Abstract
Of great importance to clinical cancer diagnosis is the use of organic biomarkers. The detection of RNA, DNA, and protein antigen are all established methods for identifying specific cancer types and instrumental in promoting greater survivorship of the patient. Despite many decades of intense cancer research, we have yet to identify a “universal” protein or nucleic acid that allows us to diagnose more than a small subset of cancers at a time. In this review, we examine the use of localized cellular acidity as a universal marker for solid tumors, outlining some successes with a small peptide we call pHLIP, a pH-sensitive biosensor that allows us to label tumor tissue in live mice.
Keywords: cancer, pHLIP, diagnosis, biosensor, biomarker, pH, inflammation, arthritis, cell translocation
One of the major forefronts in cancer research is the use of biomarkers for cancer diagnosis. Many methods have been devised in the last two decades to screen protein antigen and nucleic acid markers known to be specific to different tumor types [1]. Perhaps the most successful in this line of biomarker technologies has been the development of assays sensitive to changes in Her2/Neu expression, the aggressive breast tumor proto-oncogene [2]. Protein and RNA expression assays have been in use clinically for some time, and the recent advent of DNA-based techniques allows clinicians the opportunity to monitor specific mutations over time by tracking susceptible genes in patients with established genetic risk [3]. Although these methods are crucial for the early detection of disease, they are all limited by the fact that each method detects only one or, at best, a minimal subset of cancer types. Despite more than a half century of intense cancer research, we still have not found a protein or nucleic acid that functions as a “universal” biomarker for all cancers.
Interestingly, there is a universal marker for virtually all solid tumors often overlooked and lost in the vast wealth of cancer literature. This universal marker is localized cellular acidity [4]. It has long been observed that cancer tissues in vivo are more acidic than normal tissues. This observation is attributed to the effects of hypoxia and called the Warburg effect [5] and from the heightened metabolic activity of cancer cells. A variety of conjectures and observations exist in the literature about how this localized acidity may facilitate certain cancer traits such as longevity, growth, and metastasis [6,7,8]. Several methods, including magnetic resonance imaging [9] and direct in vivo measurements with glass probes [10], have been used in the last few decades to confirm the observation. Despite a good deal of information on the phenomenon, little systematic effort has been devised until recently to try to exploit this fundamental characteristic for use as a cancer biomarker.
pHLIP as a Cancer Biosensor
On average, tumor tissues have been found to be roughly a half pH unit more acidic than their normal tissue counterparts [4]. This observation has been made for both mouse and human tumors and seems to be a universal trait for all solid tumors examined thus far in the literature. The degree of the pH difference is, of course, variable depending on tumor type with adenocarcinomas typically giving the largest pH differences [9,10], sometimes upwards of 0.7 to 0.8 pH units. Quite by accident, our lab has discovered a small pH-sensitive peptide called pHLIP (pH-Low Insertion Peptide) that seems to be sensitive enough to detect these small pH differences in vivo, allowing us to label areas of localized acidity in mice.
pHLIP was first reported in 1997 [11] when we were using bacteriorhodopsin to study the two-stage model of membrane protein assembly [12]. At this time, our lab was interested in testing the possibility that the cleaved helices of bacteriorhodopsin could spontaneously reassemble into a functional protein in the presence of lipids. During this process, it was found that one of the helices, the C3 helix, despite being quite hydrophobic, would not insert into membranes unless the pH was reduced. This C3 helix is the peptide we now call pHLIP (Figure 1).
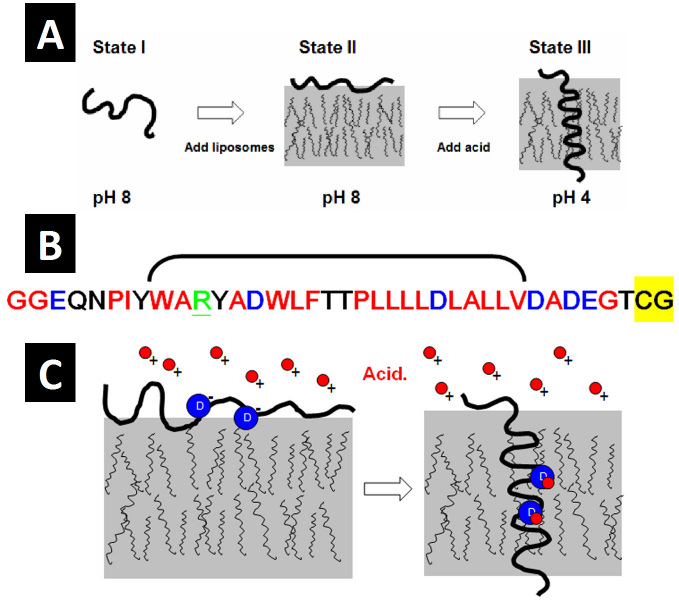
Figure 1.
