Abstract
The structures of the ligand-binding domains (LBD) of the wild-type androgen receptor (AR) and the T877A mutant corresponding to that in LNCaP cells, both bound to dihydrotestosterone, have been refined at 2.0 Å resolution. In contrast to the homodimer seen in the retinoid-X receptor and estrogen receptor LBD structures, the AR LBD is monomeric, possibly because of the extended C terminus of AR, which lies in a groove at the dimerization interface. Binding of the natural ligand dihydrotestosterone by the mutant LBD involves interactions with the same residues as in the wild-type receptor, with the exception of the side chain of threonine 877, which is an alanine residue in the mutant. This structural difference in the binding pocket can explain the ability of the mutant AR found in LNCaP cells (T877A) to accommodate progesterone and other ligands that the wild-type receptor cannot.
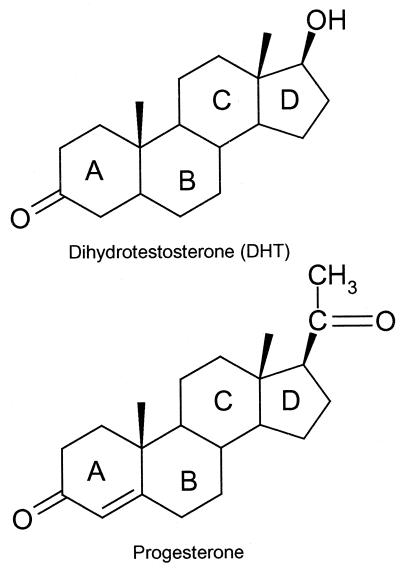
The androgen receptor (AR) is a member of the steroid nuclear-receptor superfamily of ligand-dependent transcription factors (refs. 1 and 2 and refs. therein). The binding of androgens 5α-dihydrotestosterone (DHT) or testosterone (Fig. 1) to the AR initiates male sexual development and differentiation (3). Because the gene coding for the AR is located on the X chromosome, mutations of the AR gene cannot be compensated for by a second allele. Thus, a large number of AR mutants, many affecting male development, have been described in the literature (4).
Figure 1.
Chemical structures of DHT and progesterone.
The presence of androgens is also an important factor in prostate cancer. Castration, by eliminating a major source of steroidal products, has been shown to reduce prostate size (5). Some current treatments for prostate cancer are based on surgical or chemical castration or on the use of AR antagonists (ref. 6 and refs. therein). A number of mutations of the AR have been identified in prostate cancer cells, both cultured and from patient samples. The replacement of threonine 877† with alanine is the most frequent mutation in prostate cancer patients and corresponds to the mutation found in LNCaP cells (7–9). Significantly, this mutation has not been found among the mutations associated with Androgen Independent Syndrome (AIS), although the number of mutant receptors associated with AIS that have been sequenced far exceeds the number from prostate cancer patients that have been so analyzed (The Androgen Receptor Gene Mutations Database World Wide Web Server, http://www.mcgill.ca/androgendb/and refs. therein). There is evidence indicating that the T877A mutation allows the AR to be activated by binding to cortisol or other steroids and even antiandrogens such as flutamide, thereby promoting prostate cancer cell growth (10–12). However, in flutamide-treated patients, the therapeutic effect seen with flutamide withdrawal (withdrawal syndrome) is suggestive of changes in the AR as seen in the LNCaP mutant cell line (13). The relative frequencies of occurrence of the several AR mutations in prostate cancer have not yet been fully evaluated.
As with other members of the steroid receptor family, AR has modular functional domains, including two transactivation functions (AF1 and AF2), a DNA-binding domain, and a ligand-binding domain (LBD) (14). Interactions of each of the domains with coactivators, corepressors, and DNA are well characterized (15). In addition, an intramolecular interaction between the NH2-terminal AF1 and LBD domains of AR has been reported (16, 17). Structural changes resulting from the binding of androgens to the LBD, located at the C-terminal end of the molecule, are responsible for turning on the androgen function.
Structures have been determined for the LBDs of the nuclear receptors for thyroid hormone (TR) (18), retinoic acid RAR-γ (19), retinoid-X RXR-α (20), estrogen (ER) (21), peroxisome PPARγ (22), progesterone (PR) (23), vitamin D (VitD) (24), and androgens (25). They share a similar structural motif consisting of 10–12 α-helices in an antiparallel sandwich motif (Fig. 2). In particular, given that the LBD of the PR has a 50% sequence homology to AR LBD, it was anticipated that the structure of AR LBD would follow a similar folding pattern. We report here the crystal structures of the 261-residue AR LBD (molecular weight = 30,245 Da) and the T877A mutant corresponding to the T877A mutation prevalent in prostate cancer, both in complex with the natural agonist DHT, at 2.0 Å resolution.
