa1561r primers (McDonald et al., 1995). All PCR amplification was checked through electrophoresis on agarose gel (1.5% w/v agarose) and UV visualization of the ethidium bromide stained gels, after which, PCR products were purified (PureLink, Invitrogen). The 16S rDNA fragments were sequenced using internal primers for both strains in an automated sequencer (MegaBACE 1000), whereas mxaF gene fragments were sequenced with two primers (mxa1003f and mxa1561r).
Sequence analysis
All the cromatograms were first trimmed for high quality bases (80% of bases with quality > 20) by means of Phred software and the trimmed sequences used for comparison in the Ribosomal Data Project (for 16S rRNA gene) and the GenBank database (nr/nt) (for the mxaF gene). The best hits of well-characterized strains of the Methylobacterium genus were retrieved from the databases, and subsequently used for alignment and phylogeny analysis with MEGA 4.0 version software (Tamura et al., 2007). Evolutionary history was inferred through the Neighbor-Joining method (Saitou and Nei, 1987) and evolutionary distances were computed by the Kimura 2-parameter method (Kimura, 1980). All the sequences obtained here were assigned to operational taxonomic units (OTUs) using MOTHUR (Schloss et al., 2009), at the frequency of 97% sequence similarity. Furthermore, Venn diagrams were constructed for 16S rRNA and mxaF gene analysis to cross-compare and visualize the distribution of these OTUs in plant species.
Nucleotide sequence accession numbers
120 DNA sequences of partial 16S rRNA and mxaF genes were deposited in the GenBank database under accession numbers EU789466 to EU789518 and EU789406 to EU789465, respectively.
Results
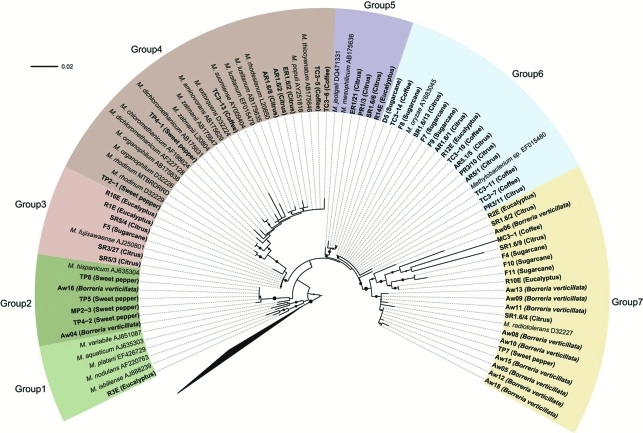
Phylogenetic analysis was carried out with partial 16S rRNA and partial mxaF gene sequences from isolates obtained in both the present study and from the GenBank and RDP databases. In the present study, phylogeny based on the 16S rRNA partial gene sequence with V6 and V7 regions generated 7 groups (Figure 1 and Table 1). Of these, group 1 presented only one eucalyptus isolate, similar to sequences from M. isbiliense and M. nodulans, whereas group 7, comprised of isolates obtained from all the hosts used here, was similar to those from M. radiotolerans. The other groups (2, 3, 4, 5 and 6) consisted of isolates from two to four different hosts. Although group 7 was close to M. radiotolerans, analysis revealed certain isolates, such as R2E, SR1.6/2, Aw06, MC3-1, SR1.6/9, F4, F10, F11 and R10E, to be divergent from the main group, thus possibly indicating the occurrence of species, as yet not described for this genus.
Figure 1.
Phylogenetic analysis of the 16S rRNA gene. Bootstrap values (1000 repetitions) above 50% are represented by solid circles next to tree branches. There were 642 nucleotide positions in the final dataset. Beijerinckia mobilis, Methylocella silvestris and Methylosinus trichosporium were used as outgroup. The layout of trees was designed using the online application “Interactive Tree Of Life” (iTOL) (http://itol.embl.de/).