A) pHLIP exists in three biophysical states depending on context: State I, as a largely unstructured monomer in aqueous solution at slightly basic or neutral pH; State II, as a largely unstructured monomer partitioned to the membrane surface in the presence of liposomes; and State III, as an inserted, bilayer-spanning α-helix at low pH. All states are reversible as denoted by the double arrows. Adapted from Biophys J. 2007;93(7):2363-72 [17]. B) The primary sequence of pHLIP. Red indicates hydrophobic residues, black indicates polar residues, blue indicates acidic residues, and underlined green indicates basic residues. The bracket denotes residues at positions 9-30 containing the two aspartic acids (blue D) at positions 14 and 25. The highlighted yellow residues comprise the cysteine tag attached to the inserting end of the peptide for use in conjugating cargo molecules for translocation into cells. C) The mechanism of pHLIP insertion at low pH: Insertion is largely dependent on the titration of the two aspartic acids (blue circles) at positions 14 and 25. Acidic protons (red circles) bind to the basic aspartic side chains at low pH, neutralizing the charge and converting the side chains into a more hydrophobic form. This thermodynamic conversion facilitates insertion by lowering the energy barrier (by about 3 kCal/mol [13]) for stable interaction with the membrane bilayer.
Using a variety of biophysical methods, including tryptophan fluorescence [13], circular dichroism [14], and Fourier-transformed infrared spectroscopy [11], our lab has extensively characterized the behavior of pHLIP in synthetic liposomes to determine that pHLIP exists in three different biophysical states depending on environmental context (Figure 1A). These three states are as follows: 1) in the absence of lipids and at neutral or slightly basic pH, pHLIP exists as a relatively unstructured monomer in aqueous solution; 2) in the presence of lipid membranes at neutral or slightly basic pH, unstructured pHLIP partitions very favorably to the membrane surface; and 3) at low pH, pHLIP inserts spontaneously (and reversibly) into the membrane as a spanning α-helix. It is this latter insertion and the transition between states 2 and 3 that allows us to use pHLIP as a pH-sensitive biosensor. This is because local acidic milieu in vivo is enough to trigger the pH-dependent insertion of pHLIP (Figure 1C), allowing the peptide to insert and be retained in the membranes of cells found in areas of localized acidity.
pHLIP In Vivo Labeling
By covalently attaching fluorophore molecules to the N-terminal, non-inserting end of pHLIP, in a collaboration with groups at the University of Rhode Island, we have been able to intravenously and peritoneally inject pHLIP-fluorophore conjugates into live mice harboring human tumors and observe preferential retention of the construct in cancerous tissue (Figure 2A-B) [15]. This retention was very pronounced for a variety of tumor sizes [15], even 5 days after an initial implantation of only 105 cancer cells subcutaneously in nude mice. As mentioned above, we attribute this retention to the well-characterized pH-dependent insertion of the peptide: The peptide senses localized acidity and responds by inserting into cell membranes as a spanning α-helix, anchoring the fluorophore in place.
Figure 2.
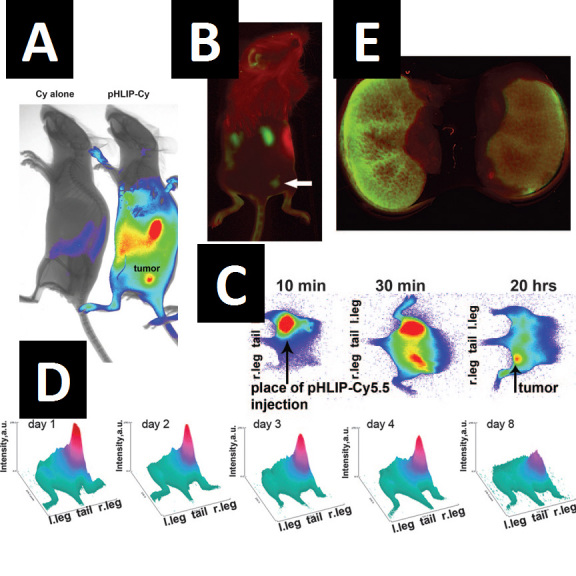
Intravenously and intraperitoneally injected pHLIP-infrared dye conjugates localize preferentially to tumors. A) Overlay of NIR fluorescence and X-ray images obtained the day after injection of 500µg/kg pHLIP-Cy5.5 conjugate (right) or an equivalent amount of free Cy5.5 (left) into mice bearing tumors in the right flank. Peptide injection was performed on the sixth day after an initial cancer-cell implantation of 5x104 cells subcutaneously. Localization of the fluorophore is seen in the tumor, kidneys, and spleen. B) pHLIP-Alexa750 fluorescence (green) in a mouse bearing a tumor in the right flank (6 days after 106 cell implant) imaged the day after peptide injection. Reflectance is denoted in red. Localization of the fluorophore is seen in the tumor (arrow), kidneys, and spleen. C) pHLIP-Cy5.5 given as a single injection (200µg/kg) into the left side of mice initially diffuses, accumulating in the right flank tumor by 20 hours. Blue represents background and red represents the highest fluorescence. D) 3D representations of pHLIP-Cy5.5 fluorescence from a tumor-bearing mouse at different days after injection of 500µg/kg peptide. Peptide injection was done on the seventh day after subcutaneous tumor implantation. The height (z axis) and intensity of red indicates the strength of NIR signal, showing that pHLIP retention lasts for several days. E) The NIR fluorescence of kidneys 2 days after injection of 500µg/kg pHLIP-Alexa750 into mice fed water (left) or 80 mM NaHCO3 at pH 8.2 (right). The alkaline treatment resulted in an approximate 50 percent reduction of pHLIP kidney retention without affecting tumor localization. Figures reprinted from [15]: Andreev OA, et al. