Abstract
Abnormal genome hypermethylation participates in the tumorigenesis and development of prostate cancer. Prostate cancer cells highly express DNA methyltransferase 3 (DMNT3) family genes, essential for maintaining genome methylation. In the present study, multi-target siRNA, based on the homologous region of the DNMT3 family, was designed for the in vitro investigation of its effects on the proliferation, migration, and invasion of TSU-PR1 prostate cancer cells. The consequential cell-cycle derangement, through DNMT3A/B or only DNMT3B silencing, was partially efficient, without affecting apoptosis. DNMT3A silencing had absolutely no effect on changing TSU-PR1 cell biological behavior. Hence, DNMT3B alone apparently plays a key role in maintaining the unfavorable behavior of prostate-cancer cells, thereby implying its potential significance as a promising therapeutic target, with DNMT3A simply in the role of helper.
Keywords: prostate cancer, DNA methylation, DNMT3, RNA interference
Introduction
Prostate cancer is the most common solid tumors in men in developed countries. In China prevalence is continuously on the rise, due to changes in lifestyle and an aging population. Abnormal regulation of the enzymes involved in DNA methylation, and incorrect methylation of CpG islands, are all common phenomena in human tumors (Sharma et al., 2010). Specifically, abnormal genome hypermethylation in the tumorigenesis and development of prostate cancer, with higher expression of DNMT proteins compared to normal prostate tissue, has been established (Benbrahim-Tallaa et al., 2007; Morey Kinney et al., 2008; Lin et al., 2011). The DNMT family, including DNMT1, DNMT2, DNMT3, etc., plays a critical role in the initiation and maintenance of DNA methylation. The DNMT3 subfamily includes DNMT3A and DNMT3B, crucial in the process of de novo genome methylation (Das and Singal, 2004; Siedlecki and Zielenkiewicz, 2006; Patra et al., 2008). The catalytic domain of DNMT3 resides in its C terminal, which is highly conserved among mammals (Cheng and Blumenthal, 2008). As the specific function of the DNMT3 subfamily in the initiation and development of prostate cancer remains elusive, it was decided to apply novel multi-target small interfering RNA (siRNA) targeting at the conservative region of human DNMT3 family proteins to the human prostate cancer cell line TSU-PR1, and then record and analyze subsequent changes in biological behavior. The results could be of help in enlightening the roles played by the observed high-level DNMT expression in prostate cancer cells. Furthermore, our Multi-target siRNA design will also be useful in discerning the importance of the two subfamily members.
Material and Methods
Multi-target siRNA construction
The homologous sequences of DNMT3A (NM175629.1NM022552.3NM175630.1) and DNMT3B (NM066892NM175848.1NM175849.1NM001207055.1N M00120756.1) genes were determined through 2 Sequence-Blast. Three pairs of siRNAs (siDNMT3AB-1, siDNMT3AB-2 and siDNMT3AB-3) for targeting at the conservative sequence coding catalytic domain of both DNMT3A and DNMT3B were designed online at the website of Ambion Inc. The principals, as presented in Table 1, were strictly observed for siRNA optimization. siRNA-NC served as negative control (5′-GACTTCATA AGGCGCATGCTTCAAGACGGCATGCGCCTTATGA-3′) The sequences of siRNA for DNMT3A and DNMT3B were taken from a previous study (Robert et al., 2003: DNMT3A 5′-GCCTCAGAGCTATTACCCA-3′, DNMT3B 5′-GAAGATCAAGCTCGCGACT-3′). After careful checking to assure specificity across the whole genome, all the siRNAs, were synthesized by Wuhan Genesil Biotechnology Co., Ltd. A FAM – labeled siRNA was also synthesized to evaluate transfection efficiency.
Table 1.
