Abstract
Hydroxyurea (HU) is a chemotherapeutic agent commonly used for various malignancies and hematological disorders, including chronic myelogenous leukemia and sickle cell anemia. We show here that chronic, low-level treatment with HU induces a variety of defects in telomere replication and maintenance. HU treatment preferentially decreased the rate of telomere DNA synthesis and altered the cell cycle timing of telomere replication. HU reduced the expression levels of telomere repeat RNA (TERRA). In some cells, HU caused a rapid loss of telomere restriction fragment length. Chromatin immunoprecipitation (ChIP) assay indicated that telomere repeat binding factors TRF1 and TRF2 dissociate from telomere DNA after HU treatment. TRF2 protein purified from HU treated cells showed a modest reduction in DNA binding activity and a change in isoelectric point as measured by 2D gel electrophoresis. However, chronic low level HU treatment did not evoke a DNA replication checkpoint response, suggesting that the mechanism of action is distinct from the well-characterized S-phase checkpoint pathway. We conclude that therapeutic doses of HU preferentially effects telomere replication and maintenance, through a mechanism that may involve the direct modification of TRF2. These findings provide new insight into the potential mechanisms of action of HU at telomeres and in cancer chemotherapies.
Keywords: hydroxyurea, telomere, TRF2, replication, TERRA
Introduction
The molecular underpinnings of human carcinogenesis have been studied in great detail in the last several decades. Among the critical events for cellular transformation are the acquisitions of several genetic alterations that promote the cancer phenotype.1 It is thought that early mutations in “caretaker” genes can promote the genetic instability that allows for the higher rates of mutation and selection responsible for cancer cell evolution. Recently, it has been proposed that intrinsic replication stress found in many early neoplastic tissues may also drive genetic instability and cancer cell evolution.2,3 Replication stress may occur when cellular division signals are confronted with suboptimal DNA replication conditions, like excess DNA damage or reduced levels of nucleotide pools, which restrict the completion of DNA synthesis. Some regions of the genome are thought to be hypersensitive to this kind of genetic instability, including the common fragile sites and complex repetitive sequences, like telomeres and centromeres, which are prone to recombination and replication fork stalling.
The telomeres at the ends of each chromosome are important in maintaining genome stability.4,5 In human cells telomeres consist of hexameric T2AG3 repeats that can span from 4–10 kB depending on the chromosome, cell type and genetic variation.6 Telomeres progressively shorten during each round of DNA replication due to the end replication problem, and critically short telomeres can lead to the activation of a DNA damage response and cellular senescence. In self-renewing stem cell populations and germ line tissues telomere length shortening is reversed by telomerase, the reverse transcriptase that can extend the DNA repeats at the end of telomeres. Telomerase is also activated in ~90% of human cancer and represents a mechanism of immortalization as well as a potential selective target of cancer cells.
To maintain the structure of the telomere a six-protein complex know as shelterin resides at telomeres and acts in concert with numerous other factors to facilitate protection, replication and processing of chromosome ends after DNA replication.7 This complex interacts with components of the DNA damage response proteins, including Mre11, NBS1, Rad50 and ATM, to prevent the recognition of chromosome ends as DNA damage.8,9 Telomere repeat factor 2 (TRF2) is a shelterin component that binds directly to the TTAGGG repeat DNA and can also interact with components of the DNA damage response pathway. Germ-line deletion of TRF2 results in early embryonic lethality, while transient deletion causes chromosomal end-to-end fusions between telomeres and chromosomal fusion-breakage cycles.10 Ectopic expression of a dominant negative allele (TRF2ΔBΔM), which lacks that amino terminal basic domain and the C-terminal myb DNA binding domain, and causes end to end fusions and telomere dysfunction similar to the TRF2 knock-out.11 A second allele of TRF2 (TRF2ΔB) that has a deletion in the N-terminal basic domain causes the rapid loss of telomere repeat signals and the formation of telomere circles.12 This allele lacks the ability to stabilize recombination intermediates, and cannot recruit the Origin Recognition Complex (ORC) to telomeres.13,14 ORC is an essential component of the DNA replication initiation machinery and can also contribute to chromatin organization.15 The precise function of TRF2 interaction with ORC is not completely understood.
The interaction between TRF2 and components of the DNA replication machinery, as well as components of the DNA damage response, suggests that TRF2 plays a central function in coordinating telomere length regulation with DNA replication and DNA replication stress.16,17 In this study we explore the impact of low doses of the chemotherapeutic agent hydroxyurea (HU) on human telomeres. HU is an inhibitor of ribonucleotide reductase and can effectively reduce the dNTP pool.18 High levels of HU lead to cell cycle arrest and accumulation of single and double strand breaks due to replication fork collapse.19 However, lower levels of HU commonly used in therapeutic dosages have a more subtle effect on DNA replication and cell cycle progression. Previous studies have revealed that low doses of HU can induce the loss of telomere repeat containing double minute chromosomes.14 HU treatment has also been used to eliminate extrachromosomal copies of Epstein-Barr virus (EBV) from latently infected B-lymphocytes. In earlier studies, we found that TRF2 bound the EBV origin of plasmid replication (OriP) and regulated its replication and maintenance function.20 More recently, we found that addition of HU caused a partial loss of TRF2 from OriP and a change in OriP replication timing.21 Here, we explore the effects of HU on telomere DNA structure and replication. We show that HU reduces telomere repeat length, and inhibits the in vivo DNA binding properties of TRF2. This mechanism does not involve the activation of a canonical DNA damage checkpoint pathway, but does appear to act through post-translational modification of TRF2 protein.
