Full text
PDF





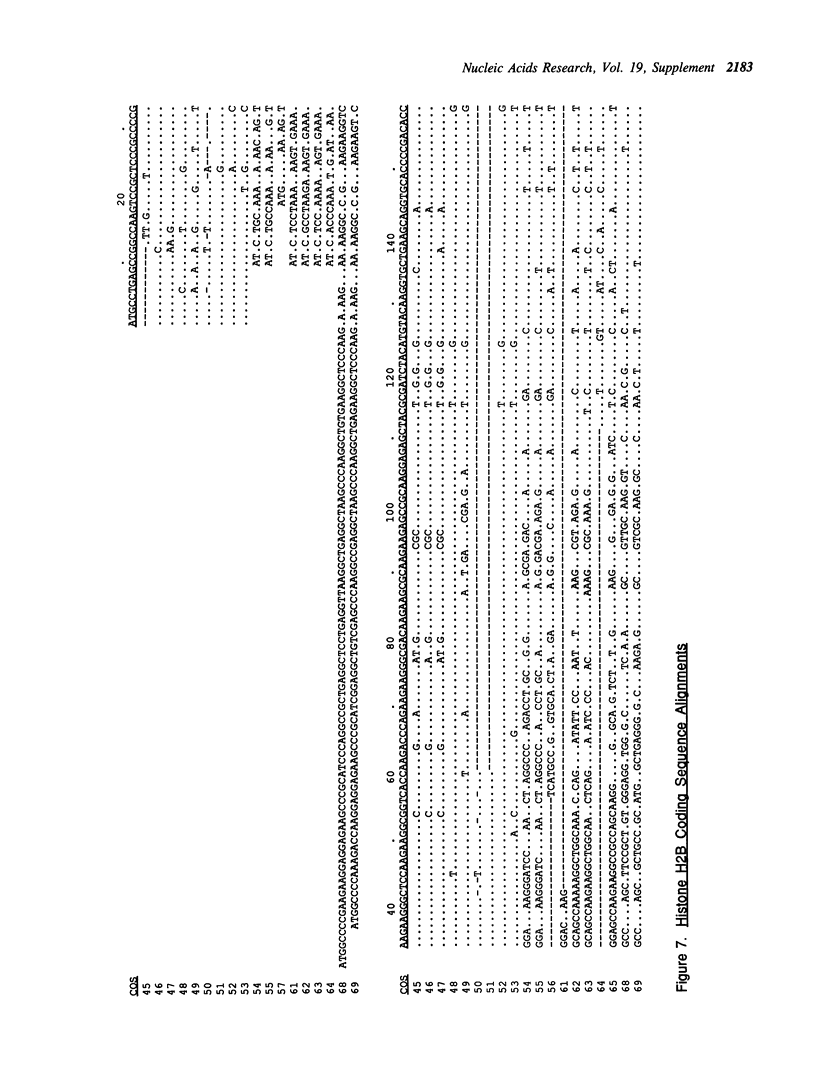
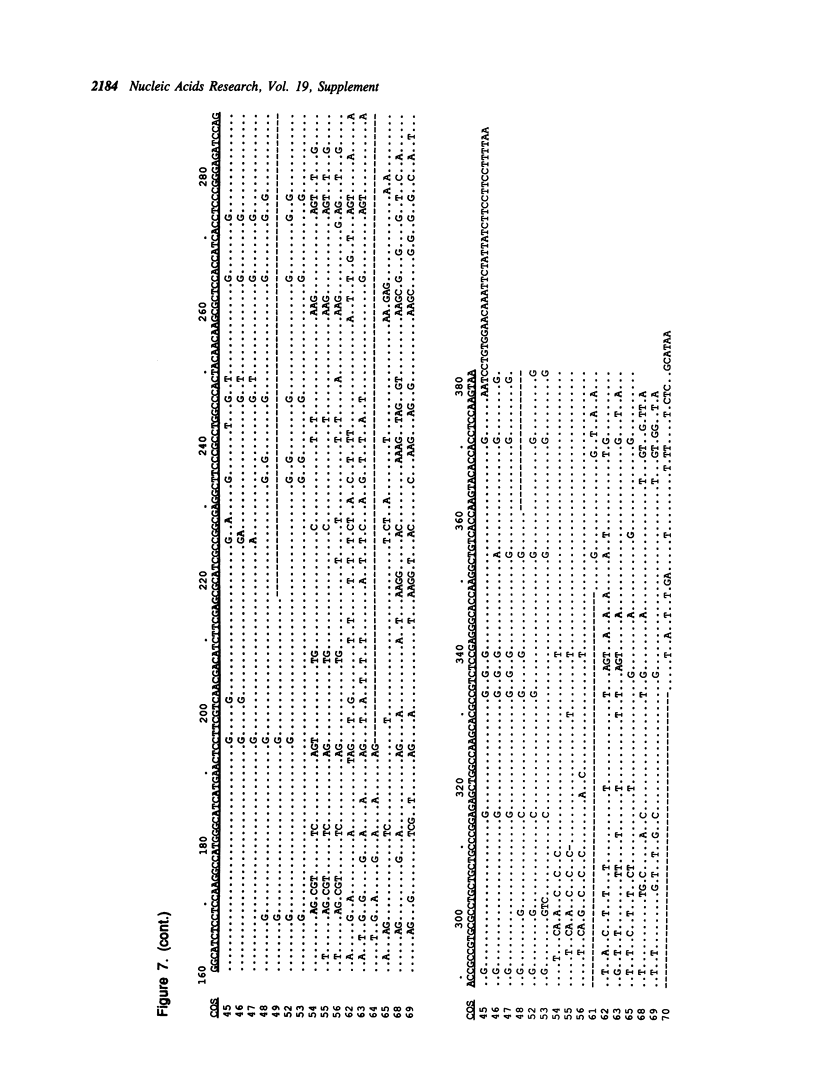
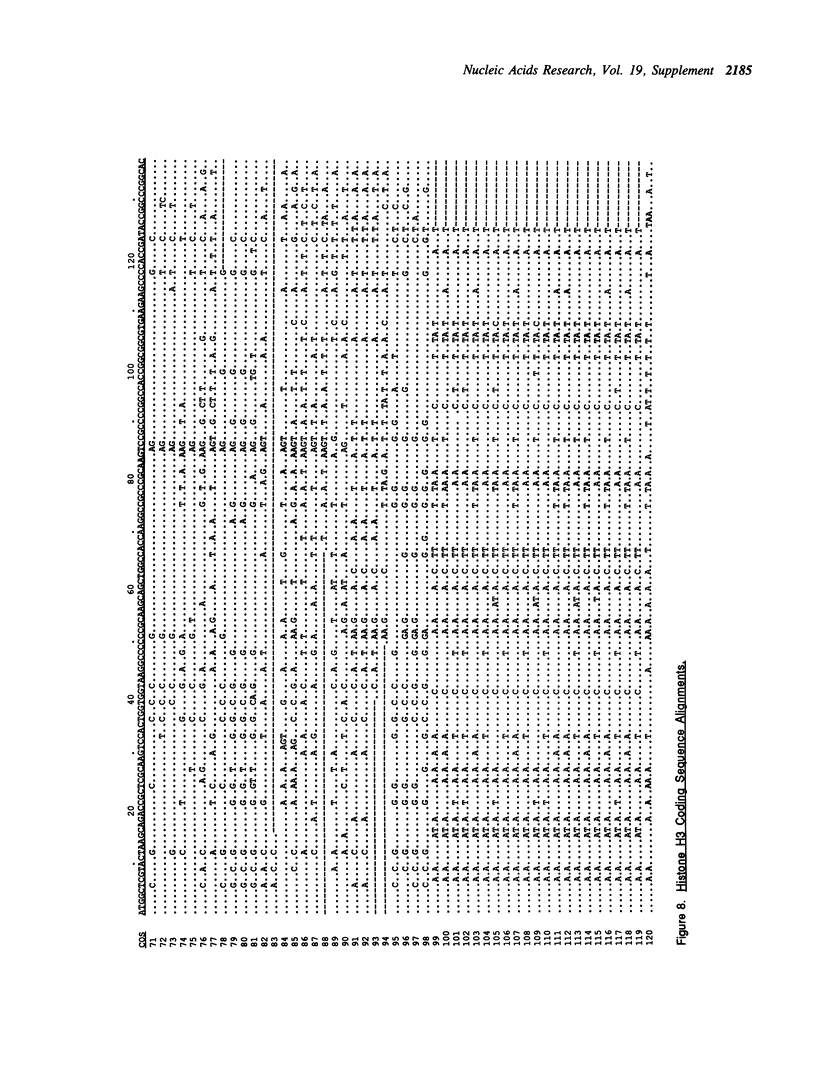
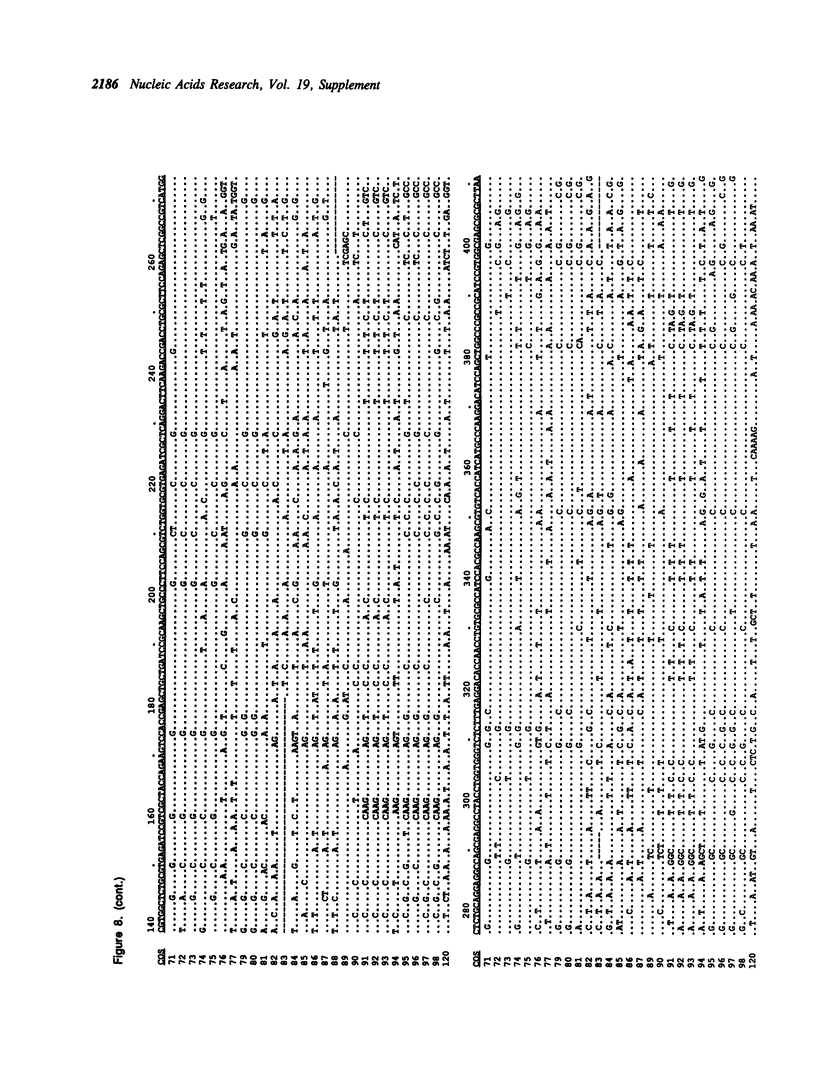
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso A., Breuer B., Bouterfa H., Doenecke D. Early increase in histone H1(0) mRNA during differentiation of F9 cells to parietal endoderm. EMBO J. 1988 Oct;7(10):3003–3008. doi: 10.1002/j.1460-2075.1988.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artishevsky A., Wooden S., Sharma A., Resendez E., Jr, Lee A. S. Cell-cycle regulatory sequences in a hamster histone promoter and their interactions with cellular factors. 1987 Aug 27-Sep 2Nature. 328(6133):823–827. doi: 10.1038/328823a0. [DOI] [PubMed] [Google Scholar]
- Brandt W. F., de Andrade Rodrigues J., von Holt C. The amino acid sequence of wheat histone H2B(2). A core histone with a novel repetitive N-terminal extension. Eur J Biochem. 1988 May 2;173(3):547–554. doi: 10.1111/j.1432-1033.1988.tb14033.x. [DOI] [PubMed] [Google Scholar]
- Brunk C. F., Kahn R. W., Sadler L. A. Phylogenetic relationships among Tetrahymena species determined using the polymerase chain reaction. J Mol Evol. 1990 Mar;30(3):290–297. doi: 10.1007/BF02099999. [DOI] [PubMed] [Google Scholar]
- Chalmers M., Wells D. Extreme sequence conservation characterizes the rabbit H3.3A histone cDNA. Nucleic Acids Res. 1990 May 25;18(10):3075–3075. doi: 10.1093/nar/18.10.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G. H., Nandi A., Clerk S., Skoultchi A. I. Different 3'-end processing produces two independently regulated mRNAs from a single H1 histone gene. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7002–7006. doi: 10.1073/pnas.86.18.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole K. D., Kandala J. C., Kremer E., Kistler W. S. Isolation of a genomic clone encoding the rat histone variant, H1d. Gene. 1990 May 14;89(2):265–269. doi: 10.1016/0378-1119(90)90015-j. [DOI] [PubMed] [Google Scholar]
- Cool D., Banfield D., Honda B. M., Smith M. J. Histone genes in three sea star species: cluster arrangement, transcriptional polarity, and analyses of the flanking regions of H3 and H4 genes. J Mol Evol. 1988;27(1):36–44. doi: 10.1007/BF02099728. [DOI] [PubMed] [Google Scholar]