Location of candidate siRNA target sequences in different DNMT3A and DNMT3B transcripts.
| DNMT3AB-1 | DNMT3AB-2 | DNMT3AB-3 | ||
|---|---|---|---|---|
| Accession No. | 5′-CCAGTGATGATTGATGCCA-3′ | 5′-GATACTTCTGGGGCAACCT-3′ | 5′-ATAAACTCGAGCTGCAGGA-3′ | |
| DNMT3A | NM175629.1 | 2670–2685(EXON 23) | 2715–2731(EXON 23) | 2778–2788(EXON 24) |
| NM022552.3 | 2589–2604(EXON 23) | 2634–2650(EXON 23) | 2697–2707(EXON 24) | |
| NM175630.1 | No similarity | 814–821(EXON 5b) | 1177–1185(EXON 5b) | |
| DNMT3B | NM066892 | 2473–2491(EXON 21) | 2519–2537(EXON 21) | 2576–2594(EXON 22) |
| NM175848.1 | 2413–2431(EXON 21) | 2459–2477(EXON 86) | 2516–2534(EXON 22) | |
| NM175849.1 | 2413–2431(EXON 21) | 2459–2477(EXON 21) | 398–405(EXON 3) | |
| NM001207055.1 | 2287–2305(EXON 21) | 2333–2351(EXON 21) | 398–405(EXON 3) | |
| NM00120756.1 | 2185–2203(EXON 21) | 2231–2249(EXON 21) | 398–405(EXON 3) | |
Cell line screening
Human prostate cancer cell LNCaP and its sublines (C4, C42, C4-2B), ARCaP cell lines IA-8, IF-11, and PC-3, DU-145, TSU-PR1 were provided by Professor Leland WK Chuang (Emory University, USA), and preserved at the Institute of Urology, Medicine College of Xi’an Jiaotong University.
Total RNA was isolated with Trizol and quantitated by absorbance at 260 nm. 2 μg RNA was reverse-transcribed into cDNA by using a RevertAidtm First Strand cDNA Synthesis Kit (MBI Fermentas, St Leon-Rot, Germany). Primer sequences for DNMT3A, DNMT3B and β-actin are presented in Table 2. The PCR amplification procedure was as follows: 94 °C for 5 min, 32 cycles of 90 °C for 30 s, 58 °C for 30 s, 72 °C for 45 s, followed by a final extension of 72 °C for 7 min. PCR products were run on a 2% agarose gel (120 V, 45 min) and visualized using ethidium bromide staining (Benbrahim-Tallaa et al.). The mean density of each band was analyzed using Glyko BandScan software (Glyko, Novato, USA).
Table 2.
Primer sequences for DNMT3A, DNMT3B and β-actin.
| Primer | Sense (5′-3′) | Antisense (5′-3′) | Product length (bp) |
|---|---|---|---|
| DNMT3A | CACACAGAAGCATATCCAGGAGTG | AGTGGACTGGGAAACCAAATACCC | 551 |
| DNMT3B | AATGTGAATCCAGCCAGGAAAGGC | ACTGGATTACACTCCAGGAACCGT | 190 |
| β-actin | ATCATGTTTGAGACCTTCAACA | CATCTCTTGCTCGAAGTCCA | 330 |
All the cells were washed twice with PBS (0.01 M, pH 7.4), and then lysed by an ice-cold radio immuno-precipitation assay RIPA buffer (Beyotime Institute of bio-technology, China) containing 1% Cocktail and 1 mmol/L Phenylmethanesulfonyl fluoride PMSF (Beyotime Institute of biotechnology, China). Purified lysates (150 μg) of each sample were separated by 12% SDS-PAGE and transferred to a nitrocellulose membrane at 4 °C (25 V, 12 h). The membrane was incubated with TBST (50 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 7.6)-diluted primary antibodies against desired molecules (Santa Cruz, CA, USA) overnight at 4 °C, and then incubated with HRP coupled rabbit anti-goat IgG (1:3000) or mouse IgG (1:3000) (Invitrogen, Life Technologies Corporation, USA) for 1 h at room temperature. Finally, the membrane was visualized by ECL (Beyotime Institute of biotechnology, China). Loading difference was normalized using a human GAPDH antibody (Invitrogen, Life Technologies Corporation, USA). The mean density of each band was analyzed using Glyko BandScan software (Glyko, Novato, USA).