Results
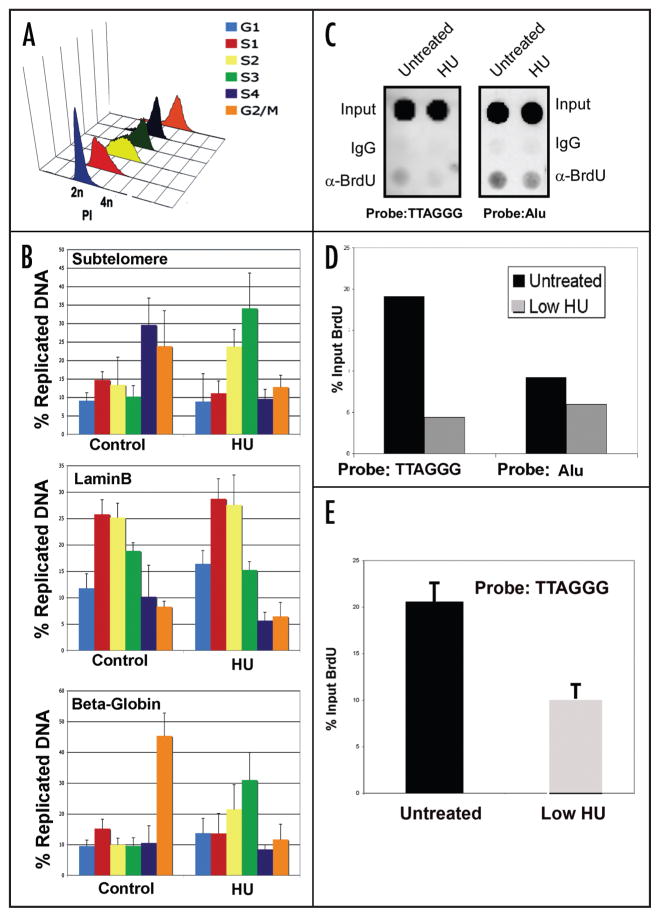
Therapeutic doses of HU alters replication timing and efficiency at telomere repeats
Relatively low doses of HU (50–100 μM) have been used to eliminate EBV minichromosomes from latently infected B cells.22,23 We recently found that HU-associated loss of EBV correlates with a change in the replication timing of the EBV genome.21 Since telomere repeats were implicated in the control of EBV replication timing, we tested cellular telomere repeat DNA was similarly affected by HU treatment (Fig. 1). Raji cells were treated with 50 μM HU for 6 d and then pulse labeled with BrdU for 45 min. At the end of the pulse label, cells were fractionated by centrifugal elutriation to isolate different stages of the cell cycle.21 Cell cycle separation was confirmed by propidium iodide staining and FACS analysis of each elutriated fraction (Fig 1A). BrdU specific ChIP assays were then used to quantify the amount of BrdU incorporated into telomere DNA at each stage of the cell cycle (Fig. 1B). We have previously used this method to assay cell cycle timing of DNA replication for EBV and several cellular loci.21 Telomere replication was measured using real-time PCR for a primer specific for subtelomere sequence adjacent to the telomere repeat tracts. The lamin B2 origin was used as a control for an early replicating region, and the β-globin locus was used as a control for a late replication region (Fig. 1B). In untreated Raji cells, subtelomere replication occurred predominantly in late S phase (fractions S4 and G2/M) (Fig. 1B). In contrast, subtelomere replication in HU-treated Raji cells occurred in S2 and S3 fraction of the cell cycle (Fig. 1B). HU-treatment had no effect on lamin B replication timing, but did accelerate the late replication timing of the β-globin locus. This suggests that low HU treatment may deregulate controls for replication timing at multiple regions of the genome, including telomere repeats.
Figure 1.
Therapeutic dose of HU alter telomere replication timing and efficiency. (A) Raji cells were either mock-treated or treated with low levels of HU for 6 d. On the last day of mock or HU-treatment, BrdU was added to the cultures (50 μM) for 45 minand then cells were subject to centrifugal elutriation. A characteristic cell cycle profile of fractions after centrifugal elutriation is shown. Cell cycle fraction was determined by propidium iodine DNA content flow cytometry. (B) DNA was extracted from cells in each elutriated fraction and used in a BrdU immunoprecipitation reaction. The resulting DNA was used in real-time PCR to determine the efficiency of replication of telomeres in each fraction using a sub-telomere primer set. Primers were the laminB2 and β-globin coding regions were used as controls. Average values are plotted (n = 3) and error bars represent the s.d. (C) Raji cells were treated with low dose HU for one day and cells were pulse-labeled with BrdU for 30 min prior to DNA extraction. The purified DNA was used in a BrdU immunoprecipitation and the resulting DNA was used in a dotblot. A (TTAGGG)6 probe was used to assess replication at the telomere and an Alu probe was used as a control. (D) Quantification of the dot blot shown in (C). (E) Summary of three independent BrdU incorporation assays, as shown in (C), and quantified for (TTAGGG)6.
To investigate whether HU was altering the replication efficiency at telomere repeats, we measured the percent incorporation of BrdU during a short 30 min pulse label (Fig. 1C). Replication rates were measured by BrdU immunoprecipitation followed by dot blotting with probes specific for telomere repeat DNA (left) or Alu repeat DNA (right) and quantified relative to total input DNA. In untreated Raji cells, we found that BrdU is incorporated into a smaller percentage of total telomere repeat DNA than into Alu repeats. After 24 h of 50 μM HU treatment we observed a significant reduction in replication efficiency at both repeat regions, although the effects at telomeres were more pronounced (Fig. 1C, lower). HU treatment caused a ~78% reduction in telomere BrdU incorporation, while only a 21% reduction was observed at Alu repeats (Fig. 1D and E). These results suggest that low levels of HU preferentially inhibit replication rates through telomere repeat regions.
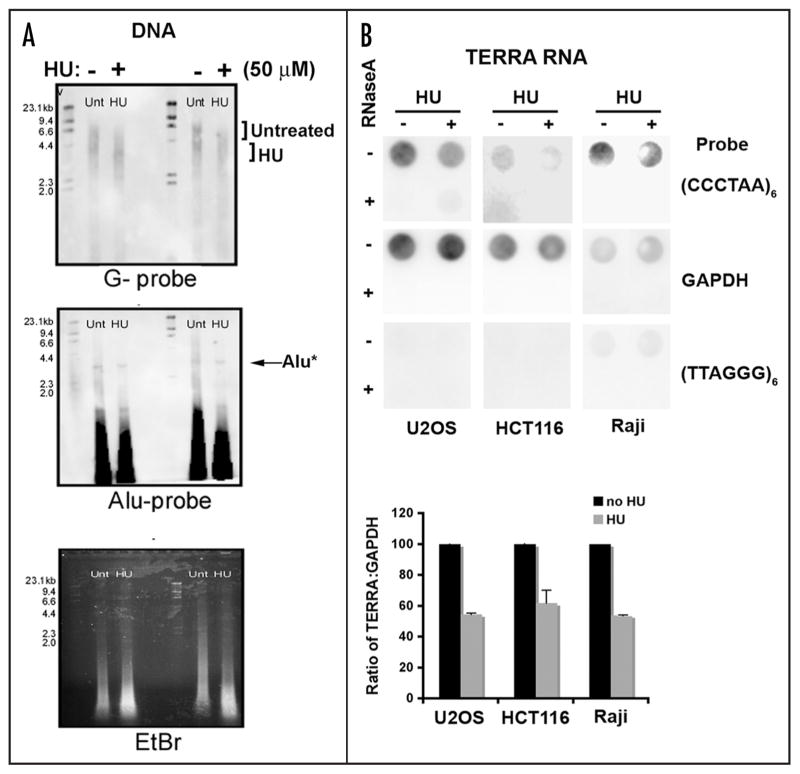
HU alters telomere structure
HU treatment has also been found to induce telomere circle formation.14,17 We therefore tested whether low doses of HU altered telomere length (Fig. 2A). Raji cells were treated with 50 μM of HU for 6 days and then assayed by restriction fragment length assay for potential changes in average telomere length. We found that this treatment led to a significant decrease (~1 kB) in average telomere length (Fig. 1A, top). HU treatment did not affect the average length of an Alu resistant restriction fragment (Alu*). These results were consistent with a previous observation that HU treatment causes an increase in telomere circle formation, which may correspond to the sudden loss of telomere length observed in these experiments.14
Figure 2.