Figure 2.
Structure-based alignment of the sequences of AR, PR, ER, retinoic acid receptor (RAR), RXR, thyroid hormone receptor (TR), PPAR, and vitamin D (VitD) receptors based on the position of the helices. Regions not included in the crystal structures are signified by small letters.
Materials and Methods
Cloning, Expression, and Purification of the AR LBD.
The rat AR (rAR) LBD cDNA, from amino acids 646–901 (rat numbering) was cloned from a rat prostate cDNA library (CLONTECH) by PCR. The primers used were CATATGATTGAAG GCTATGAATGTCAACCTATCTTT and TCACTGTGTGTGGAAATAGATGGG. The rAR LBD was expressed as a fusion protein driven by the T7 promoter of pET28b vector (Novagen) to include an N-terminal polyhistidine tag and a thrombin cleavage site. The replacement of T877 for A in this rAR LBD expression construct was performed with the QuickChange Site-Directed Mutagenesis kit (Stratagene). DHT was included in the Escherichia coli (BL21-DE3) fermentation medium at a concentration of 0.05 mM. Induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside was allowed to proceed for 16 h at 20°C in M9 minimal media supplemented with casamino acids (Difco) and trace minerals, and pellets were stored at −70°C. A total of 6–9 mg of recombinant AR LBD was isolated from a 15-g cell pellet after sonication and chromatography on a nickel–chelate resin. Polyhistidine-tagged AR LBD of ≈90% purity eluted at 0.45 M imidazole in a gradient of 0.05–1.0 M imidazole. This material was quantitatively cleaved at an engineered site for thrombin recognition, followed by chromatography on benzamidine Sepharose (Amersham Pharmacia) to remove the serine protease, with a 70% recovery. The final sample containing the sequence Gly-Ser-His-Met at the N terminus followed by residues 646–901 of the rAR (664–919 in human) LBD protein was concentrated for crystallography to 2 mg/ml in 20 mM Tris (pH 7.5)/0.5 M NaCl/10% glycerol/1 mM EDTA/1 mM DTT.
Crystallization and Data Collection.
The AR LBD–DHT complex was crystallized at 20°C by vapor diffusion in the hanging-drop mode. In the initial trial, Crystal Screens 1 and 2 (Hampton Research, Riverside, CA) were used. For each crystallization trial, a 2-μl drop was prepared by mixing 1 μl of purified protein (2 mg/ml) with an equal volume of reservoir solution. The reservoir contained 1.0 ml of the precipitating solution.
Small crystals were obtained in 2 days from six of the drops [solutions 16, 29, and 30 of Crystal Screen (CS) 1; 20, 32, 42 of CS 2]. The most promising results were obtained in solution 29 of CS 1 (0.8 M Na/K tartrate/0.1 M Na Hepes, pH 7.5), and these conditions were optimized on a Cyberlab C-200 automated crystallization workstation (Cyberlab, Brookfield, CT). The largest crystal, rod-shaped with dimensions 0.09 × 0.09 × 0.20 mm, was obtained at 0.887 M Na tartrate. A similar procedure was used to prepare crystals of the complex of the T877A mutant with DHT.
A crystal of the native complex was flash-cooled by dipping it in a cryoprotectant solution consisting of the precipitating solution with 250 mM NaCl and 20% (vol/vol) glycerol, and then placing it in a cold stream at 100 K. X-ray diffraction data were collected at the Industrial Macromolecular Crystallography Association Collaborative Access Team beam line (sector 17-ID) at the Advanced Photon Source synchrotron on a Bruker (Billerica, MA) 2 × 2 mosaic charge-coupled device detector. The crystal diffracted to better than 2.0 Å resolution. Indexing and processing of the measured intensity data were carried out with the HKL2000 software package (26). The data collection and processing statistics for both data sets, native and mutant, are summarized in Table 1.
Table 1.