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc Natl Acad Sci USA. 2007;104(19):7893-8, reused according to permissions guidelines.
The kinetics of pHLIP localization are such that most of the conjugate clears from mice within 24 hours (Figure 2C), leaving an obvious retention in the tumor that persists for more than a week (Figure 2D). Although we have not yet performed in-depth toxicology on the mice used in these studies, careful monitoring of the animals showed that the conjugates resulted in no obvious adverse physiological or behavioral side effects. As far as we could detect, the conjugates seemed to label the entirety of the tumors with no adjacent tissue being labeled at all [21].
The pHLIP conjugate did, however, exhibit some off-site localization [15]. The most prominent off-site localization was found in the kidneys (Figure 2A-B), where the proximal tubule region is particularly well labeled (Figure 2E). This labeling is consistent with the fact that our conjugates are small enough to enter through the glomerular fenestrae and be treated as filtrate. Since the proximal tubules of the kidneys are acidified to drive sodium ions across the membrane, our conjugates are likely responding to this acidity, inserting into kidney cells and being retained in the tubule tissue. We do not know exactly in what part of the cells this retention occurs, but we did find that feeding the mice carbonated water helps to alleviate kidney labeling (Figure 2E) without changing tumor labeling, in agreement with the interpretation.
pHLIP’s Insertion Mechanism
The mechanism of pHLIP’s pH-dependent insertion intimately involves the side chain topology of the peptide [11,17]. pHLIP is a hydrophobic peptide with a long stretch of hydrophobic residues from positions 9 to 30 (Figure 1B). From a number of experiments, we have determined that pHLIP’s pH-dependence is largely due to the titration of the two aspartic residues at positions 14 and 25. When acid is added, these two residues become protonated, converting from charged forms to polar, uncharged forms. This protonation effectively converts each residue into a more hydrophobic form, facilitating and stabilizing peptide insertion thermodynamically (Figure 1C), with the two aspartates likely participating directly in spanning the membrane. In addition to the two aspartic residues in the spanning domain, it is likely the three charged residues at the C-terminal, inserting end of the peptide (Figure 1B) and the free carboxyl at the C-terminus also need to be protonated (perhaps only transiently) to facilitate insertion [18].
Molecular Cargo Translocation Using pHLIP
One of the implications of the insertion process is that the C-terminal (inserting end) of the peptide makes contact with the other side of the bilayer after insertion (Figure 1C). We were interested to see if this translocation of the C-terminal could be used to translocate cargo molecules from one side of the membrane bilayer to the other. By adding a cysteine residue to the C-terminus, we have, in fact, been able to attach molecules such as dansyl dye and PNAs by disulfide bonding or bonds and translocate them across synthetic liposome bilayers and cell plasma membranes [14]. Since disulfide bonds are cleaved when they make contact with the highly reducing environment of the cytosol, our pHLIP-conjugate platform offers a cleavable, releasable cargo translocation that results in retention of cargo molecules in cells even after the peptide has been washed away (Figure 3A-B) [14]. This platform has been used to translocate phalloidin toxin into cells and stimulate cell death in culture (Figure 3C-D) [19], a very direct demonstration of the high potential pHLIP has for use in translocating and targeting drug molecules to tumor tissues.