Telomere dysfunction in cells treated with low dose hydroxyurea (HU). (A) Raji cells were treated with low dose HU (50 μM) for 6 days and genomic DNA was extracted and digested with Alu/MboI. A Southern hybridization with a DIG-labeled (TTAGGG)6 probe was used to assess telomere length. An Alu probe and ethidium bromide stained were used as loading controls. A DNA ladder is located on the left hand side of each gel and the numbers correspond to kilobases of DNA. (B) U20S, HCT116, and Raji cells were treated with low dose HU and total RNA was extracted. Samples were treated with (+) or without (−) RNAse. Dot-blots of RNA samples were used to assess the TERRA levels in treated cells using a DIG-labeled (CCCTAA)6 probe. Control probes for GAPDH or TERRA-complementary strand RNA (TTAGGG)6 probe were used as specificity controls.
It has recently been shown that telomeres are actively transcribed by RNA polymerase II to produce a heterogeneous RNA transcript containing telomere repeats.24,25 This telomere repeat containing RNA, referred to as TERRA, can be observed to varying extents in different cell types and contributes to telomere stability. We found that HU treatment led to a significant decrease in TERRA RNA in all cell types where TERRA RNA is readily detectable (Fig. 2B). U2OS cells, which have very high levels of TERRA RNA, had a 63% reduction in TERRA levels when cultured in 50 μM HU treatment for 4 d. Similarly, HCT116 and Raji cells, which have lower levels of detectable TERRA, had ~38% and 54% reduction, respectively, in TERRA RNA abundance after HU treatment. TERRA RNA levels were measured by RNA dot blotting (top), and quantitated (lower). TERRA RNA was sensitive to RNase A treatment indicating that the signal was specific for RNA, and not telomere repeat DNA. Furthermore, HU treatment had no detectable effect on GAPDH RNA. These findings indicate that low levels of HU reduce TERRA RNA abundance may lead to telomere defects in vivo.
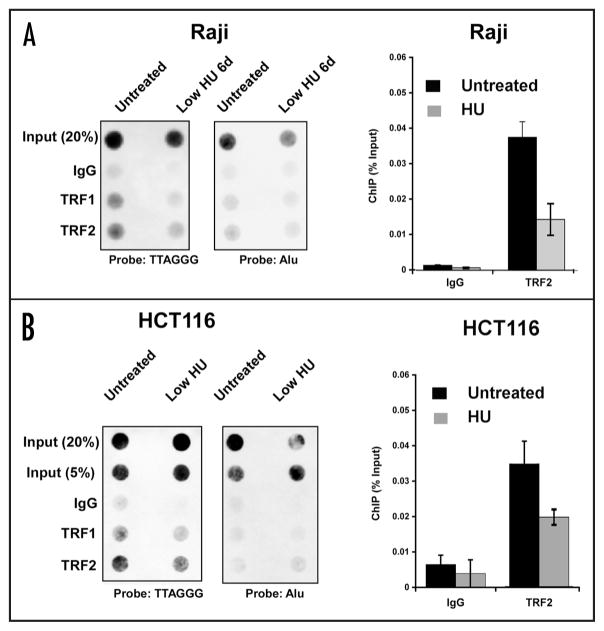
HU reduces TRF binding to telomere repeats by chromatin immunoprecipitation
To explore the molecular mechanism underlying the HU-induced loss of telomere repeat DNA and TERRA RNA, we examined the relative binding of telomere repeat factors to telomere repeat DNA in response to HU treatment (Fig. 3). TRF1 and TRF2 binding was assayed by chromatin immunoprecipitation (ChIP) assay and compared to control IgG for specific binding to telomere repeat DNA (left) or to control Alu repeats (right). We found that HU treatment caused a small, but consistent reduction of TRF1 and TRF2 binding in both Raji (Fig. 3A) and HCT116 (Fig. 3B) cells. TRF1 and TRF2 binding was not enriched at Alu repeats relative to IgG control, indicating that these proteins bind specifically to telomere repeat DNA. Quantification of these dot blots revealed that HU treatment caused an ~86% reduction in TRF1 and a ~40% reduction in TRF2 binding to telomere repeat DNA. Using real-time PCR primers that recognize subtelomere DNA regions in at least 17 human chromosomes we found similar results, in that both TRF1 and TRF2 protein levels were also reduced at these regions of the region (data not shown). These results indicate that low level HU treatment causes a loss of TRF1 and TRF2 binding to telomere repeats in multiple cell types in vivo.
Figure 3.
Dissociation of telomere repeat factors from telomeres after low HU treatment. (A) Raji cells were treated with low HU and then assayed by ChIP for TRF1, TRF2, and control IgG binding to telomeres. ChIP DNA was visualized by dot blot and probing with a (TTAGGG)6 (left) or Alu (right) probe. Bar graph is a summary of three independent ChIP assays as shown (A), quantified by PhosphorImager analysis and presented bound DNA relative to input DNA. (B) HCT116 cells were assayed, as in (A), by ChIP assay for TRF1 and TRF2 binding to telomere repeats (left) or Alu repeats (right). Bar graph shows quantification of three independent ChIP assays in HCT cells for TRF2 and IgG, and presented as percentage bound relative to input.