Summary of data collection and processing
| Native | T877A mutant | |
|---|---|---|
| Source/detector | IMCA/APS 17ID | IMCA/APS 17ID |
| Detector | Bruker 2 × 2 CDD | MarCCD (165 mm) |
| Wavelength | 1.00 Å | 1.00 Å |
| Temperature | 100 K | 100 K |
| Frames | 400 | 180 |
| ΔΦ | 0.5° | 0.5° |
| Crystal distance | 135 mm | 140 mm |
| Time/frame | 10 sec | 10 sec |
| Reduction program | HKL2000 | HKL2000 |
| Unique reflections | 17,329 | 17,291 |
| Resolution | 2.0 Å (2.1–2.0 Å) | 2.0 Å (2.1–2.0 Å) |
| Completeness | 94.8% (73.0%) | 92.8% (53.1%) |
| Multiplicity | 7.3 | 3.2 |
| Mosaicity | 0.33 | 0.57 |
| Rsym (on I) | 10.1% (25.6%) | 4.0% (22.3%) |
| Space group | P212121 | P212121 |
| a | 56.08 Å | 56.01 Å |
| b | 65.76 Å | 67.71 Å |
| c | 70.51 Å | 70.77 Å |
| Wilson B value | 29.26 Å2 | 27.70 Å2 |
Values for data in the last resolution shell are given in parentheses.
Initial Structure Determination.
The structure of the native complex was determined by molecular replacement with the program amore (27) and the PR LBD structure, which has 55% sequence identity and 77% sequence similarity to AR LBD, as the search model. The atomic coordinates of PR LBD (ref. 22, ref. code 1A28) obtained from the Protein Data Bank (28) were unmodified except for the removal of the ligand and solvent molecules. The PR LBD structure gave a solution 1.7σ above background with no molecular interpenetration.
Refinement of the Wild-Type Structure.
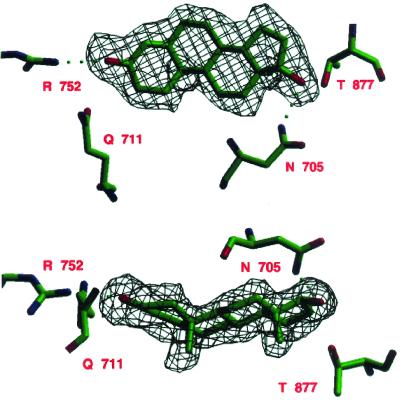
The wild-type structure was refined with program x-plor (29) in eight cycles to an R factor of 24.2%. Each refinement cycle consisted of least-squares minimization, simulated annealing at 3,000°, and individual isotropic B-factor refinement. The first cycle with the PR LBD molecular replacement search model gave an R factor of 33.8%. The model was then rebuilt to introduce the AR amino acid sequence, and a second refinement cycle gave an R factor of 29.6%. At this stage of refinement, the DHT molecule could be clearly seen in the difference electron density map (Fig. 3).
Figure 3.
Two orthogonal views of the omit electron density map of DHT in the hormone-binding site of AR LBD (3σ level) after the final refinement cycle. There are hydrogen bonds between the steroid and the side chains of R752 and N705. Figs. 3 and 6 were drawn by using setor (42).
After each cycle, the structure was carefully examined with the help of the molecular computer graphics program chain (30), and modifications were made to the structure as needed. Several residues, from both the N and C termini of the molecule, which were not seen in the electron density maps, were removed from the model. After the second cycle of refinement, the DHT was added to the model. Solvent molecules were added where there were 3σ peaks in both the 2Fo − Fc and Fo − Fc electron density maps with appropriate hydrogen-bond interactions and were later removed if their B factor went above 60 Å2 in refinement. After eight cycles of x-plor refinement, the electron density is clear except for some of the loop regions, particularly the loop between the first two helices, which was also poorly modeled in the PR LBD structure. At the end of refinement, the structure has an R factor of 24.2% with a total of 2,118 atoms (1,991 protein atoms, 21 atoms corresponding to the ligand, and 106 solvent molecules). The final refinement statistics for the wild-type structure are presented in Table 2.
Table 2.
Final refinement parameters
| Native | T877A mutant | |
|---|---|---|
| Refinement program | XPLOR-99 | CNX 99.0 |
| Refinement cycles | 5 | 5 |
| Completeness | 82.6% | 84.9% |
| Reflections used | 15,067 | 15,455 |
| R value | 0.242 | 0.280 |
| Free R value | 0.312 | 0.345 |
| Number of atoms | 2,118 | 2,091 |
| Protein | 1,991 | 1,994 |
| Ligand (DHT) | 21 | 21 |
| Solvent | 106 | 76 |
| Residues | 246 (672–917) | 247 (672–918) |
| Mean B value | 25.2 Å2 | 34.5 Å2 |
| Resolution range | 10.0–2.0 Å | 10.0–2.0 Å |
| Estimated errors | ||
| Coordinates (Luzzati) | 0.34 Å | 0.33 Å |
| Bond lengths | 0.014 Å | 0.008 Å |
| Bond angles | 1.6° | 1.2° |
| Dihedral angles | 21.5° | 20.1° |
| Improper angles | 1.56° | 0.84° |
Refinement of the T877A Mutant.