- Domier L. L., Rivard J. J., Sabatini L. M., Blumenfeld M. Drosophila virilis histone gene clusters lacking H1 coding segments. J Mol Evol. 1986;23(2):149–158. doi: 10.1007/BF02099909. [DOI] [PubMed] [Google Scholar]
- Fecker L., Ekblom P., Kurkinen M., Ekblom M. A genomic clone encoding a novel proliferation-dependent histone H2A.1 mRNA enriched in the poly(A)+ fraction. Mol Cell Biol. 1990 Jun;10(6):2848–2854. doi: 10.1128/mcb.10.6.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt J. S., Key J. L. Molecular cloning of a pea H1 histone cDNA. Eur J Biochem. 1987 Jul 1;166(1):119–125. doi: 10.1111/j.1432-1033.1987.tb13490.x. [DOI] [PubMed] [Google Scholar]
- Gargiulo G., Razvi F., Ruberti I., Mohr I., Worcel A. Chromatin-specific hypersensitive sites are assembled on a Xenopus histone gene injected into Xenopus oocytes. J Mol Biol. 1985 Feb 5;181(3):333–349. doi: 10.1016/0022-2836(85)90223-2. [DOI] [PubMed] [Google Scholar]
- Grimes S., Weisz-Carrington P., Daum H., 3rd, Smith J., Green L., Wright K., Stein G., Stein J. A rat histone H4 gene closely associated with the testis-specific H1t gene. Exp Cell Res. 1987 Dec;173(2):534–545. doi: 10.1016/0014-4827(87)90293-x. [DOI] [PubMed] [Google Scholar]
- Gruber A., Streit A., Reist M., Benninger P., Böhni R., Schümperli D. Structure of a mouse histone-encoding gene cluster. Gene. 1990 Nov 15;95(2):303–304. doi: 10.1016/0378-1119(90)90377-4. [DOI] [PubMed] [Google Scholar]
- Harper D. S., Jahn C. L. Actin, tubulin and H4 histone genes in three species of hypotrichous ciliated protozoa. Gene. 1989 Jan 30;75(1):93–107. doi: 10.1016/0378-1119(89)90386-7. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Thomas J. O. Core histone-DNA interactions in sea urchin sperm chromatin. The N-terminal tail of H2B interacts with linker DNA. Eur J Biochem. 1990 Jan 12;187(1):145–153. doi: 10.1111/j.1432-1033.1990.tb15288.x. [DOI] [PubMed] [Google Scholar]