Transfection
The optimal transfection concentration was determined by siRNA-FCM. The cell line which expressed DNMT3A and DNMT3B at the highest level was transfected with siDNMT3AB-1, siDNMT3AB-2, siDNMT3AB-3 and siRNA-NC respectively, according to the standard lipofectamintm 2000 protocol (Invitrogen, CA, USA). The culture medium was replaced by fresh DMEM (Gibco, Life Technologies Corporation, United States) supplemented with 10% fetal bovine serum (FBS) 4 h after the transfection process. Total RNA and proteins were extracted 48 h later for assaying DNMT3A and DNMT3B expression.
MTT assay
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT, Sigma, St Louis, MO, USA) proliferation assays were performed to assess changes in TSU-PR1 cell viability after transfection. After the respective transfection process of siDNMT3AB-1 (Group 1), siDNMT3B (Group 2), siDNMT3A (Group 3) and siRNA-NC (Negative Control) for 48 h, cells were seeded at a density of 1 × 103/well in 96-well plates. After culture for 24 h, 48 h, 72 h and 96 h, 20 μL of MTT (5 mg/mL in PBS) was added to each well, and kept for 4 h, whereupon the medium was removed and 150 μL DMSO added to solubilize formazan crystals. Absorbance (OD) was measured at 590 nm by a Microplate Autoreader (Bio-Tek Instruments, VT, USA). The growth inhibitory rate was calculated according to the formula: growth inhibitory rate = (average OD value in the control group - average OD value in the treatment group)/average OD value in the control groupx100%.
Scratch assay
Cells from each group were inoculated into 6-well dishes at a concentration of 5 × 104/well at 48 h after transfection. When cellular fusion reached 80%, a 20 μL tip was used to draw a vertical line across the dishes, after which cell debris was washed twice with sterile PBS, and fresh medium added. Photos were taken immediately after scratching, as well as 24 and 48 h later. Wound closure was monitored with a UOP DSZ500X inverted microscope (UOP Microscope, Chongqing, China). Wound sizes were checked against the scale of the images to ensure that all the respective widths were the same at the start. Migration inhibitory rate was calculated according to the formula: migration inhibitory rate = (average migration distance in the control group – average migration distance in the treatment group)/average migration distance in the control group × 100.
Invasive assay
Matrigel (Sigma, St Louis, MO, USA) and serum-free RPMI-1640 medium (Gibco, Life Technologies Corporation, USA) were mixed at a ratio of 1:5. 50 μL of the mixture was placed into the upper chamber of a Transwell permeable support (Corning, NY, USA) with an 8 μm pore size, prior to incubating at 37 °C for 4 h. 100 μL suspension of each group, containing 5 × 103 transfected cells were inoculated into the upper chamber of the Transwell permeable support for 48 h. Simultaneously, a 600 μL RPMI-1640 medium containing 20% FBS was added to the lower chamber. Both chambers were left in an incubator for 24 h, prior to washing with PBS, fixing in formaldehyde for 5 min, and final staining with 0.1% crystal purple for 3 min. The number of penetrated cells was counted in three random fields (100x) per filter.
Cell cycle apoptosis assay
48 h after transfection, the TSU-PR1 cells of each group were collected and labeled with propidium iodide (Sigma, St Louis, MO, USA). 1 × 104 cells per group were counted by flow cytometry (FACSCalibur, BD Inc, USA) to determine both changes in DNA content throughout the cell-cycle, and the percentage of cells in each cell stage. Cell-cycle regulatory protein Cyclin D and PCNA were tested by western blotting using multiclonal human Cyclin D (1:500) and PCNA (1:300) antibodies (Santa Cruz, CA, USA), respectively.
Apoptosis assay
48 h after transfection, TSU-PR1 cells of each group were collected and labeled with Annexin V Kit (Sigma, St Louis, MO, USA). Flow cytometry was used to detect FITC and propidium iodide fluorescence of each group. The number of apoptotic cells was also calculated. Apoptosis regulatory protein Cleaved Caspase 3 was tested by western blotting using polyclonal human Caspase 3 (1:500) (Santa Cruz, CA, USA).