Data to 2.0 Å resolution were collected for the complex of the T877A mutant of AR LBD with DHT at the Industrial Macromolecular Crystallography Association Collaborative Access Team beamline at Advanced Photon Source. They were collected with a marCCD area detector (marUSA, Evanston, IL) and reduced with the hkl2000 software package (26) (Table 1). The T877A mutant crystal is isostructural with the wild-type AR LBD–DHT complex (R factor = 32.8% before refinement). The structure was refined by the method of Maximum Likelihood with program cnx (31). The first cycle excluded ligand from the calculation (R factor = 31.3%). The protein structure was then manually refitted with program chain. The difference electron density clearly showed the position of the DHT, which was modeled into the binding site, and the structure was then subjected to four additional cycles of cnx refinement with the ligand present. A total of 76 water molecules were added in positions that had difference electron density in both 2Fo −Fc and Fo − Fc maps of greater than 2.5σ. The refined mutant structure gives an R factor = 28.0% (Rfree = 34.5%) and contains 2,091 atoms (1,994 protein atoms, 21 ligand atoms, and 76 solvent molecules).
Results and Discussion
Description of the Molecule.
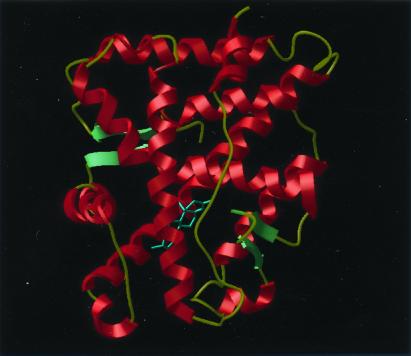
As expected, the structure of the AR LBD has the same overall three-dimensional structure as those of the other nuclear hormone receptor LBDs (Fig. 4). The structure is complete from residues 672 through 917 for the wild-type and 672 to 918 for the T877A mutant. An analysis of the structures with the program procheck (32) showed only minor exceptions to the allowed geometry. In the wild-type structure, the first eight residues of the chain (664) are not seen in the electron density and are probably disordered. This leaves only one residue before the initial residue of the first α-helix (H1). On the C-terminal end, the last two residues (918) are not seen in the electron density of the wild-type structure, but only the last is missing in the mutant. In addition, the loop between helices 9 and 10 (residues 845–850) is poorly defined and has been modeled as polyalanine.
Figure 4.
Ribbon-style drawing of the AR LBD. The substrate DHT and the side chains that interact with it are shown as ball-and-stick figures. Figs. 5, 6, and 8 were drawn by using icm Ver. 2.7 (Molsoft, San Diego, CA)
Comparison with PR.
Although there is only 55% sequence identity between AR LBD and PR LBD, there is a 77% sequence similarity, and as expected, the three-dimensional structures of these two LBDs are very similar, with an rms deviation of 1.3 Å between corresponding Cα atom positions. As with PR, AR LBD has no helix 2, but its helix 12 is longer than those of RXR or the thyroid hormone receptor. In the case of AR, whereas helices 10 and 11 are nearly contiguous, there is a proline residue at position 868 that causes a kink between the two helices. As seen in the PR LBD structure, there is a very short β sheet, formed by the loop between helices 8 and 9 and the C terminus, which holds helix 12 in the closed agonist conformation (21), close to and capping the ligand-binding site.
Comparison with the Complex of R1881 with AR LBD.
A comparison with the previously reported structure of AR LBD with R1881 (25) shows that these structures are very similar. The most notable difference is the continuity of helix 12 versus the split into two shorter helical segments found in the other structure. Because the precise positioning of helix 12 is required for the activation of AR (21), differences in this helix may be caused by the presence of the synthetic ligand R1881 in the steroid-binding site (25). Several other reported features, such as the presence of a cis-peptide bond at P849 and a possible disulfide bond between C669 and C844, are not seen in our higher-resolution structure. These discrepancies may be caused by interpretations of the structure at the limit of experimental resolution.
Lack of Dimer Formation.