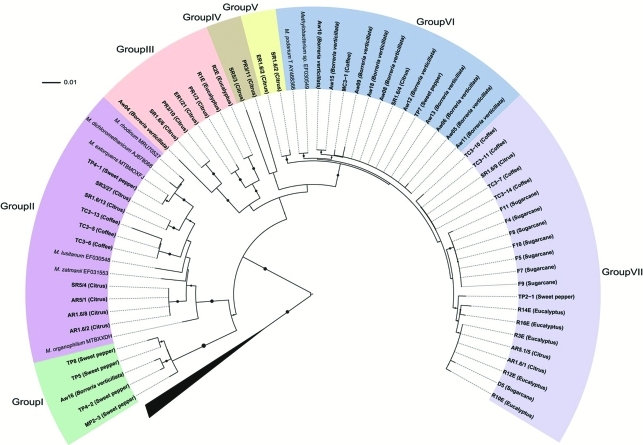
Congruency between the 16S rRNA and mxaF phylogenetical trees was incomplete. Comparative analysis of mxaF partial gene sequences by BLASTn against the nr/nt database at GenBank, classified most isolates as “uncultured methylotrophic bacterium or Methylobacterium sp.” (Table 1 and Figure 2). This was a possible outcome of the limited number of mxaF sequences available in the database. In addition, phylogenetic analysis with the mxaF gene sequences also revealed the formation of seven groups (Figure 2). Groups I, II, III and IV presented isolates from two or three hosts, groups IV and V only from citrus and group VI mainly from B. verticillata (except for TP7 and MC3-1). On the other hand, group VII contained isolates from all the hosts, with the exception of B. verticillata.
Figure 2.
Phylogenetic analysis of the mxaF gene. Bootstrap values (1000 repetitions) above 50% are represented by solid circles next to tree branches. There were 423 nucleotide positions in the final dataset. Beijerinckia mobilis, Methylocella silvestris and Methylosinus trichosporium were used as outgroup. The layout of trees was designed using the online application “Interactive Tree Of Life” (iTOL) (http://itol.embl.de/).
We observed that the clusters obtained by mxaF gene sequence analysis, revealed a certain association with host plants, since isolates from B. verticillata were located in group VII, those from sugarcane mainly in group VI (only two belonged to groups I and III), those from eucalyptus mainly in group VII (only two in group III), and those from sweet pepper mainly in group I (three in groups II, VI and VII). However, the bacterial population isolated from citrus plants was found in four of the seven groups (II, IV, V, VII).
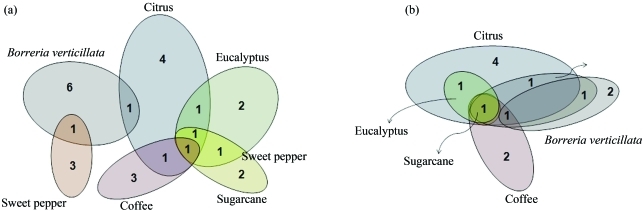
This was confirmed by a Venn diagram, obtained using 97% similarity in 16S rRNA gene sequences (Figure 3a). The analysis showed that 74% (20) of OTUs were found to be exclusive to one host plant (six to B. verticilata, four to citrus, three to sweet pepper, three to coffee, two to eucalyptus, and two to sugarcane). Additionally, only 26% (7) of OTUs were found in two host plants, and only one in four. A similar analysis, using mxaF gene sequences (Figure 3b), revealed 13 OTUs, of which, 61.5% (eight) were exclusive to only one host plant, and 38.5% (5) to two.
Figure 3.
Venn diagrams of operational taxonomic units (OTUs) assigned at 97% sequence similarity. (a) Venn diagram for 16S rRNA gene analysis with 27 OTUs, and (b) Venn diagram for mxaF gene analysis with 13 OTUs.