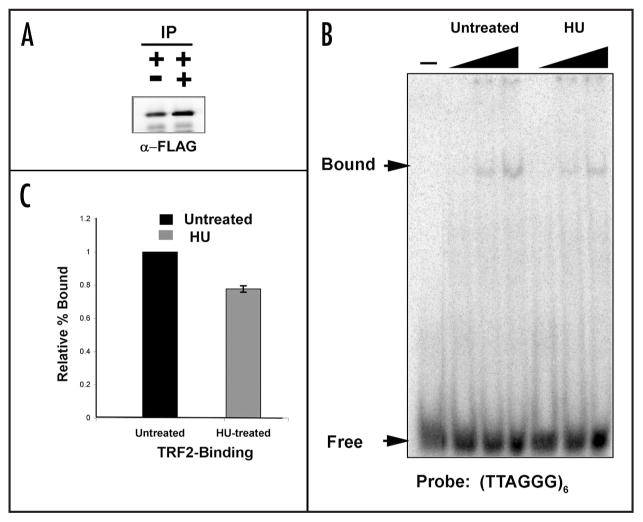
HU reduces TRF2 binding in EMSA
Previous studies have specially implicated the TRF2 protein in the regulation of EBV replication and plasmid maintenance. We therefore focused on the effects of HU on the biochemical properties of TRF2. FLAG-tagged TRF2 protein was expressed transiently in HCT116 cells and then treated with or without 50 μM HU for 24 h post-transfection. FLAG-TRF2 protein was purified from whole cell extracts derived from transfected HCT116 cells. FLAG-TRF2 protein was eluted from anti-FLAG-agarose resin using FLAG-peptide and visualized by western blot analysis (Fig. 4A). Similar levels of FLAG-TRF2 were recovered from untreated and HU-treated cells. The proteins were then assayed for their ability to bind a telomere repeat duplex DNA probe using EMSA (Fig. 4B). We found that affinity purified preparations of FLAG-TRF2 derived from HU-treated cells were reduced for DNA binding to ~78% of untreated TRF2 protein (Fig. 4C). We conclude that a 50 μM HU treatment has an inhibitory effect on TRF2 DNA binding activity.
Figure 4.
Low HU treatment decreases TRF2-DNA binding activity. (A) HCT116 cells were transfected with a FLAG-TRF2 plasmid and treated with low HU for one day. Western blots of the inputs of the transfection are shown to the left and the FLAG-eluted protein is shown the right. (B) Increasing concentrations of FLAG-TRF2 protein (1, 5 and 9 μl) were mixed with a radio-labeled duplex telomere probe and electrophoresed on a 6.0% polyacrylamide gel and binding was assessed via PhosphoImage analysis of the dried gel. (C) PhosphorImager quantification of the percent of (TTAGGG)6 probe bound by TRF2 without or with HU treatment from three independent EMSA assays. Untreated TRF2 binding was normalized to 1% for each experiment. Errror bars represent standard deviation from the mean.
HU alters TRF2 protein-interactions and post-translational modifications
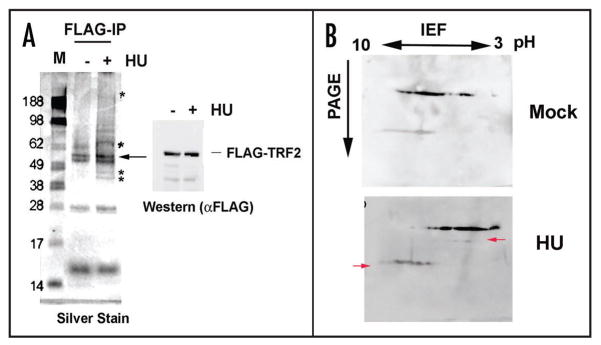
To better understand the molecular basis of the decrease in TRF2 DNA binding after HU treatment, we first examined the proteins associated with FLAG-TRF2 preparations (Fig. 5A). Affinity purified FLAG-TRF2 proteins were assayed by silver-staining (Fig. 5A, left) and by western blot (Fig. 5A, right). We observed that HU-treated TRF2 contained several additional species. We have attempted to obtain identity of these proteins by mass spectrometry and no significant differences in protein composition could be readily confirmed by western blot. TRF2-associated protein hRap1 was identified in both preparations, but no additional proteins were confirmed to be enriched in the HU treated samples (data not shown). We therefore tested whether some of these additional species may reflect changes in TRF2 post-translational modifications (Fig. 5B). To this end, we examined affinity purified FLAG-TRF2 proteins by 2D isoelectric focusing, followed by western blotting with anti-FLAG antibody. We observed that FLAG-TRF2 derived from HU-treated cells was significantly altered in its isoelectric properties. Notably, the bulk of TRF2 was more acidic and a new species appeared at higher molecular masses (Fig. 5B, red arrows). The precise identity of the post-translational modifications is not known, but the increase in protein acidity is consistent with an increase in protein phosphorylation. TRF2 phosphorylation has been reported in other studies with UV and gamma-irradiation sufficient to cause DNA damage checkpoint responses.26
Figure 5.
Low HU alters TRF2-associated proteins and the isoelectric point of TRF2. (A) FLAG-TRF2 eluted protein was electrophoresed onto a 4–12% NuPage gel and silver stained. * represent unique bands and the arrow indicates the position of the TRF2 protein. A western blot of the input material is shown to the right of the silver stained gel. (B) FLAG-TRF2 eluted protein was used in 2D protein gel electrophoresis to determine a difference in isoelectric point of the protein after treatment. The protein was first run on pH 3–10 (non-linear) strips and then subjected to second-dimension protein electrophoresis according to molecular weight. The protein was transferred to a nitrocellulose membrane and used in western blotting. Arrows indicate changes in TRF2 mobility after treatment with HU.