Studies have indicated that the ERs, PRs, and ARs all function as homodimers (33), and that AR LBD forms dimers in solution (34). Thus it could be expected that the AR LBD domains might form homodimers in the crystal similar to those previously seen in the RXR-α and ER LBD crystal structures (20, 21, 35, 36). In the PR LBD structure (23), the two monomers in the asymmetric unit are related by a dyad, but the 2-fold symmetric configuration is strikingly different from that of the RXR and ER homodimers, and the area buried in this configuration is much smaller than would be expected for stable dimer formation. In the AR LBD crystal, the LBDs are unmistakably monomeric, and there is no 2-fold axis that relates the domains. Moreover, the homodimer interaction seen in the structures of ER and RXR LBDs is not possible for the AR LBD, as the C-terminal tail is bound to the groove formed by helices 9 and 10, thereby obstructing the contact region between monomers in RXR and ER homodimers. Whether this observation reflects a nondimeric state of the AR LBD in the functional AR dimer or is an artifact of the conditions used for AR LBD crystallization remains to be determined. It is noteworthy that the ER LBD constructs used for crystallization have been truncated to remove an analogous C-terminal extension (21, 35, 36).
Binding of DHT.
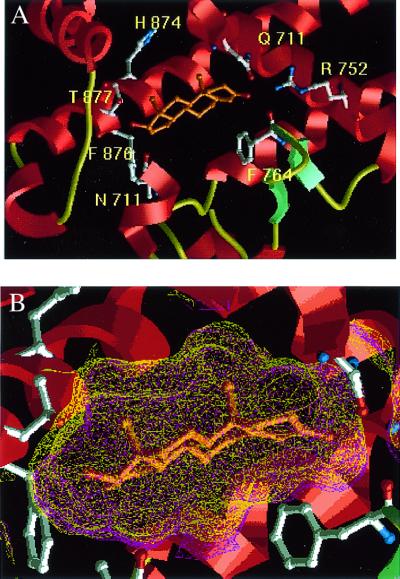
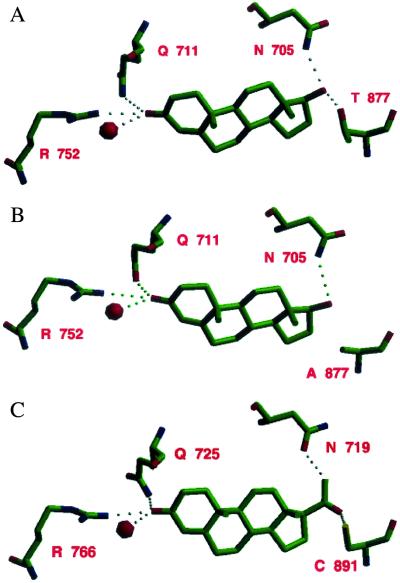
At the end of the molecular replacement procedure with the PR LBD structure (23) without progesterone as search model, the largest piece of difference electron density, at approximately the 3σ level, was found at the progesterone-binding site. Replacing the bound progesterone agonist (which has an acetyl group at the 17-position) with a model of DHT (which has a hydroxyl group at the 17-position) produced an even better fit to the difference electron density, indicating that DHT binds to AR LBD (Fig. 5A) in an almost identical fashion to the way progesterone binds to PR LBD (Table 3). Both agonists interact with helices 3, 5, and 11 of their respective LBDs. As shown in part in Fig. 6, the interactions of ring A of each hormone with its receptor are similar, specifically those with the side chains of Q711, M745, and R752 in AR LBD, corresponding to those with Q725, M759, and R766 in PR LBD, and a conserved water molecule (Table 4). The interactions of AR and PR with ring C are also similar, with close contacts to the main-chain of L704 (L718 in PR LBD) and sidechain of N705 (N719 in PR LBD). The contact between C18 of DHT and the Oγ1 of T877 is unique to the wild-type AR LBD, as the corresponding cysteinyl side chain is pointed away from the steroid in the PR LBD structure.
Figure 5.
(A) Model of DHT in the binding site of AR LBD. (B) Comparison of the ligand contact surface area for DHT and progesterone.
Table 3.