Discussion
The genus Methylobacterium is commonly found in natural environments, such as soil, air, dust, ocean and lake waters, and sediments, as well as urban environments (Van Aken et al., 2004). A remarkable niche of this group is its association with plants, where it is capable of colonizing leaf surfaces (Chanprame et al., 1996; Madhaiyan et al., 2011), inner tissues (Pirttilä et al., 2000; Araújo et al., 2001, 2002; Andreote et al., 2006; Yates et al., 2007), and nodules (Sy et al., 2001, Yates et al., 2007). These features could possibly have arisen from an intimate co-evolution process between Methylobacterium spp. and host plants. An example of this co-evolutive process is the bacterial capacity to mediate high photosynthetic activity in the host, by the induction of a higher number of stomata, increased chlorophyll concentration and greater amount of malic acid (Cervantes-Martinez et al., 2004). Moreover, mxaF gene associated with methylotrophic metabolism is responsible for increasing M. extorquens fitness during plant epiphytic colonization under competitive conditions (Sy et al., 2005). All together, it is assumed that plants are the main niche for assessing the diversity of the genus Methylobacterium.
As diversity in the genus Methylobacterium has not been fully explored, e.g. 17 new species of Methylobacterium were only described quite recently (Gallego et al., 2005a, b, 2006; Aslam et al., 2007; Kang et al., 2007; Madhaiyan et al., 2007; Wang et al., 2007; Kato et al., 2008; Weon et al., 2008), the present study constitutes a significant contribution to the description of diversity in this ubiquitous bacterial group.
The mxaF phylogeny analysis suggests the role of plant species in the selection of Methylobacterium species for establishing an endophytic interaction. As previously described, epiphytic colonization is the first stage towards developing such an association (Andreote et al., 2006). Under like circumstances, the methylotrophic metabolism state is advantageous for M. extorquens under competitive conditions (Sy et al., 2005). This advantage is associated to the ability to use, as a carbon source, methanol produced during plant-growth. However, some isolates affiliated by 16S rRNA genes to the Methylobacterium genus, through not having mxaF genes, were incapable of colonizing or nodulating Lotononis spp. (Ardley et al., 2009), thereby implying that the capacity to use methanol produced by the plant itself is an important characteristic determining selection.
All the groups containing isolates from two or more different hosts (except group 1, with only one isolate) show species ability in colonizing various hosts. Thus, the host plant is not able to completely select the bacterial genotypes. Controversially, Borreria verticillata isolates were found mainly in group 7 (except for two isolates in group 2), thus indicating that part of Methylobacterium spp. diversity inside the host plant could be determined by specific association, although random events may occur.
Notably, all the isolates observed in group I (from mxaF phylogeny) are present in group 2 (16S rRNA phylogeny), whereas isolates in group VI (mxaF phylogeny) are so in group 7 (16S rRNA phylogeny). However, exceptions occurred, such as eventual changes in positioning. On comparing the two phylogenetic trees, this variable allocation could be attributed to (i) ecological differentiation of the isolate in the environment where it develops (Konstantinidis et al., 2006), or (ii) the occurrence of horizontal gene transfer (HGT) (Heyer et al., 2002).
The results obtained in the present work show the genetic diversity of the Methylobacterium spp. community associated with plants, with the inference that this specific diversity inside the host plant could be impelled not only by the host plant itself, but also by the generalist behavior of some strains for using certain plant compounds, such as alcohols produced during plant metabolism. If so, B. verticillata is the strongest plant species when selecting Methylobacterium spp. endophytes. It can also be concluded that it is possible to acquire additional knowledge on Methylobacterium spp. phylogeny through studies using distinct plant species. In summary, it is assumed that, although, in a first step of plant colonization, the generalist behavior of Methylobacterium species plays a pivotal role in niche occupation, afterwards, niche-specific-association may be driven by the host plant.
Acknowledgments
This work was supported by a grant from the Foundation for Research Assistance, São Paulo State, Brazil (Proc. 2010/07594-5). We thank FAPESP (Proc. 04/15414-6) and CNPq for Fellowships to M.N.D. and F.D.A., respectively.