Low levels of HU does not evoke a canonical double DNA damage response
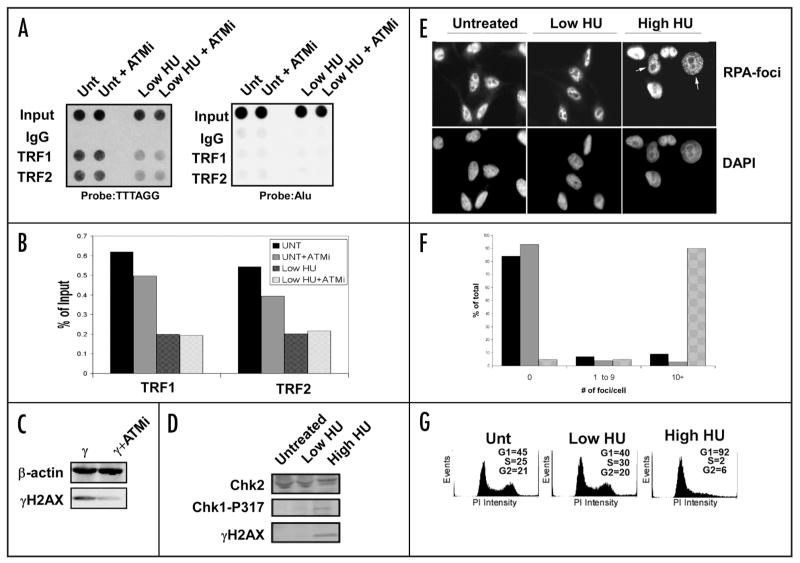
High levels of HU (~1 mM) are known to cause G1/S phase arrest in most cell types, and this arrest is mediated by the ATM S-phase checkpoint kinase. Low levels of HU (50 μM) do not cause cell cycle arrest, but may induce subthreshold DNA damage that might evoke some of the known S-phase checkpoint signaling pathways. We therefore tested whether a known pharmacological inhibitor of ATM (ATMi) blocks the HU-induced alterations on TRF1 or TRF2 binding to telomeres (Fig. 6A). Raji cells were untreated or treated with 50 μM HU for 24 hr, in the presence or absence of ATMi. We observed that HU treatment induced a significant loss of TRF2 and TRF1 binding to telomere repeats, and that ATMi had no effect on this loss of DNA binding (Fig. 6A, left, quantified in B). The effectiveness of the ATMi was monitored by assaying its ability to block the accumulation of γH2AX in γ-irradiated Raji cells (Fig. 6C). The effects of low and high levels of HU were also assessed by monitoring accumulation of γH2AX and phospho-Chk1 (P317) which are hallmarks of the S phase checkpoint response (Fig. 6D). While high levels of HU induced detectable levels of γH2AX and phospho-Chk1 (P317), low levels of HU had no detectable induction of these checkpoint modifications. Similarly, high levels (1 mM), but not low levels (50 μM), generated RPA-associated foci typical of recruitment of proteins to stalled replication forks (Fig. 6E and F). As expected, high levels of HU causes a G1/S cell cycle arrest, while low levels had a cell cycle profile indistinguishable from untreated cells (Fig. 6G). These findings indicate that low level HU treatment induces changes at telomeres that are independent of the ATM-mediated intra-S phase DNA damage checkpoint pathway and importantly, does not result in cell cycle arrest.
Figure 6.
Lack of DNA damage response after low dose HU treatment. (A) Raji cells were treated with the ATM inhibitor KU-55933 (10 μM, 24 h) with or without HU and used in ChIP experiments to determine the efficiency of TRF1 and TRF2 binding to telomeres. An Alu probe was used as a control. (B) Quantification of the data collected in (A). (C) Western blot of the common DNA damage marker and ATM target γH2AX after treatment with the ATM inhibitor. The cells were irradiated with 5 Gy γIR and harvested 15 min after treatment. β-actin is shown as a loading control. (D) Western blot of Raji cell extracts treated with low HU show little induction of common replication stress or damage markers. A high dose of HU (1 mM) is used as a positive control. A Chk2 western blot is used as a loading control. (E) Low HU treatment does not induce RPA foci after treatment. HeLa cells were treated with low HU for 1 d and then used for immunofluoresence to determine if RPA is recruited to sites of replication stress after treatment. A high dose (1 mM) sample is included as a control. All nuclei were stained with DAPI as a counterstain. White arrows indicate cells containing >10 RPA-foci. (F) Quantification of the data generated in (E). One hundred nuclei were counted for each condition shown. Untreated cells (black bars), low HU (gray bars), or high HU (patterned bars) were scored for 0, 1–9 or >10 foci per cell, as indicated. (G) Propidium iodide staining of HU-treated cells shows no drastic shift in cell cycle profile after treatment. A high dose HU sample (1 mM) is included as a positive control. Quantification of cells in each phase of the cell cycle is presented for each panel.
Discussion
HU is commonly used in chemotherapeutic regimens for a variety of malignancies and hematological disorders, including chronic myelogenous leukemia and sickle cell anemia.27,28 Long-term use of HU has been linked to mucocutaneous complications and formation of secondary carcinomas.29,30 While high doses of HU are known to cause cell cycle arrest by inhibiting ribonucleotide reductase and the production of sufficient nucleotides for S phase completion, little is known about the effects of lower doses commonly used in chemotherapies. We show here that low doses of HU can induce telomere dysfunction through a mechanism that correlates with the post-translational modification of TRF2 and loss of TRF2 DNA binding activity. The telomere dysfunction observed in cells treated with low HU may partly explain some of the complications observed in patients after long-term HU chemotherapy.
Low doses of HU induced several different defects at telomeres. We found that DNA replication timing at telomere repeats was accelerated by HU treatment (Fig. 1A) and replication efficiency was reduced (Fig. 1B). HU caused a decrease in the average telomere restriction fragment length (Fig. 2A), consistent with other published reports using low HU treatment.17 HU also caused a reduction in TERRA RNA levels (Fig. 2B). The molecular basis for these telomere defects was investigated by examining the DNA binding properties of the primary telomere repeat binding factors, TRF1 and TRF2. HU consistently reduced TRF2 and TRF1 binding to telomere repeat DNA in cell-based assays (ChIP) (Fig. 3) and in nuclear extracts as measured by EMSA (Fig. 4). HU treatment altered the post-translational modification of TRF2 and changed the composition of TRF2-associated polypeptides (Fig. 5). We suggest that the HU-induced telomere defects are driven by changes in the properties of telomere repeat binding factors. Altered TRF2 binding activity or interacting proteins may account for the changes in telomere maintenance and replication observed after treatment with chronic low levels of HU.
There are several known mechanisms that control TRF2 stability at the telomere. The DNA damage kinase ATM phosphorylates TRF2 on at least two sites, correlated with its removal from chromatin and potential role as an early signal of DNA double strand breaks.26 Other modes of regulation include the poly (ADP) ribosylation of TRF2 by PARP2,31 and the control of TRF2 interaction with telomeric chromatin via the nuclease XPF.32 While the association of TRF2 with the telomere is relatively stable during the cell cycle, there is a decrease in association at the G1/S transition, as well as at the G2/M phase of the cell cycle. 33–35 The data presented here suggests that a post-translational modification of TRF2 is involved in its response to low HU treatment; however it appears to be independent of an ATM-dependent checkpoint response and may represent a novel mode of regulation. Previous studies have demonstrated that radiation-induced checkpoint response is deactivated after pre-treatment with high dose HU that prevents S-phase.36 The doses used in the experiments described here clearly do not elicit a checkpoint response, and it would important to understand how this could affect subsequent radiation responses in a clinical setting.