Dihydrotestosterone contacts, 3.4 Å
| DHT | Protein | Native | T877A | Progresterone |
|---|---|---|---|---|
| Possible hydrogen bonds | ||||
| O20 | Asn-705 Nδ2 | 2.80 Å | 3.04 Å | |
| O3 | Gln-711 Nɛ2 | 3.36 Å | 3.44 Å | Gln-725 Nɛ2 3.21–3.28 Å |
| O20 | Thr-877 Oγ1 | 2.70 Å | N/A | N/A |
| O3 | Arg-752 Nη2 | 2.89 Å | 3.02 Å | Arg-766 Nη2 2.77–2.78 Å |
| O3 | Water O | 3.46 Å | 3.26 Å | W1004 3.09–3.40 Å |
| Possible close contacts | ||||
| C11 | Leu-704 O | 3.31 Å | 3.48 Å | |
| C12 | Leu-704 O | 3.49 Å | (3.74 Å) | |
| C12 | Asn-705 Nδ2 | 3.07 Å | 3.17 Å | Asn-719 Oδ1 3.38–3.40 Å |
| C17 | Asn-705 Nδ2 | 3.34 Å | 3.43 Å | |
| C2 | Gln-711 Oɛ1 | 3.47 Å | 3.28 Å | Gln-725 Nɛ2/Oɛ1 3.20–3.34 Å |
| O3 | Gln-711 Oɛ1 | 4.05 Å | 3.27 Å | Gln-725 Nɛ2/Oɛ1 3.20–3.48 Å |
| C19 | Met-745 Sδ | 3.38 Å | 3.39 Å | Met-759 Sδ 3.46–3.47 Å |
| C4 | Phe-764 Cδ2 | 3.47 Å | 3.50 Å | |
| C18 | Ala-877 Cβ | 3.45 Å | 3.38 Å | |
| C18 | Thr-877 Oγ1 | 3.07 Å | N/A | |
Figure 6.
Comparison of the binding of (A) DHT to AR LBD, (B) DHT to the AR LBD T877A mutant, and (C) progesterone to PR LBD.
Table 4.
Comparison of AR and PR steroid binding
| AR | PR | |
|---|---|---|
| Ring A | ||
| O3 | H-bond to R752 NH2 (2.9 Å) | H-bond to R766 NH2 (2.8 Å) |
| H-bond to water (3.5 Å) | H-bond to water (3.1/3.4 Å) | |
| SC of Q711 in different rotomer distance to O3 is 3.4 Å and 4.13 Å | Contact to SC of Q725 distance to O3 is 3.2 and 3.3 Å | |
| C19 | Contact to M745 SD (3.4 Å) | Similar orientation to M759 (3.5 Å) |
| C2 | SC of Q711 (3.5 Å) | Different rotomer (3.2 and 3.3 Å) distance to C4 is 4.1 Å |
| Ring C | ||
| C11 | L704 O (3.3 Å) | L718 O (3.5 Å) |
| C12 | Contact to N705 Nδ2 (3.1 Å) | Contact to N719 Oδ1 (3.4 Å) |
| C18 | Contact T877 Oγ1 (3.1 Å) | SC of C891 pointing away distance to Sγ is 3.8 Å |
| Ring D | ||
| O20 | O20 in AR is close to C21 in PR | (Possible flip of acetyl in PR?) |
| N/A | O20: Contact to C891 Ca (3.2 Å) | |
| O20 | H-bond N705 Nδ2 (2.8 Å) | C21: Contact to N719 OD1 (3.2 Å) |
| O20: Contact T877 Cγ1 (2.7 Å) | SC of C891 pointing away | |
| C17 | Contact N705 Nδ2 (3.3 Å) | Ring in slightly different orientation; distance to N719 Oδ1 is 4.7 Å |
Because progesterone and DHT differ in the substituent on ring D, it is expected that interactions with respective receptors will differ in this region. In the AR structure, the hydroxyl of DHT makes a hydrogen bond to the Nδ2 of N705. Although rings A are in similar orientations in the two structures, the double bond unique to progesterone causes a slightly different conformation of the two steroids resulting in ring D being offset by over 1.0 Å, bringing the 17-hydroxyl in DHT close to the position corresponding to that of the methyl in the acetyl group in progesterone. It is noteworthy that a hydrogen bond could be made between progesterone and the PR LBD similar to that between the 17-hydroxyl and the side chain of N705 if both the steroid acetyl group and the side chain of N719 were flipped relative to current positions. This would place the acetyl oxygen ≈3.2 Å from the Nδ2 atom of N719 (Fig. 6). The ligand contact surface area is slightly smaller for DHT in AR than for progesterone in PR (448 vs. 483 Å2), but they are both considerably smaller than the ligand contact surface area in the thyroid hormone receptor (TR) (559 Å2), peroxisome proliferator activated receptor (PPARγ) (583 Å2), or the vitamin D receptor (VitD) (677 Å2).