Figure 3.
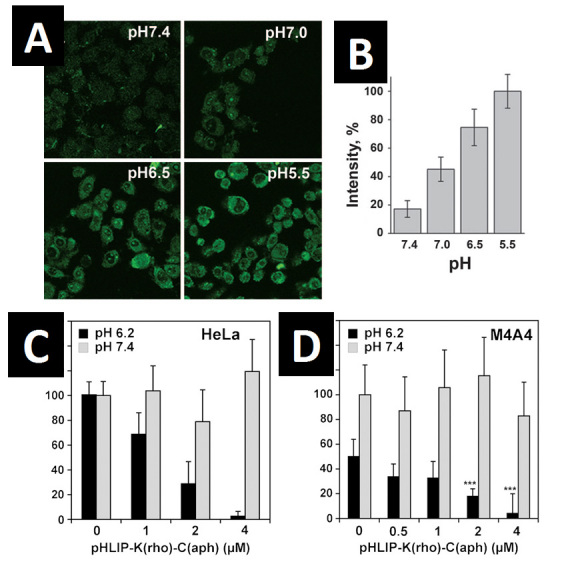
pHLIP can translocate cargo molecules across a lipid bilayer at low pH. A) HeLa cells were incubated for 15 minutes with 7 µM cleavable pHLIP-S-S-dansyl at extracellular pH of 5.5, 6.5, 7.0, or 7.4, followed by many pH 7.4 PBS washes to remove inserted peptide. The observed fluorescence is from the dansyl dye retained in the cells after disulfide cleavage and peptide removal. pH treatment values more acidic than the typical pH of an in vivo solid tumor were used to accelerate the translocation process. B) Quantification of the retained dye fluorescence in (A). The signal from cells at pH 5.5 was taken as 100 percent. Dye retention strongly decreases with an increase in extracellular pH. C) HeLa cell growth is strongly inhibited with incubation of increasing concentrations of a cleavable pHLIP-S-S-phalloidin construct at low extracellular pH. HeLa cells were incubated with 1, 2, or 4 μM pHLIP-S-S-phalloidin for 3 hours at pH 6.2 (black bars) or 7.4 (gray). After 4 days growth, the number of remaining viable cells was determined using the Promega AQueous One assay. All readings are normalized to a 0 μM peptide, pH 7.4 control. Errors of the mean were estimated using the two-tailed Student t distribution. D) Same experiment as in (C) using M4A4 cells instead of HeLa. Relabeled figures reprinted according to permissions guidelines: A-C: Reshetnyak YK, et al. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc Natl Acad Sci USA. 2006;103(17):6460-5 [14]; D: An M, et al. pH-(low)-insertion-peptide (pHLIP) translocation of membrane impermeable phalloidin toxin inhibits cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107(47):20246-50 [19].
Significance and Outlook
In addition to solid tumors, areas of inflammation such as arthritis also have been shown to exhibit a locally restricted acidic milieu [16]. To see if pHLIP could be used as a probe for these other areas of localized acidity, pHLIP conjugates were tested in a rat arthritis model [15], where inflammation was induced using a solution of Freud’s adjuvant and BSA injected into the rat’s knee. Much to our satisfaction, we found pHLIP-fluorophore conjugates preferentially localized to the areas of induced inflammation with similar kinetics as for tumors [15], opening the door for pHLIP’s use in a wider range of diagnostic and drug-delivery applications.
We have reviewed here some of the key characteristics and properties of pHLIP, which uses acidity as a marker for disease tissue. Not only does pHLIP allow for specific localization of fluorophores to tumors and sites of inflammation, but it also offers immense potential as a translocation and drug delivery platform. The obvious noninvasive nature of the technology is a key attraction, along with the fact that pHLIP appears to be benign even when injected into mice in moderate quantities. Unlike most other diagnostic methods dependent on specific protein antigens such as CA-2 (prostate cancer) or CA-125 (ovarian cancer), pHLIP offers the potential for use as a broader-spectrum diagnostic tool that does not simply identify proteins specific to a limited subset of cancer types but actually localizes in vivo to the site of the tumor, giving pHLIP power in not only diagnosing the presence of solid tumors but also in labeling their location and size. We expect our ongoing efforts in conjugating pHLIP to positron emission tomography (PET) [20] and MRI probes to further develop this technology.
It should be noted that the pHLIP technology is still in its infancy. We have thus far only demonstrated proof-of-principle applications. We have not yet conducted any head-to-head comparisons with existing cancer diagnostic techniques to determine relative efficacy, nor have we performed an exhaustive labeling survey of more than a few tumor types. We are also aware of the possibility of false-positives for cancer detection, particularly in older patients with chronic inflammatory disorders. Nevertheless, pHLIP shows great promise as a diagnostic tool, and we hope continuing work will yield new interest and ideas in the use of pH as a universal biomarker for cancer.