- Hraba-Renevey S., Kress M. Expression of a mouse replacement histone H3.3 gene with a highly conserved 3' noncoding region during SV40- and polyoma-induced Go to S-phase transition. Nucleic Acids Res. 1989 Apr 11;17(7):2449–2461. doi: 10.1093/nar/17.7.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt M. M., Chodchoy N., Marzluff W. F. The mouse histone H2a.2 gene from chromosome 3. Nucleic Acids Res. 1989 Nov 11;17(21):8876–8876. doi: 10.1093/nar/17.21.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosciessa U., Doenecke D. Nucleotide sequences of mouse histone genes H2A and H3.1. Nucleic Acids Res. 1989 Nov 11;17(21):8861–8861. doi: 10.1093/nar/17.21.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer H., Hennig W. Isolation and characterization of a Drosophila hydei histone DNA repeat unit. Nucleic Acids Res. 1990 Mar 25;18(6):1573–1580. doi: 10.1093/nar/18.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber T., Angerer L. M., Angerer R. C., Childs G. A histone H1 protein in sea urchins is encoded by a poly(A)+ mRNA. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4123–4127. doi: 10.1073/pnas.85.12.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. J., Liu L., Marzluff W. F. Mouse histone H2A and H2B genes: four functional genes and a pseudogene undergoing gene conversion with a closely linked functional gene. Nucleic Acids Res. 1987 Apr 10;15(7):3023–3039. doi: 10.1093/nar/15.7.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannironi C., Bonner W. M., Hatch C. L. H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3' processing signals. Nucleic Acids Res. 1989 Nov 25;17(22):9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y., Yamazaki T. tRNA derived insertion element in histone gene repeating unit of Drosophila melanogaster. Nucleic Acids Res. 1989 Jan 11;17(1):225–238. doi: 10.1093/nar/17.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxson R., Mohun T., Gormezano G., Kedes L. Evolution of late H2A, H2B, and H4 histone genes of the sea urchin, Strongylocentrotus purpuratus. Nucleic Acids Res. 1987 Dec 23;15(24):10569–10582. doi: 10.1093/nar/15.24.10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier V. S., Böhni R., Schümperli D. Nucleotide sequence of two mouse histone H4 genes. Nucleic Acids Res. 1989 Jan 25;17(2):795–795. doi: 10.1093/nar/17.