References
- Andreote FD, Lacava PT, Gai CS, Araújo WL, Maccheroni W, Jr, van Overbeek LS, van Elsas JD, Azevedo JL. Model plants for studying the interaction between Methylobacterium mesophilicum and Xylella fastidiosa. Can J Microbiol. 2006;52:419–426. doi: 10.1139/w05-142. [DOI] [PubMed] [Google Scholar]
- Andreote FD, Carneiro RT, Salles JF, Marcon J, Labate CA, Azevedo JL, Araújo WL. Culture-independent assessment of Rhizobiales-related Alphaproteobacteria and the diversity of Methylobacterium in the rhizosphere and rhizoplane of transgenic eucalyptus. Microbial Ecol. 2009;57:82–93. doi: 10.1007/s00248-008-9405-8. [DOI] [PubMed] [Google Scholar]
- Araújo WL, Saridakis HO, Barroso PAV, Aguilar-Vildoso CI, Azevedo JL. Variability and interactions between endophytic bacteria and fungi isolated from leaf tissues of citrus rootstocks. Can J Microbiol. 2001;47:229–236. doi: 10.1139/w00-146. [DOI] [PubMed] [Google Scholar]
- Araújo WL, Marcon J, Maccheroni W, Jr, van Elsas JD, Azevedo JL. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plant. Appl Environ Microbiol. 2002;68:4906–4914. doi: 10.1128/AEM.68.10.4906-4914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardley JK, O’Hara GW, Reeve WG, Yates RJ, Dilworth MJ, Tiwari RP, Howieson JG. Root nodule bacteria isolated from South African Lotononis bainesii, L. listii and L. solitudinis are species of Methylobacterium that are unable to utilize methanol. Arch Microbiol. 2009;191:311–318. doi: 10.1007/s00203-009-0456-0. [DOI] [PubMed] [Google Scholar]
- Aslam Z, Lee CS, Kim KH, Im WT, Ten LN, Lee ST. Methylobacterium jeotgali sp. nov., a non-pigmented, facultatively methylotrophic bacterium isolated from jeotgal, a traditional Korean fermented seafood. Int J Syst Evol Microbiol. 2007;57:566–571. doi: 10.1099/ijs.0.64625-0. [DOI] [PubMed] [Google Scholar]
- Cao YR, Wang Q, Jin RX, Tang SK, Jiang Y, He WX, Lai HX, Xu LH, Jiang CL. Methylobacterium soli sp. nov. a methanol-utilizing bacterium isolated from the forest soil. Antonie Van Leeuwenhoek. 2011;99:629–634. doi: 10.1007/s10482-010-9535-0. [DOI] [PubMed] [Google Scholar]
- Cervantes-Martinez J, Lopez-Diaz S, Rodriguez-Garay B. Detection of the effects of Methylobacterium in Agave tequilana Weber var. azul by laser-induced fluorescence. Plant Sci. 2004;166:889–892. [Google Scholar]
- Chanprame S, Todd JJ, Widholm JM. Prevention of pink-pigmented methylotrophic bacteria (Methylobacterium mesophilicum) contamination of plant tissues cultures. Plant Cell Rep. 1996;16:222–225. doi: 10.1007/BF01890872. [DOI] [PubMed] [Google Scholar]
- Ferreira A, Quecine MC, Lacava PT, Oda S, Azevedo JL, Araújo WL. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol Lett. 2008;287:8–14. doi: 10.1111/j.1574-6968.2008.01258.x. [DOI] [PubMed] [Google Scholar]
- Gallego V, Garcia MT, Ventosa A. Methylobacterium hispanicum sp. nov. and Methylobacterium aquaticum sp. nov., isolated from drinking water. Int J Syst Evol Microbiol. 2005a;55:281–287. doi: 10.1099/ijs.0.63319-0. [DOI] [PubMed] [Google Scholar]
- Gallego V, Garcia MT, Ventosa A. Methylobacterium isbiliense sp. nov., isolated from the drinking water system of Sevilla, Spain. Int J Syst Evol Microbiol. 2005b;55:2333–2337. doi: 10.1099/ijs.0.63773-0. [DOI] [PubMed] [Google Scholar]
- Gallego V, Garcia MT, Ventosa A. Methylobacterium adhaesivum sp. nov., a methylotrophic bacterium isolated from drinking water. Int J Syst Evol Microbiol. 2006;56:339–342. doi: 10.1099/ijs.0.63966-0. [DOI] [PubMed] [Google Scholar]
- Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer J, Galchenko VF, Dunfield PF. Molecular phylogeny of type II methane-oxidizing bacteria isolated from various environments. Microbiology. 2002;148:2831–2846. doi: 10.1099/00221287-148-9-2831. [DOI] [PubMed] [Google Scholar]
- Inagaki F, Tsunogai U, Suzuki M, Kosaka A, Machiyama H, Takai K, Nunoura T, Nealson KH, Horikoshi K. Characterization of C1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, Southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA and 16S rRNA genes. Appl Environ Microbiol. 2004;70:7445–7455. doi: 10.1128/AEM.70.12.7445-7455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayashree S, Lalitha R, Vadivukkarasi P, Kato Y, Seshadri S. Cellulase production by pink pigmented facultative methylotrophic strains (PPFMs) Appl Biochem Biotechnol. 2011;164:666–680. doi: 10.1007/s12010-011-9166-6. [DOI] [PubMed] [Google Scholar]
- Kang YS, Kim J, Shin HD, Nam YD, Bae JW, Jeon CO, Park W. Methylobacterium platani sp. nov., isolated from a leaf of the tree Platanus orientalis. Int J Syst Evol Microbiol. 2007;57:2849–2853. doi: 10.1099/ijs.0.65262-0. [DOI] [PubMed] [Google Scholar]
- Kato Y, Asahara M, Goto K, Kasai H, Yokota A. Methylobacterium persicinum sp. nov., Methylobacterium komagatae sp. nov., Methylobacterium brachiatum sp. nov., Methylobacterium tardum sp. nov. and Methylobacterium gregans sp. nov., isolated from freshwater. Int J Syst Evol Microbiol. 2008;58:1134–1141. doi: 10.1099/ijs.0.65583-0. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Ramette A, Tiedje JM. Toward a more robust assessment of intraspecific diversity, using fewer genetic markers. Appl Environ Microbiol. 2006;72:7286–7293. doi: 10.1128/AEM.01398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacava PT, Araújo WL, Marcon J, Maccheroni W, Jr, Azevedo JL. Interaction between endophytic bacteria from citrus plants and the phytopathogenic bacteria Xylella fastidiosa, casual agent of Citrus Variegated Chlorosis. Lett Appl Microbiol. 2004;39:55–59. doi: 10.1111/j.1472-765X.2004.01543.x. [DOI] [PubMed] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid-determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:120–130. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhaiyan M, Poonguzhali S, Sundaram SP, Sa TM. A new insight into foliar applied methanol influencing phylloplane methylotrophic dynamics and growth promotion of cotton (Gossypium hirsutum L.) and sugarcane (Saccharum officinarum L.) Environ Exp Bot. 2006a;57:168–176. [Google Scholar]
- Madhaiyan M, Reddy BVS, Anandham R, Senthilkumar M, Poonguzhali S, Sundaram SP, Sa TM. Plant growth-promoting Methylobacterium induces defense responses in groundnut (Arachis hypogaea L.) compared with rot pathogens. Curr Microbiol. 2006b;53:270–276. doi: 10.1007/s00284-005-0452-9. [DOI] [PubMed] [Google Scholar]
- Madhaiyan M, Kim BY, Poonguzhali S, Kwon SW, Song MH, Ryu JH, Go SJ, Koo BS, Sa TM. Methylobacterium oryzae sp. nov., an aerobic, pink-pigmented, facultatively methylotrophic, 1-aminocyclopro-pane-1-carboxylate deaminase-producing bacterium isolated from rice. Int J Syst Evol Microbiol. 2007;57:326–331. doi: 10.1099/ijs.0.64603-0. [DOI] [PubMed] [Google Scholar]
- Madhaiyan M, Poonguzhali S, Kwon SW, Sa TM. Methylobacterium phyllosphaerae sp. nov., a pink-pigmented, facultative methylotroph from the phyllosphere of rice. Int J Syst Evol Microbiol. 2009;59:22–27. doi: 10.1099/ijs.0.001693-0. [DOI] [PubMed] [Google Scholar]