Structure of the Complex of DHT with the LBD of the T877A Mutant.
The mutant LBD structure has an rms deviation of 0.8 Å compared with the wild-type structure, close to the expected rms deviation caused by the estimated errors in the coordinates (37). In particular, the binding of DHT is essentially identical by wild-type and mutant LBDs except at the point of mutation (Table 4). Here the replacement of T877 by alanine leaves additional space off of the D-ring of DHT to accommodate a larger substituent on position 17 (Fig. 5B). This may explain the promiscuous ability of the T877A mutant, unlike wild-type AR, to bind to a variety of other hormones and analogs like some progestins, estrogens (38), and cortisols (12) that differ from DHT in substitution at position 17. For example, the binding of flutamide, estradiol, and progesterone to the T877A mutant can activate the mutant receptor (39, 40). Conversely, mutation of T877 to residues with larger sidechains such as aspartic acid and lysine would be expected to prevent the binding of ligands with any substituent at position 17 of the D-ring, and such mutations have been shown to eliminate androgen binding (41).
Conclusions
The structures of the complexes of wild-type and T877A-mutant AR LBD with DHT provide clear explanations for the differential affinities of these nuclear receptors for androgens and for the promiscuity of the T877A mutant of AR in its ability to respond to other ligands. In addition, the binding of the extended C terminus of the AR LBD to the homodimerization interface used by the RXR and ER LBDs prevents formation of that homodimer in the crystal. Interestingly, the failure of protease digestion experiments with the AR LBD to remove or truncate the extended C terminus (J.E.S. and S.E.K., unpublished work) suggests that the C terminus remains tightly bound to the AR LBD monomer in solution. We speculate that the ability of an RXR- or ER-LBD-like homodimeric interaction to contribute to the interactions stabilizing the functional AR homodimer is prevented unless the C termini are displaced by intervention of other macromolecules or by other portions of the AR itself. The importance of this interaction of C-terminal domains to the integrity of the functional receptor is highlighted by the effects of mutations in the C terminus, such as P916L and H917R, which in men result in an inactive receptor leading to development as a female.
The availability of the structure of the AR LBD will help in our understanding of the mechanism of action of androgen agonists and antagonists, coactivators, and corepressors, in the development of treatments for prostate cancer and other AR diseases.
Acknowledgments
We thank Steven Sheriff, Bruce Jacobson, Herbert Klei, and the staff of the Industrial Macromolecular Crystallography Association Collaborative Access Team (IMCA-CAT) at the Advanced Photon Source for assistance in data collection. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. Data were collected at IMCA-CAT beamline 17-ID at the Advanced Photon Source. These facilities are supported by the companies of the IMCA through a contract with Illinois Institute of Technology, executed through the Center for Synchrotron Radiation Research and Instrumentation.
Abbreviations
- AR
androgen receptor
- LBD
ligand-binding domain
- DHT
5α-dihydrotestosterone
- PR
progesterone receptor
- ER
estrogen receptor
- RXR
retinoid-X receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1I37 and 1I38).
Because the protein sequences for rat and human AR LBD are identical, the human numbering system was used throughout this report, although the protein was expressed from a rat cDNA construct.