HU treatment has also been used to induce replication stress. Our findings suggest that TRF1 and TRF2 association with telomeres may be altered during HU-induced replication stress. Conditions of replication stress may require increase access of the replication machinery to telomere repeat DNA. This is consistent with the dissociation of TRF1 and TRF2 after HU treatment (Fig. 3), and the advances cell cycle timing of telomere DNA replication in HU treated cells (Fig. 1A). While reduction in TRF2 binding may increase accessibility to DNA replication machinery, it also comes at a cost to the structural stability of the telomere repeat DNA. HU-associated telomere dysfunctions included the rapid loss of telomere repeats (Fig. 2A), loss of TERRA RNA (Fig. 2B), and generation of telomere repeat circles.14 These broad effects of HU suggest that DNA replication stress induced by chronic HU treatment leads to telomere maintenance defects. These findings suggest that chronic HU treatment in patients may produce genetic instability through deregulation of TRF2 and normal telomere control mechanisms. Future studies will be required to determine whether the telomere-dysfunction induced by HU is a direct consequence of TRF2 post-translational modification, and if the telomere-dysfunction is a primary mechanism of HU-associated anti-neoplastic activity.
Materials and Methods
Cell culture and transfection
HeLa, U20S, HCT116 cells and Raji cells were grown at 37°C incubator with 5% CO2 in DMEM or RPMI media supplemented with antibiotics and 10% heat-inactivated fetal bovine serum. For transfection experiments, HCT116 cells were seeded one day prior to transfection in antibiotic-free media. Three μg of FLAG-TRF2 (full-length) plasmid was transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) onto 10 cm plates with cells at 50% confluence. After 6 h the media was removed and the cells were expanded. To induce replication stress, hydroxyurea (Sigma) was added to logarithmically growing cultures at a concentration of 50 μm (low dose) or 1 mM (high dose). ATMi (KU55933) was obtained from Kudos (UK) and was used at 10 μM in culture media for 24 h.
Western blotting
Whole cell extracts were prepared by either scraping adherent cells on ice in PBS or directly spinning down suspension cultures at 300 xg. After washing with PBS, cells were lysed in RIPA buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease and phosphatase inhibitors (Sigma). After a sonication step the cellular debris was cleared from the whole cell extract by centrifugation at 4°C for 15 min at 15,000 rpm. Cell extracts were loaded onto 8–16% tris-glycine gels or 4–12% NuPage bis-tris gels and electrophoresed. The protein was transferred to nitrocellulose membrane and pre-hybridized with tris-buffered saline with Tween-20 (TBST) containing 5% dry milk. The following antibodies were used in the experiments described here: TRF2,20 γH2AX (Upstate), Chk1pS317 (Bethyl), β-actin (Sigma) and FLAG (Sigma). AP-linked Secondary antibodies were purchased from GE Amersham and used at a concentration of 1:5,000 and a chemiluminescent substrate solution was added to the blot for development. Images were captured and analyzed using the Fuji Imaging System and accompanying Multi-Gauge software package.
Cell cycle profile analysis
To determine the cell cycle profiles of cells, cultures were either treated or left untreated and fixed in ice-cold 70% ethanol for at least 30 min. After fixation, cells were stained with staining solution (0.5 mg/mL propidium iodide, 100 mg/mL RNAse A) for 30 min in the dark. Samples were analyzed using an EPICS XL (Beckman-Coulter, Inc., Miami, FL), and 50,000 events were recorded. For all flow cytometry experiments the WINMDI software program (The Scripps Institute) was used to analyze the data.
Indirect immunofluoresence
Cells were grown on sterile coverslips place in culture dishes and either treated with HU or left untreated. The coverslips were removed from the dishes, rinsed with PBS and fixed with 100% methanol at −20°C for 10 min. After washing in 1X PBS, 0.2% Triton-X was added and the coverslips were incubated for 10 min at 4°C. The coverslips were washed and a RPA antibody (Genetex) was added to PBS and incubated for 1 h. The primary antibody was washed off, and an Alexa fluor 568 secondary antibody was added and incubated for 30 min. After washing, the coverslips were placed in mounting media containing DAPI and foci were visualized using a Nikon E600 microscope with the 100X objective lens. The ImagePro software package (Media Cybernetics, Bethesda, MD) was used for image acquisition.
FLAG-TRF2 purification
FLAG-TRF2 transected or mock transfected cells were scraped from culture dishes in cold PBS and pelleted. After lysis in RIPA buffer containing protease and phosphatase inhibitors the mixtures were centrifuged at 15,000 rpm for 15 min at 4°C. Two hundred μl of whole cell extract was added to 30–50 μl of FLAG-bead resin (Sigma) and incubated on a rotating platform at 4°C overnight. Beads were washed twice in low salt wash buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 0.1% Triton-X 100) and once in high salt buffer (50 mM Tris (pH 7.4), 250 mM NaCL, 2 mM EDTA, 2 mM EGTA and 0.1% Triton-X). FLAG-TRF2 protein complexes were eluted at 4°C off of the beads by using the FLAG peptide (0.4 μg/mL). Expression and purity was verified with western blotting. In experiments to determine TRF2 interacting proteins, silver staining was performed using the SilverQuest kit (Invitrogen).
Chromatin immunoprecipitation (ChIP)
Treated or untreated cells were crosslinked in 1% formaldehyde at room temperature with mild agitation. After 15 min, 0.125 M glycine was added and the cultures were rotated for another 5 min. Adherent cells were scraped off of the culture dishes with cold PBS and pelleted at 300 xg. Cells were washed in cold PBS and resuspended in lysis buffer 1% SDS, 10 mM EDTA, 50 mM Tris (pH 8.0); 1 mL buffer/1 × 107 cells. The cells were lysed on ice and then sonicated to shear the chromosomal DNA. Sonication efficiency was determined by agarose gel electrophoresis. Lysates were pre-cleared with protein A sepharose beads for thirty min and the antibody was added to the lysate plus IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.0], 167 mM NaCl), and the tubes were rotated overnight at 4°C. Protein A beads were added to the tubes for two hours, centrifuged and then liquid was aspirated. The beads were washed with successive solutions of low salt wash buffer (0.1% SDS, 1% Triton-X100, 2 mM EDTA, 20 mM Tris [pH 8.0] and 150 mM NaCl), high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris [pH 8.0], 500 mM NaCl), LiCl2 wash buffer (250 mM LiCl2, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA and 10 mM Tris [pH 8.0]) and TE. Protein complexes were eluted off of the beads with a 1% SDS/TE solution at 65° and de-crosslinked overnight at 65°C with a separate 20% input sample. After de-crosslinking, 2 μl of RNAse (0.5 mg/mL) was added to each sample and incubated for 1 hour at 37°C. Samples were passed through a Qiagen PCR cleanup column and eluted in dH2O.