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss S. B., Challoner P. B., Groudine M. Expression of a novel histone 2B during mouse spermiogenesis. Dev Biol. 1989 May;133(1):83–92. doi: 10.1016/0012-1606(89)90299-6. [DOI] [PubMed] [Google Scholar]
- Palla F., Casano C., Albanese I., Anello L., Gianguzza F., Di Bernardo M. G., Bonura C., Spinelli G. Cis-acting elements of the sea urchin histone H2A modulator bind transcriptional factors. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6033–6037. doi: 10.1073/pnas.86.16.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D. C., Moy G. W., Vacquier V. D. The amino terminal sequence of sea urchin sperm histone H1 and its phosphorylation by egg cytosol. Comp Biochem Physiol B. 1989;92(2):381–384. doi: 10.1016/0305-0491(89)90296-4. [DOI] [PubMed] [Google Scholar]
- Rutledge R. G., Neelin J. M., Seligy V. L. Isolation and expression of cDNA clones coding for two sequence variants of Xenopus laevis histone H5. Gene. 1988 Oct 15;70(1):117–126. doi: 10.1016/0378-1119(88)90110-2. [DOI] [PubMed] [Google Scholar]
- Sellos D., Krawetz S. A., Dixon G. H. Organization and complete nucleotide sequence of the core-histone-gene cluster of the annelid Platynereis dumerilii. Eur J Biochem. 1990 May 31;190(1):21–29. doi: 10.1111/j.1432-1033.1990.tb15540.x. [DOI] [PubMed] [Google Scholar]
- Smith R. C., Dworkin-Rastl E., Dworkin M. B. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 1988 Oct;2(10):1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- Spiker S., Weisshaar B., da Costa e Silva O., Hahlbrock K. Sequence of a histone H2A cDNA from parsley. Nucleic Acids Res. 1990 Oct 11;18(19):5897–5897. doi: 10.1093/nar/18.19.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J., Milner J. S. Cloning and analysis of the Kluyveromyces lactis TRP1 gene: a chromosomal locus flanked by genes encoding inorganic pyrophosphatase and histone H3. Yeast. 1989 Jan-Feb;5(1):35–50. doi: 10.1002/yea.320050106. [DOI] [PubMed] [Google Scholar]