References
- 1.Tenbaum S, Baniahmad A. Int J Biochem Cell Biol. 1997;29:1325–1341. doi: 10.1016/s1357-2725(97)00087-3. [DOI] [PubMed] [Google Scholar]
- 2.Whitfield G K, Jurutka P W, Haussler C A, Haussler M R. J Cell Biochem. 1999;Suppl. 32–33:110–122. doi: 10.1002/(sici)1097-4644(1999)75:32+<110::aid-jcb14>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.McPhaul M J, Marcelli M, Tilley W D, Griffin J E, Wilson J D. FASEB J. 1991;5:2910–2915. doi: 10.1096/fasebj.5.14.1752359. [DOI] [PubMed] [Google Scholar]
- 4.Gottlieb B, Beitel L K, Lumbroso R, Pinsky L, Trifiro M. Hum Mutat. 1999;14:103–114. doi: 10.1002/(SICI)1098-1004(1999)14:2<103::AID-HUMU2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 5.Huggins C. Cancer Res. 1965;25:1163–1167. [PubMed] [Google Scholar]
- 6.Denis L J, Griffiths K. Semin Surg Oncol. 2000;18:52–74. doi: 10.1002/(sici)1098-2388(200001/02)18:1<52::aid-ssu8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Taplin M E, Bubley G J, Shuster T D, Frantz M E, Spooner A E, Ogata G K, Keer H N, Balk S P. N Engl J Med. 1995;332:1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 8.Veldscholte J, Ris-Stalpers C, Kuiper G G, Jenster G, Berrevoets C, Claassen E, van Rooij H C, Trapman J, Brinkmann A O, Mulder E. Biochem Biophys Res Commun. 1990;173:534–540. doi: 10.1016/s0006-291x(05)80067-1. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki H, Akakura K, Komiya A, Aida S, Akimoto S, Shimazaki J. Prostate. 1996;29:153–158. doi: 10.1002/1097-0045(199609)29:3<153::aid-pros2990290303>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang C, Young W J, Chang C. Endocrine. 2000;12:69–76. doi: 10.1385/ENDO:12:1:69. [DOI] [PubMed] [Google Scholar]
- 11.Taplin M E, Bubley G J, Ko Y J, Small E J, Upton M, Rajeshkumar B, Balk S P. Cancer Res. 1999;59:2511–2515. [PubMed] [Google Scholar]
- 12.Zhao X-Y, Malloy P J, Krishnan A V, Swami S, Navone N M, Peehl D M, Feldman D. Nat Med. 2000;6:703–706. doi: 10.1038/76287. [DOI] [PubMed] [Google Scholar]
- 13.McDonald S, Brive L, Agus D B, Scher H I, Ely K R. Cancer Res. 2000;60:2317–2322. [PubMed] [Google Scholar]
- 14.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schuetz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 16.He B, Kemppainen J A, Voegel J J, Gronemeyer H, Wilson E M. J Biol Chem. 1999;274:37219–37225. doi: 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- 17.He B, Kemppainen J A, Wilson E M. J Biol Chem. 2000;275:22986–22994. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- 18.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. Nature (London) 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 19.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 20.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 21.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engström O, Öhman L, Greene G L, Gustafsson J A, Carlquist M. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 22.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 23.Williams S P, Sigler P B. Nature (London) 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 24.Rochel N, Wurtz J M, Mitschler A, Klaholz B, Moras D. Mol Cell. 2000;5:173–179. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- 25.Matias P M, Donner P, Coelho R, Thomaz M, Peixoto C, Macedo S, Otto N, Joschko S, Scholz P, Wegg A, et al. J Biol Chem. 2000;275:26164–26171. doi: 10.1074/jbc.M004571200. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. In: Macromolecular Crystallography. Carter W C Jr, Sweet R M, editors. Vol. 276. New York: Academic; 1997. pp. 307–326. [Google Scholar]
- 27.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 28.Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brünger A T. Nature (London) 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 30.Sack J S. J Mol Graphics. 1988;6:224–225. [Google Scholar]
- 31.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges N, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 33.Liao M, Zhou Zx, Wilson E M. Biochemistry. 1999;38:9718–9727. doi: 10.1021/bi990589i. [DOI] [PubMed] [Google Scholar]
- 34.Nemoto T, Ohara-Nemoto Y, Shimazaki S, Ota M J. Steroid Biochem Mol Biol. 1994;50:225–233. doi: 10.1016/0960-0760(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 35.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 36.Pike A C W, Brzozowski A M, Hubbard R E, Bonn T, Thorsell A-G, Engstrom O, Ljunggren J, Gustafsson J-A, Carlquist M. EMBO J. 1999;18:4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luzatti V. Acta Crystallogr. 1953;6:142–152. [Google Scholar]
- 38.Veldscholte J, Ris-Stalpers C, Kuiper G G J M, Jenster G, Berrevoets C, Claassen E, Van Rooij H C J, Trapman J, Brinkmann A O, Mulder E. Biochem Biophys Res Commun. 1990;173:534–540. doi: 10.1016/s0006-291x(05)80067-1. [DOI] [PubMed] [Google Scholar]
- 39.Doesburg P, Kuil C W, Berrevoets C A, Steketee K, Faber P W, Mulder E, Brinkmann A O, Trapman J. Biochemistry. 1997;36:1052–1064. doi: 10.1021/bi961775g. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Young W J, Chang C. Endocrine. 2000;12:69–76. doi: 10.1385/ENDO:12:1:69. [DOI] [PubMed] [Google Scholar]
- 41.Ris-Stalpers C, Verleun-Mooijman M C T, Trapman J, Brinkmann A O. Biochem Biophys Res Commun. 1993;196:173–180. doi: 10.1006/bbrc.1993.2231. [DOI] [PubMed] [Google Scholar]
- 42.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]