For dot blotting a 0.4 N NaOH/10 mM EDTA solution was added to each sample and the DNA was denatured at 90°C for 10 min. The samples were added to the membrane (Bio-Rad) using a dot-blotting apparatus. The membrane was UV cross-linked and pre-hybridized with DIG Easy Hyb solution (Roche). After thirty min a DIG-labeled telomeric G-probe (TTAGGG6) was added to the hybridization buffer and incubated overnight at 42°C. The blot was washed in 2X SSC/0.1% SDS solution for 15 min then 0.1X SSC/0.1% SDS for 30 min at 42°C. The hybridization signal was developed using the DIG detection system (Roche) and detected using the Fuji Imager. Blots were stripped in 0.2 N NaOH/1% SDS solution and re-hybridized with a probe to the Alu regions of the genome as a control. For real-time PCR detection of the ChIP signal at the sub-telomere, two primer sets were used separately (5′-TCC ATG ATT TAG CAG GAA TGC A-3′/5′-TTC AAA GAA TGG CCT TGG TTT C-3′ and 5′-GCA GCC TGT AGC TCC TGA A-3′/5′-TGG GCA GTG CCT CCT CAT-3′) with SYBR green (Applied Biosystems) in standard real-time PCR reactions in an ABI 7000 real time detection system. These primers amplify DNA adjacent to a large (about 700–800 bp) internal (TTAGGG)n-like island on a duplicated segment of subtelomeric DNA; the sequences are complementary with the subtelomeres of 7p, 9q, 16p, 20p and 16q; and have 98% sequence identity with 8p, 11p, 1p and 19p. There is lower similarity (92–95%) with 19q, 22q, 10q, 5p, 13q, 4q and 4p subtelomeres, and a non-telomeric site with 93% similarity in an internal evolutionarily recent telomere fusion site on 2q. All reactions were done in triplicate and values were obtained using the standard curve method for each primer pair. As a control a primers to the laminB2 coding region were used.
All dot blots were quantified using a Luminescent Image analyzer LAS-3000 (Fujifilm) and quantified with MultiGauge software (Fujifilm). ChIP assays were quantified as percentage of input for each probe evaluated.
Electrophoretic mobility shift assays (EMSA)
FLAG-tagged TRF2 protein was purified from the transfected starting material (see above) was mixed with D100 KCl buffer and then added to 20 μl of hybridization mix (5 mM MgCl2, 5 mM β-mercaptoethanol, 500 μg/mL BSA, 40 μg/mL dI:dC)) in increasing concentrations. A 32P-labeled telomere duplex probe was added to each sample and left at room temperature for 30 min. Samples were added to a 6% acrylamide gel and electrophoresed at 150 V. Gels were dried on a piece of Whatman filter paper and exposed to a phophoimaging screen overnight. The images were retrieved with a phosphoimager (Molecular Dynamics) and the density of each band was quantitated using ImageQuant software (Molecular Dynamics).
Telomere length assays
Genomic DNA was harvesting from cells that were treated with 50 μM HU for 6 d or left untreated using the QiaAMP Genome DNA Extraction Kit (Qiagen). Five μg of genomic DNA was digested with MboI/AluI overnight at 37°C. The DNA was electrophoresis on a 0.7% agarose gel at 80 V, transferred to a nylon membrane (Bio-Rad) and UV cross-linked. A (TTAGGG)6 oligo was used to probe for used to determine telomere length and an Alu probe was used a control. Hybridization was detected using the DIG labeling system (Roche) and images were captured and analyzed using the Fuji LAS-3000 imager and accompanying Multi-Gauge software.
Telomere replication/BrdU IP
Logarithmically growing cells were pulsed labeled with 50 μM BrdU for 30 min and collected by centrifugation. Cells were lysed in a buffer of 50 mM Tris (pH 8.0), 1 M NaCl, 10 mM EDTA 0.5% SDS and 0.2 mg/mL proteinase K for 2 h at 50°C. The DNA was extracted with phenol:chloroform and precipitated with ethanol. After re-dissolving the DNA in TE, the tubes were sonicated and then denatured at 95°C for 5 min. 10X IP buffer was then added to the sample (100 mM phosphate buffer [pH 7.0], 1.4 M NaCl and 0.5% Triton X-100). BrdU antibody (Becton Dickinson) was then added to each tube and rotated for 1 h at room temperature. Rabbit anti-mouse IgG (Sigma) was then added to each tube and incubated again for 30 min with rotation. Samples were washed twice with 1X IP buffer and then centrifuged at 15,000 rpm for 5 min. Pellets were resuspended in 200 μl lysis buffer II (10 mM EDTA, 50 mM Tris [pH 8.8], 0.5% SDS and 0.25 mg/mL proteinase K) overnight at 37°C. The next day samples were treated with another 100 μl of lysis buffer II and incubated at 50°C for 1 h. Finally the samples were collected and the DNA was purified with a PCR column and used in dot blotting as described above.
2D gel protein electrophoresis
A FLAG-tagged TRF2 vector was transfected into HCT116 cells and whole cell protein extracts were made as described above. To recover FLAG-TRF2 from treated and untreated cells, extracts were incubated with FLAG resin beads overnight and washed as above. Beads were incubated with Protein Solubilizer 2 (Invitrogen) and boiled for 10 min to remove the protein from the FLAG resin. The FLAG-protein was reduced and alkalyted per the manufacturer’s instructions, and loaded onto IPG pH 3–10 non-linear strips (Invitrogen) and the strips were re-hydrated overnight. IPG strips were loaded onto the IPG ZOOM runner cassette (Invitrogen) at electrophoresed at 175 V for 15 min, 175–2,000 V ramp for 45 min, and finally at 2,000 V for 1 hr. Strips were equilibrated at loaded onto 4–12% NuPage gels and electrophoresed. Proteins were transferred to a nitrocellulose membrane and used in western blotting as described above.