- Sturm R. A., Dalton S., Wells J. R. Conservation of histone H2A/H2B intergene regions: a role for the H2B specific element in divergent transcription. Nucleic Acids Res. 1988 Sep 12;16(17):8571–8586. doi: 10.1093/nar/16.17.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Wellman S. E., Marzluff W. F. Sequences of four mouse histone H3 genes: implications for evolution of mouse histone genes. J Mol Evol. 1986;23(3):242–249. doi: 10.1007/BF02115580. [DOI] [PubMed] [Google Scholar]
- Thomas G., Padayatty J. D. Restriction map & partial sequence of a rice DNA fragment carrying histone genes H2A, H2B & H4. Indian J Biochem Biophys. 1984 Feb;21(1):1–6. [PubMed] [Google Scholar]
- Tönjes R., Doenecke D. Structure of a duck H3 variant histone gene: a H3 subtype with four cysteine residues. Gene. 1985;39(2-3):275–279. doi: 10.1016/0378-1119(85)90323-3. [DOI] [PubMed] [Google Scholar]
- Tönjes R., Munk K., Doenecke D. Conserved organization of an avian histone gene cluster with inverted duplications of H3 and H4 genes. J Mol Evol. 1989 Mar;28(3):200–211. doi: 10.1007/BF02102477. [DOI] [PubMed] [Google Scholar]
- Vanfleteren J. R., Van Bun S. M., De Baere I., Van Beeumen J. J. The primary structure of a minor isoform (H1.2) of histone H1 from the nematode Caenorhabditis elegans. Biochem J. 1990 Feb 1;265(3):739–746. doi: 10.1042/bj2650739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren J. R., Van Bun S. M., Van Beeumen J. J. The histones of Caenorhabditis elegans: no evidence of stage-specific isoforms. An overview. FEBS Lett. 1989 Nov 6;257(2):233–237. doi: 10.1016/0014-5793(89)81541-8. [DOI] [PubMed] [Google Scholar]
- Vanfleteren J. R., Van Bun S. M., Van Beeumen J. J. The primary structure of histone H4 from the nematode Caenorhabditis elegans. Comp Biochem Physiol B. 1987;87(4):847–849. doi: 10.1016/0305-0491(87)90400-7. [DOI] [PubMed] [Google Scholar]
- Vanfleteren J. R., Van Bun S. M., Van Beeumen J. J. The primary structure of the major isoform (H1.1) of histone H1 from the nematode Caenorhabditis elegans. Biochem J. 1988 Oct 15;255(2):647–652. [PMC free article] [PubMed] [Google Scholar]