Statistical analysis
All assaying was three-fold. Data were presented as means ± SEM. One-way ANOVA was applied for multiple comparisons between groups. In all cases, p < 0.05 was considered statistically significant. All statistical analyses were performed using SPSS 13.0 (SPSS Inc, Chicago, IL, USA).
Results
DNMT3A and DNMT3B expression in prostate cancer cell-lines
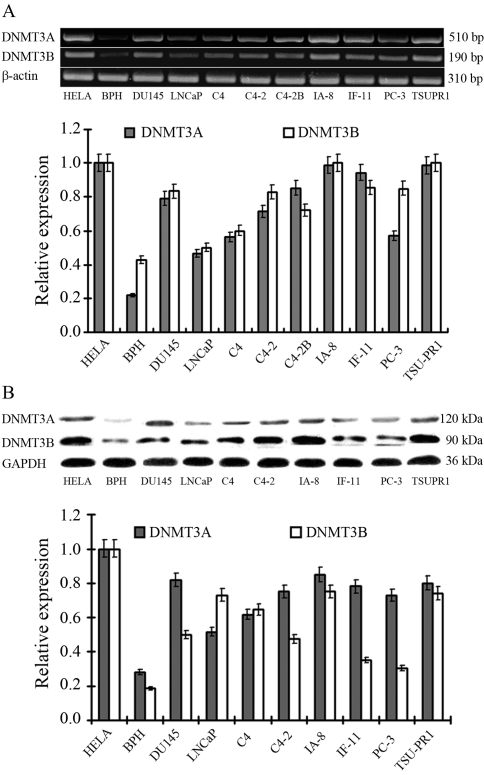
Compared with negative control BPH cells, all prostate cancer cell lines expressed DNMT3A and DNMT3B at much higher levels (Figure 1). As revealed by semi-quantative PCR and western blot analysis, TSU-PR1 and IA-8 were the two cell lines which showed the the highest levels of DNMT3A and DNMT3B expression. Since IA-8 transfection efficiency was extremely low, TSU-PR1 entered into subsequent studies.
Figure 1.
DNMT3A and DNMT3B expression in prostate cancer cell lines assayed by semi-quantative PCR (A) and western blotting (B). The Hela cell line served as positive control and the BPH as negative.
Inhibition efficiency
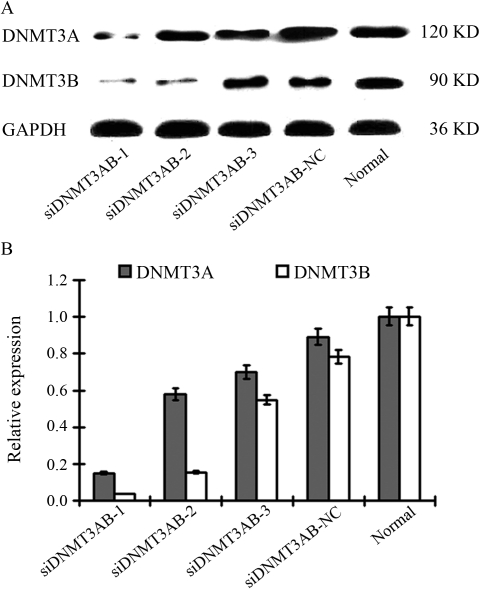
As determined by siRNA-FCM, the optimal transfection concentration was 140 nM. In comparison with the negative control, siDNMTAB-1 could effectively knock down both DNMT3A and DNMT3B, whereas siDNMTAB-2 only inhibited DNMT3B expression, and siDNMT3AB-3 had absolutely no effect on the expression of either (Figure 2).
Figure 2.
DNMT3A and DNMT3B expression in TSU-PR1 cells after siRNA transfection detected by western blot analysis.