Acknowledgments
We thank NIH and DOD for their support, as well as our collaborators Yana Reshetnyak and Oleg Andreev for their essential contributions to this work.
Abbreviations
- pHLIP
pH-Low Insertion Peptide
- PNA
peptide nucleic acid
References
- Simon E. Biological and chemical sensors for cancer diagnosis. Meas Sci Technol. 2010;21(11):112002. [Google Scholar]
- Press MF, Slamon DJ, Flom KJ, Park J, Zhou JY, Bernstein L. Evaluation of HER-2/neu Gene Amplification and Overexpression: Comparison of Frequently Used Assay Methods in a Molecularly Characterized Cohort of Breast Cancer Specimens. J Clin Oncol. 2002;20(14):3095–3105. doi: 10.1200/JCO.2002.09.094. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free Nucleic Acids as Biomarkers in Cancer Patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- Gillies RJ, Raghunand N, Garcia-Martin ML, Gatenby RA. pH imaging. A review of pH measurement methods and applications in cancers. IEEE Eng Med Biol Mag. 2004;23(5):57–64. doi: 10.1109/memb.2004.1360409. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the Origin of Cancer Cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wike-Hooley JL, Haveman J, Reinhold HS. The relevance of tumour pH to the treatment of malignant disease. Radiother Oncol. 1984;2(4):343–366. doi: 10.1016/s0167-8140(84)80077-8. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66(10):5216–5223. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- Rofstad EK, Mathiesen B, Kindem K, Galappathi K. Acidic extracellular pH promotes experimental metastasis of human melanoma cells in athymic nude mice. Cancer Res. 2006;66(13):6699–6707. doi: 10.1158/0008-5472.CAN-06-0983. [DOI] [PubMed] [Google Scholar]
- Stubbs M, McSheehy PM, Griffiths JR. Causes and consequences of acidic pH in tumors: a magnetic resonance study. Adv Enzyme Regul. 1999;39:13–30. doi: 10.1016/s0065-2571(98)00018-1. [DOI] [PubMed] [Google Scholar]
- Kallinowski F, Vaupel P. pH distributions in spontaneous and isotransplanted rat tumours. Br J Cancer. 1988;58(3):314–321. doi: 10.1038/bjc.1988.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt JF, Rath P, Rothschild KJ, Engelman DM. Spontaneous, pH-dependent membrane insertion of a transbilayer alpha-helix. Biochemistry. 1997;36(49):15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- Popot JL, Engelman DM. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990;29(17):4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- Reshetnyak YK, Andreev OA, Segala M, Markin VS, Engelman DM. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proc Natl Acad Sci USA. 2008;105(40):15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshetnyak YK, Andreev OA, Lehnert U, Engelman DM. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc Natl Acad Sci USA. 2006;103(17):6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev OA, Dupuy AD, Segala M, Sandugu S, Serra DA, Chichester CO. et al. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc Natl Acad Sci USA. 2007;104(19):7893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H. Cellular pH regulators: potentially promising molecular targets for cancer chemotherapy. Cancer Treat Rev. 2003;29(6):541–549. doi: 10.1016/s0305-7372(03)00106-3. [DOI] [PubMed] [Google Scholar]
- Reshetnyak YK, Segala M, Andreev OA, Engelman DM. A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers. Biophys J. 2007;93(7):2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev OA, Karabadzhak AG, Weerakkody D, Andreev GO, Engelman DM, Reshetnyak YK. pH (low) insertion peptide (pHLIP) inserts across a lipid bilayer as a helix and exits by a different path. Proc Natl Acad Sci USA. 2010;107(9):4081–4086. doi: 10.1073/pnas.0914330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An M, Wijesinghe D, Andreev OA, Reshetnyak YK, Engelman DM. pH-(low)-insertion-peptide (pHLIP) translocation of membrane impermeable phalloidin toxin inhibits cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107(47):20246–20250. doi: 10.1073/pnas.1014403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavere AL, Biddlecombe GB, Spees WM, Garbow JR, Wijesinghe D, Andreev OA. et al. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Cancer Res. 2009;69(10):4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segala J, Engelman DM, Reshetnyak YK, Andreev OA. Accurate analysis of tumor margins using a fluorescent pH low insertion peptide. Int J Mol Sci. 2009;10(8):3478–3487. doi: 10.3390/ijms10083478. [DOI] [PMC free article] [PubMed] [Google Scholar]