- Madhaiyan M, Poonguzhali S, Senthilkumar M, Lee JS, Lee KC. Methylobacterium gossipiicola sp. nov., a pink-pigmented facultative methylotrophic bacteria isolated from cotton phyllosphere. Int J Syst Evol Microbiol. 2011 doi: 10.1099/ijs.0.030148-0. (Epub) [DOI] [PubMed] [Google Scholar]
- McDonald IR, Kenna EM, Murrell JC. Detection of methanotrofic bacteria in environmental samples with the PCR. Appl Environ Microbiol. 1995;61:116–121. doi: 10.1128/aem.61.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosvi SA, Mcdonald IR, Pearce DA, Kelly DP, Wood AP. Molecular detection and isolation from Antarctica of methylotrophic bacteria able to grow with methylated sulfur compounds. Syst Appl Microbiol. 2005;28:541–554. doi: 10.1016/j.syapm.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Pirttilä AM, Laukkane H, Pospiech H, Myllylã R, Hohtola A. Detection of intracellular bacteria in buds of scoth pine (Pinus sylvestris L.) by in situ hybridization. Appl Environ Microbiol. 2000;66:3037–3077. doi: 10.1128/aem.66.7.3073-3077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovran E, Crowther GJ, Guo X, Yang S, Lidstrom ME. A systems biology approach uncovers cellular strategies used by Methylobacterium extorquens AM1 during the switch from multi-to single-carbon growth. PLoS One. 2010;24:e14091. doi: 10.1371/journal.pone.0014091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy A, Giraud E, Jourand P, Garcia N, Willems A, de Lajudie P, Prin Y, Neyra M, Gillis M, Bivin-Masson C, et al. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol. 2001;183:214–220. doi: 10.1128/JB.183.1.214-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy A, Timmers ACJ, Knief C, Vorholt JA. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl Environ Microbiol. 2005;71:7245–7252. doi: 10.1128/AEM.71.11.7245-7252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software ver. 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Toyama H, Anthony C, Lidstrom ME. Construction of insertion and deletion mxa mutants of Methylobacterium extorquens AM1 by electroporation. FEMS Microbiol Lett. 1998;166:1–7. doi: 10.1111/j.1574-6968.1998.tb13175.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Sahr F, Xue T, Sun B. Methylobacterium salsuginis sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2007;57:1699–1703. doi: 10.1099/ijs.0.64877-0. [DOI] [PubMed] [Google Scholar]
- Weon HY, Kim BY, Joa JH, Son JA, Song MH, Kwon SW, Go SJ, Yoon SH. Methylobacterium iners sp. nov. and Methylobacterium aerolatum sp. nov., isolated from air samples in Korea. Int J Syst Evol Microbiol. 2008;58:93–96. doi: 10.1099/ijs.0.65047-0. [DOI] [PubMed] [Google Scholar]
- Yates RJ, Howieson JG, Reeve WG, Nandasena KG, Law IJ, Bra UL, Ardley JK, Nistelberger HM, Real D, O’Hara GW. Lotononis angolensis forms nitrogen fixing, lupinoid nodules with phylogenetically unique, fast-growing, pink-pigmented bacteria, which do not nodulate L. bainesii or L. listii. Soil Biol Biochem. 2007;39:1680–1688. [Google Scholar]
- Van Aken B, Peres CM, Doty SL, Yoon JM, Schnoor JL. Methylobacterium populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, metane-utilizing bacterium isolated from poplar trees (Populus deltoids x nigra DN34) Microbiology. 2004;54:1191–1196. doi: 10.1099/ijs.0.02796-0. [DOI] [PubMed] [Google Scholar]