Efffects of siRNA transfection
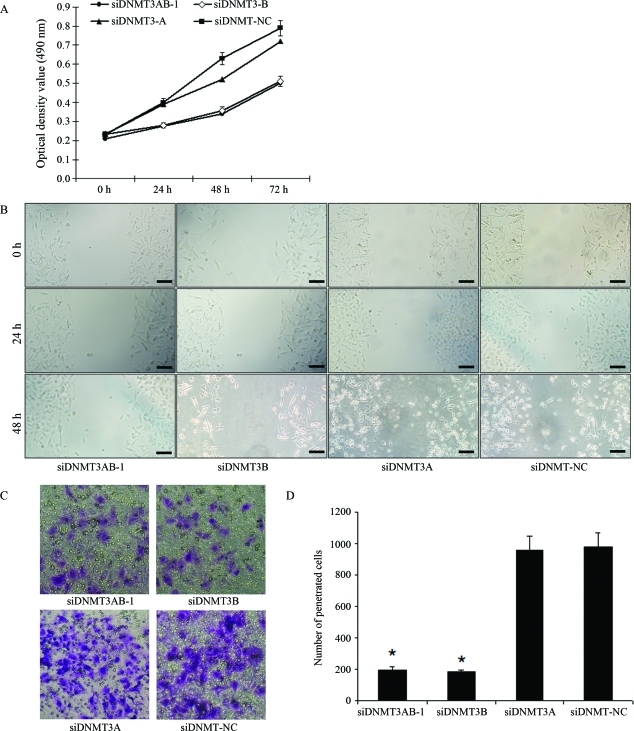
MTT assaying indicated that the knockdown of DNMT3A/B (Group1) or DNMT3B alone (Group 2) significantly inhibited the proliferation of TSU-PR1 cells at 72 h (p < 0.05), whereas silencing DNMT3A (Group 3) alone had no inhibitory effect on the growth of TSU-PR1 cells compared to the negative control (p > 0.05) (Figure 3A).
Figure 3.
Results from proliferation (A), migration (B) and invasion (C, D) assays of TSU-PR1 cells after siRNA transfection. Scale bar = 200 μm.
Migration inhibition rates of Groups 1, 2 and 3 were 10.1 ± 1.25%, 9.87 ± 0.99% and 11.67 ± 0.89% respectively, at 24 h after transfection, with no significant difference among the three. At 48 h post-transfection, the rates of Group 1, Group 2 and Group 3 were 43.2 ± 1.58%, 39.74 ± 0.72% and 14.11 ± 0.52%, respectively; hence, inhibition in Groups 1 and 2 was significantly lower than in Group 3 (p < 0.05) (Figure 3B).
According to invasive assaying, the number of TSU-PR1 cells penetrating the membrane in Groups 1 and 2 was significantly lower (p < 0.05) than in Group 3 and negative control, with no significant difference between the former two (Figure 3C, D).
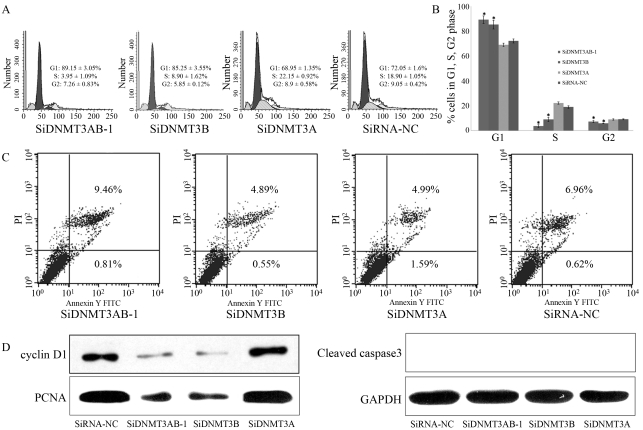
In cell cycle analysis, the percentage of cells in G1 phase in Groups 1 and 2 was much higher (p < 0.05) and the proportion of cells in S and G2 phase, lower (p < 0.05) compared to Group 3 and negative control; No differences were noted between Groups 1 and 2 (Figure 4A,B).
Figure 4.
Cell cycle distribution (A, B and C) and apoptosis changes (D) in TSU-PRI cells after siRNA transfection.