TERRA RNA analysis
Total RNA was purified with Trizol reagent (Invitrogen) as manufacturer’s instruction. RNA precipitates were treated with DNase I for 45 min at 37°C, followed by DNase I inactivation in the presence of EDTA at 65°C for 5 min. RNA was denatured, blotted to Zeta probe GT genomic membrane (BioRad), and UV crosslinked at 125 mJ. Digoxigenin (DIG)-labeled probes specific for GAPDH were generated by PCR reaction according to manufacturer’s specification (Roche). Oligonucleotide probes for TERRA RNA (6x CCCTAA) or C-rich RNA (6x TTAGGG) were labeled with DIG oligonucleotide 3′ end labeling kit (Roche). Hybridizations were performed using UltraHyb (Ambion) for 16–18 h at 42°C. The membrane was washed twice in 2x SSC, 0.1% SDS at room temperature, twice in 0.2x SSC, 0.1% SDS at 50°C, and developed by DIG-detection kit (Roche). The blots were first hybridized with a DIG-labeled (CCCTAA)6 probe, then stripped with 0.1X SSC, 40 mM Tris pH 7.5 and 1% SDS for three times 10 min each at 80°C, and probed with a DIG-labeled (TTAGGG)6 or GAPDH probe. When indicated, RNA samples were treated with RNaseA (Roche) at a final concentration of 100 μg/ml for 30 min at 37°C. Images were captured with a Luminescent Image analyzer LAS-3000 (Fujifilm) and quantified with MultiGauge software (Fujifilm).
Acknowledgments
We thank Andreas Wiedmer and Jayaraju Dheekollu for technical instruction. We acknowledge the contribution of the Wistar Institute Cancer Center Core Facilities for Flow Cytometry, Genomics, and Microscopy. This work was supported by an NIH NRSA post-doctoral fellowship to A.R.S., an LLS Special Fellowship to Z.D., an LRF post-doctoral fellowship to J.Z., and by NIH CA CA093606.
References
- 1.Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–7. doi: 10.1158/0008-5472.CAN-07-5835. [DOI] [PubMed] [Google Scholar]
- 2.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 3.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–8. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 5.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–9. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 6.Riethman H. Human telomere structure and biology. Annu Rev Genomics Hum Genet. 2008;9:1–19. doi: 10.1146/annurev.genom.8.021506.172017. [DOI] [PubMed] [Google Scholar]
- 7.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–10. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 8.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, et al. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS biology. 2004;2:240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu X-D, Kuster B, Mann M, Petrini JHJ, de Lange T. Cell cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–52. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 10.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–8. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 11.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–13. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 12.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–68. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 2006;7:716–21. doi: 10.1038/sj.embor.7400730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Z, Dheekollu J, Broccoli D, Dutta A, Lieberman PM. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr Biol. 2007;17:1989–95. doi: 10.1016/j.cub.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 15.Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–72. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 16.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–71. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 17.Narath R, Ambros IM, Kowalska A, Bozsaky E, Boukamp P, Ambros PF. Induction of senescence in MYCN amplified neuroblastoma cell lines by hydroxyurea. Gen Chrom Cancer. 2007;46:130–42. doi: 10.1002/gcc.20393. [DOI] [PubMed] [Google Scholar]
- 18.Yarbro JW. Mechanism of action of hydroxyurea. Semin Oncol. 1992;19:1–10. [PubMed] [Google Scholar]
- 19.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–55. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 20.Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, Lieberman PM. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol Cell. 2002;9:493–503. doi: 10.1016/s1097-2765(02)00476-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Snyder A, Lieberman PM. Epstein-barr virus episome stability is coupled to a delay in replication timing. J Virol. 2008;83:2154–62. doi: 10.1128/JVI.02115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chodosh J, Holder VP, Gan YJ, Belgaumi A, Sample J, Sixbey JW. Eradication of latent Epstein-Barr virus by hydroxyurea alters the growth-transformed cell phenotype. J Infect Dis. 1998;177:1194–201. doi: 10.1086/515290. [DOI] [PubMed] [Google Scholar]
- 23.Srinivas SK, Sample JT, Sixbey JW. Spontaneous loss of viral episomes accompanying Epstein-Barr virus reactivation in a Burkitt’s lymphoma cell line. J Infect Dis. 1998;177:1705–9. doi: 10.1086/517427. [DOI] [PubMed] [Google Scholar]
- 24.Azzalin CM, Reichenback P, Khoriauli L, Giulotto E, Lingner J. Telomeric Repeat Containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 25.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–36. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka H, Mendonca MS, Bradshaw PS, Hoelz DJ, Malkas LH, Meyn MS, et al. DNA damage-induced phosphorylation of the human telomere-associated protein TRF2. Proc Natl Acad Sci USA. 2005;102:15539–44. doi: 10.1073/pnas.0507915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fruchtman SM. Treatment paradigms in the management of myeloproliferative disorders. Semin Hematol. 2004;41:18–22. doi: 10.1053/j.seminhematol.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Halsey C, Roberts IA. The role of hydroxyurea in sickle cell disease. Br J Hematol. 2003;120:177–86. doi: 10.1046/j.1365-2141.2003.03849.x. [DOI] [PubMed] [Google Scholar]
- 29.Vassallo C, Passamonti F, Merante S, Ardigo M, Nolli G, Mangiacavalli S, et al. Mucocutaneous changes during long-term therapy with hydroxyurea in chronic myeloid leukaemia. Clin Exp Dermatol. 2001;26:141–8. doi: 10.1046/j.1365-2230.2001.00782.x. [DOI] [PubMed] [Google Scholar]
- 30.De Benedittis M, Petruzzi M, Giardina C, Lo Muzio L, Favia G, Serpico R. Oral squamous cell carcinoma during long-term treatment with hydroxyurea. Clin Exp Dermatol. 2004;29:605–7. doi: 10.1111/j.1365-2230.2004.01586.x. [DOI] [PubMed] [Google Scholar]
- 31.Dantzer F, Giraud-Panis MJ, Jaco I, Ame JC, Schultz I, Blasco M, et al. Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol Cell Biol. 2004;24:1595–607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y, Mitchell TR, Zhu XD. Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mech Ageing Dev. 2008;129:602–10. doi: 10.1016/j.mad.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–20. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 34.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20:551–61. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 35.Tatsumi Y, Ezura K, Yoshida K, Yugawa T, Narisawa-Saito M, Kiyono T, et al. Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres. Genes Cells. 2008;13:1045–59. doi: 10.1111/j.1365-2443.2008.01224.x. [DOI] [PubMed] [Google Scholar]
- 36.Gottifredi V, Shieh S, Taya Y, Prives C. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc Natl Acad Sci USA. 2001;98:1036–41. doi: 10.1073/pnas.021282898. [DOI] [PMC free article] [PubMed] [Google Scholar]