- Wefes I., Lipps H. J. The two macronuclear histone H4 genes of the hypotrichous ciliate Stylonychia lemnae. DNA Seq. 1990;1(1):25–32. doi: 10.3109/10425179009041344. [DOI] [PubMed] [Google Scholar]
- Wilhelm M. L., Wilhelm F. X. Histone genes in Physarum polycephalum: transcription and analysis of the flanking regions of the two H4 genes. J Mol Evol. 1989 Apr;28(4):322–326. doi: 10.1007/BF02103428. [DOI] [PubMed] [Google Scholar]
- Wolfe S. A., Anderson J. V., Grimes S. R., Stein G. S., Stein J. S. Comparison of the structural organization and expression of germinal and somatic rat histone H4 genes. Biochim Biophys Acta. 1989 Mar 1;1007(2):140–150. doi: 10.1016/0167-4781(89)90032-8. [DOI] [PubMed] [Google Scholar]
- Wu S. C., Györgyey J., Dudits D. Polyadenylated H3 histone transcripts and H3 histone variants in alfalfa. Nucleic Acids Res. 1989 Apr 25;17(8):3057–3063. doi: 10.1093/nar/17.8.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. C., Végh Z., Wang X. M., Tan C. C., Dudits D. The nucleotide sequences of two rice histone H3 genes. Nucleic Acids Res. 1989 Apr 25;17(8):3297–3297. doi: 10.1093/nar/17.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. X., Johnson L., Grunstein M. Coding and noncoding sequences at the 3' end of yeast histone H2B mRNA confer cell cycle regulation. Mol Cell Biol. 1990 Jun;10(6):2687–2694. doi: 10.1128/mcb.10.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi M., Roy C., Dove M., Seligy V. Amino acid sequence homologies between H1 and H5 histones. Biochem Biophys Res Commun. 1977 May 9;76(1):100–106. doi: 10.1016/0006-291x(77)91673-4. [DOI] [PubMed] [Google Scholar]
- Yang Y. S., Brown D. T., Wellman S. E., Sittman D. B. Isolation and characterization of a mouse fully replication-dependent H1 gene within a genomic cluster of core histone genes. J Biol Chem. 1987 Dec 15;262(35):17118–17125. [PubMed] [Google Scholar]
- van Daal A., White E. M., Elgin S. C., Gorovsky M. A. Conservation of intron position indicates separation of major and variant H2As is an early event in the evolution of eukaryotes. J Mol Evol. 1990 May;30(5):449–455. doi: 10.1007/BF02101116. [DOI] [PubMed] [Google Scholar]