In Groups 1, 2 and 3 and negative control, the early apoptosis rates were 0.81 ± 0.12%, 0.55 ± 0.11%, 1.59 ± 0.19% and 0.62 ± 0.12%, respectively, and the late apoptosisrates 9.46 ± 0.31%, 4.89 ± 0.22%, 4.49 ± 0.45% and 6.96 ± 0.35% (Figure 4C). There was no significant difference in either of the two rates among the experimental groups.
In western blot analysis (Figure 4D), whereas cyclin-D and PCNA expression in Groups 1 and 2 decreased significantly, cleaved caspase3 was not detected in either group. Consistent with the results of cell cycle and apoptosis analyses, there was no significant change in the expression of cell cycle and apoptosis-related proteins in Group 3.
Discussion
Hypermethylation of certain genes is very common in human cancers, including prostate cancer (Kim et al., 2010; Takada et al., 2010). DNMT expression is also elevated in many cancer tissues (Girault et al., 2003; Auerkari, 2006; Kanai and Hirohashi, 2007; Ding et al., 2008). High DNMT expression, which generally takes place prior to DNA methylation, might possibly be one of the causes of abnormal DNA methylation. Besides the role played by the DNMT family in DNA hypermethylation and tumorigenesis, supression of DNMT1 expression in bladder cancer cells was shown to induce demethylation and reactivation of the silenced tumor suppressor gene CDKN2A. On the other hand, the colon cancer cell HCT116 lacking DNMT1 expression still presented significant DNA methylation, whereas HCT116 clones with inactivation of both DNMT1 and DNMT3B showed much lower levels of DNA methylation (Robert et al., 2003), thereby indicating that members of the DNMT family work together complementarily or synergistically in the control of tumorigenesis,
Based on these findings, the community has come to consider DNMT as a new target for tumor treatment. 5-aza -2′-deoxycytidine (5-Aza-CdR, Decitabine), a DNMT inhibitor, is capable of suppressing tumor cells (Momparler, 2003, 2005) by combining with DNMT genes in the process of promoter methylation (Momparler, 2003, 2005; Yu and Wang, 2008). However, its short half-life in vivo, frequent inactivation caused by cytidine deaminase, neutropenia, and certain other side effects, have all limited its application in the case of solid cancers, whereby the urgent need for finding a new way of improving the stability and efficiency of these inhibitors.
In recent years, small interfering RNA (siRNA) technology has reached maturity, and is now widely used in cancer research in the characterization of gene functions (Hannon, 2002; Pai et al., 2006). The traditional design pattern is that one piece of siRNA aims at one specific sequence (Reynolds et al., 2004). Given that tumorigenesis and development generally involve dysfunction of a variety of co-regulated genes, silencing a single gene often does not necessarily lead to the expected results. The simultaneous knockdown of multiple genes can be achieved by either co-transfection (Leu et al., 2003) or duplication of transfection (Przybylski et al., 2010). However, the drawbacks are also obvious. Experimental steps are complicated, and excessive transfection reagents, by impairing target cells, may finally lead to false positive results. In contrast, multi-target siRNA is capable, not only of knocking down several genes simultaneously, with much less side effects than traditional methods. Kunze et al. (2008) when inhibiting the expression of the apoptosis suppressor protein bcl-2 family in bladder cancer cells using multi-target siRNA, showed that the proliferation of cancer cells was significantly suppressed. In the present study, the successful construction of multi-target siRNA aiming at the DNMT3 family proved its biological validity.
In-depth analysis indicated the C terminal enzyme catalytic region of the DNMT3 family to be highly conserved and homologous. The strong point of the present study was the successful synthesis of multi-target siRNAs targeting DNMT3A and DNMT3B simultaneously. Notably, silencing of the DNMT family could dramatically suppress the proliferation, invasion and migration of TSU-PR1 prostate cancer cells. Obviously, cell-cycle related protein expression also changed. The observed biological effects of suppression of DNMT3 expression may be due to the genome methylation level being down-regulated by silencing of the DNMT3 family, and thus, the genes which had been suppressed through abnormal hypermethylation in cancer cells regained activity. The present findings imply that the DNMT3 family may be a promising therapeutic target.
Unexpectedly, the apoptosis level of TSU-PR1 cells was not significantly influenced by silencing of the DNMT3 family, most possibly for two reasons. In the first place, cell-species specificity might influence the apoptosis procedure. For example, it was reported that p53 was necessary for DNMT to induce cell apoptosis (Schneider-Stock et al., 2005), whereas TSU-PR1 cells did not express p53 (Wang et al., 2009). Secondly, cell-cycle related regulators, such as the DNA repair system and anti-apoptosis genes might be activated by DNMT3 silencing, thereby preventing apoptosis of TSU-PR1 cells.
To further elucidate the function of the DNMT3 family, we separately silenced DNMT3A/B, DNMT3A and DNMT3B. Although DNMT3A/B or DNMT3B silencing significantly inhibited TSU-PR1 cell proliferation, invasion and migration, with no significant differences between the two, DNMT3A silencing lead to no observable changes. On the other hand, proliferation and apoptosis related proteins did present consistent changes. The silencing of either DNMT3A/B or DNMT3B expression significantly reduced the expression levels of cyclinD1 and PCNA, whereas no significant changes were manifest after DNMT3A silencing in TSU-PR1 cells. Similar results were reported in liver cancer cells following DNMT3B knockdown (Bai et al., 2005). According to flow cytometry, apoptosis-related protein caspase3 was not detected in any group. Therefore, although abnormal DNMT3 expression in TSU-PR1 prostate cancer cells regulated the cell cycle via cyclin D1, cell apoptosis was not visibly affected. Furthermore, DNMT3B appeared to play an overwhelming role in the process, whereas DNMT3A simply functioned as assistant, thereby presupposing that the spontaneous knocking down of both DNMT3A and DNMT3B together was in no way more advantageous than knocking down just DNMT3B.
This is a first-time elaboration of DNMT3-family functions in prostate cancer. However, due to the limited period and low efficiency of transient transfection, the effect of DNMT3 family silencing requires further validation. The establishment of a stable and efficient knock-out of the DNMT3 family in prostate cancer cells will be the focus of our future work.
In conclusion, we first demonstrated that multi-target siRNA targeting at the conservative homology region of DNMT3 family members, could effectively inhibit the proliferation, migration and invasion of TSU-PR1 prostate cancer cells. This study has thus laid the foundations for further research on the important role of DNMT3 in prostate cancer cells, and provides momentum to the clinical application of multi-target siRNA against DNMT3A and DNMT3B in the treatment of prostate cancer.
Acknowledgments
This work was supported by the Science Foundation for Young Scholars of the First Affiliated Hospital of Medical School, Xi’an Jiaotong University and National Scientific Foundation No. 30901500/H1619. We also thank Dr. Pengyuan Liu from Washington University School of Medicine, St. Louis for helpful suggestions and critical revision of the current prior version of the manuscript.
References
- Auerkari EI. Methylation of tumor suppressor genes p16(INK4a), p27(Kip1) and E-cadherin in carcinogenesis. Oral Oncol. 2006;42:5–13. doi: 10.1016/j.oraloncology.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Bai S, Ghoshal K, Datta J, Majumder S, Yoon SO, Jacob ST. DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Mol Cell Biol. 2005;25:751–766. doi: 10.1128/MCB.25.2.751-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Benbrahim-Tallaa L, Waterland RA, Dill AL, Webber MM, Waalkes MP. Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase. Environ Health Perspect. 2007;115:1454–1459. doi: 10.1289/ehp.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: A structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- Ding WJ, Fang JY, Chen XY, Peng YS. The expression and clinical significance of DNA methyltransferase proteins in human gastric cancer. Dig Dis Sci. 2008;53:2083–2089. doi: 10.1007/s10620-007-0145-2. [DOI] [PubMed] [Google Scholar]
- Girault I, Tozlu S, Lidereau R, Bieche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- Kunze D, Wuttig D, Fuessel S, Kraemer K, Kotzsch M, Meye A, Grimm MO, Hakenberg OW, Wirth MP. Multi-target siRNA inhibition of antiapoptotic genes (XIAP, BCL2, BCL-X(L)) in bladder cancer cells. Anticancer Res. 2008;28:2259–2263. [PubMed] [Google Scholar]
- Leu YW, Rahmatpanah F, Shi H, Wei SH, Liu JC, Yan PS, Huang TH. Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res. 2003;63:6110–6115. [PubMed] [Google Scholar]
- Lin J, Haffner MC, Zhang Y, Lee BH, Brennen WN, Britton J, Kachhap SK, Shim JS, Liu JO, Nelson WG, et al. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. Prostate. 2011;71:333–343. doi: 10.1002/pros.21247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momparler RL. Cancer epigenetics. Oncogene. 2003;22:6479–6483. doi: 10.1038/sj.onc.1206774. [DOI] [PubMed] [Google Scholar]
- Momparler RL. Pharmacology of 5-Aza-2′-deoxycytidine (decitabine) Semin Hematol. 2005;42:S9–16. doi: 10.1053/j.seminhematol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Morey Kinney SR, Smiraglia DJ, James SR, Moser MT, Foster BA, Karpf AR. Stage-specific alterations of DNA methyltransferase expression, DNA hypermethylation, and DNA hypomethylation during prostate cancer progression in the transgenic adenocarcinoma of mouse prostate model. Mol Cancer Res. 2008;6:1365–1374. doi: 10.1158/1541-7786.MCR-08-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SI, Lin YY, Macaes B, Meneshian A, Hung CF, Wu TC. Prospects of RNA interference therapy for cancer. Gene Ther. 2006;13:464–477. doi: 10.1038/sj.gt.3302694. [DOI] [PubMed] [Google Scholar]
- Patra SK, Patra A, Rizzi F, Ghosh TC, Bettuzzi S. Demethylation of (Cytosine-5-C-methyl) DNA and regulation of transcription in the epigenetic pathways of cancer development. Cancer Metastasis Rev. 2008;27:315–334. doi: 10.1007/s10555-008-9118-y. [DOI] [PubMed] [Google Scholar]
- Przybylski M, Kozlowska A, Pietkiewicz PP, Lutkowska A, Lianeri M, Jagodzinski PP. Increased CXCR4 expression in AsPC1 pancreatic carcinoma cells with RNA interference-mediated knockdown of DNMT1 and DNMT3B. Biomed Pharmacother. 2010;64:254–258. doi: 10.1016/j.biopha.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, Macleod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- Schneider-Stock R, Diab-Assef M, Rohrbeck A, Foltzer-Jourdainne C, Boltze C, Hartig R, Schonfeld P, Roessner A, Gali-Muhtasib H. 5-Aza-cytidine is a potent inhibitor of DNA methyltransferase 3a and induces apoptosis in HCT-116 colon cancer cells via Gadd45- and p53-dependent mechanisms. J Pharmacol Exp Ther. 2005;312:525–536. doi: 10.1124/jpet.104.074195. [DOI] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedlecki P, Zielenkiewicz P. Mammalian DNA methyltransferases. Acta Biochim Pol. 2006;53:245–256. [PubMed] [Google Scholar]
- Takada H, Wakabayashi N, Dohi O, Yasui K, Sakakura C, Mitsufuji S, Taniwaki M, Yoshikawa T. Tissue factor pathway inhibitor 2 (TFPI2) is frequently silenced by aberrant promoter hypermethylation in gastric cancer. Cancer Genet Cytogenet. 2010;197:16–24. doi: 10.1016/j.cancergencyto.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Wang JC, Wang JX, Liu HR, Zhang YG. Influence of DNA methyltransferase 3b on the expression of cyclin D1 gene and methylation of its promoters in human hepatocellular carcinoma cells. Zhonghua Yi Xue Za Zhi. 2009;89:555–558. (Abstract in English) [PubMed] [Google Scholar]
- Yu N, Wang M. Anticancer drug discovery targeting DNA hypermethylation. Curr Med Chem. 2008;15:1350–1375. doi: 10.2174/092986708784567653. [DOI] [PubMed] [Google Scholar]
Internet Resources
- 2 Sequence-Blast. http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi.