Abstract
We previously reported that C-terminal fragment of ADAMTS-18 induces platelet fragmentation through ROS release. We have shown that thrombin cleaves ADAMTS-18 and that a short form of ADAMTS-18 in in vitro translational assay. However, the exact thrombin cleavage site and whether a short form ADAMTS-18 presents in vivo are not clear. In this study, we first identified that the thrombin cleavage site is between Arg775 and Ser776 by thrombin cleavage of ADAMTS-18 peptide following mass spectrum assay. We then showed that a short form ADAMTS-18 presents in brain, kidney, lung, and testicle from C57BL/6 mouse embryo. Since alternative form of ADAMTS-18 could be a mechanism to regulate its activity, we then investigated the mechanism involves in the generation of ADAMTS-18 short form. However, neither protease inhibitors nor mutations in catalytic domain of ADAMTS-18 have any significant effect on the generation of ADAMTS-18 short form. Thus, our data demonstrate a thrombin cleavage site and confirm a short form of ADAMTS-18 presents in vivo.
Keywords: Metalloproteinase, Thrombin, Cleavage, ADAMTS-18
Introduction
The human ADAMTSs (a disintegrin and metalloproteinase with a thrombospondin type 1 motif) are a family of secreted Zn-metalloproteinases, which have multidomain structural components in common.[1; 2; 3; 4] The functions of members of the ADAMTS family include N-terminal procollagen processing (ADAMTS-2, -3, -14),[5; 6; 7; 8] spermatogenesis (ADAMTS-2),[9] inhibition of angiogenesis (ADAMTS-1, -8, and -9),[10; 11] follicular rupture and ovulation (ADAMTS- 1),[12] cleavage of matrix proteoglycans aggrecan, versican, and brevican (ADAMTS-1, -4, -5, -8, -9, -15),[13; 14; 15] degradation of cartilage oligomeric matrix protein (ADAMTS-7, ADAMTS-12), and cleavage of ultra large molecular weight von Willebrand factor (ADAMTS-13).[3] ADAMTS-18 has recently been shown to be epigenetically silenced in multiple carcinomas and to have tumor suppressor activity.[16] We have shown that C-terminal fragment of ADAMTS-18 induces platelet fragmentation through ROS (reactive oxygen species).[1] Although we reported that thrombin cleaves ADAMTS-18, the exact thrombin cleavage site and how the activity of ADAMTS-18 being regulated are still unknown.
The regulation of metalloprotease activity could be at three levels: transcriptional regulation, zymogen activation, and regulation on the level of enzymatic activity by different endogenous regulators such as protease cleavage or inhibitors.[17; 18] At the transcriptional level, it has been shown that ADAMTS-16 expression is stimulated by TGFβ in chondrocyte cell lines and by follicle-stimulating hormone (FSH) in fully differentiated luteinizing granulose cells.[19; 20] The mRNA level of ADAMTS-8 is down- regulated in brain tumor and TNFβ is able to up-regulate ADAMTS-18 mRNA level in endothelial cells.[1; 21; 22] The ADAMTSs activity can also be regulated by proteolytic process.[23] All known ADAMTSs (except 10 and 12) contain a subtilisin-like pro–protein convertase cleavage site in their prodomains that are furin recognition sequences. ADAMTS can be cleaved at the N-terminal by furin or related pro-protein convertase(s) within the trans-Golgi, resulting in secretion of mature, potentially active enzymes lacking the propeptide region.[1; 3] In addition, ADAMTS family members such as ADAMTS-1 and ADAMTS-12 have been shown to undergo proteolytic processing within their C-terminal regions, resulting in removal of domains that can bind to sulfated GAGs.[9; 24] It has been shown that C-terminal truncation enhances the aggrecanase and versicanase activities of ADAMTS-4, indicating a potential regulatory function associated with one or more domains of the ADAMTS-4 C-terminal region. [25; 26]
Alteration of ADAMTSs activity has been implicated with certain physiological conditions in vivo. It has been shown that following transient middle cerebral artery occlusion in the rat, ADAMTS-1 and -4 are up-regulated.[27] An orderly temporal expression of the metalloproteinases and ADAMTS has been shown during the progression of fracture healing.[28] We have reported that thrombin cleaves ADAMTS-18 and releases C-terminal fragment and shown that a short form of ADAMTS-18 was also present during in vitro translation of full length ADAMTS-18.[22] However, the exact thrombin cleavage site and whether the short form presents in vivo are not clear. Thus, to better understand the function of ADAMTS-18, we have investigated the thrombin cleavage site and the expression of short form ADAMTS-18 in vivo.
Materials and Methods
Reagents and plasmid
All reagents were purchased from Sigma unless otherwise designed. ADAMTS-18 peptide was synthesized by Bio-Synthesis (Lewisville, TX). The in vitro translation kit was purchased from Promega (Madison, WI USA). Full-length ADAMTS-18 cDNA coding sequence was purchased from ATCC (Manassas, VA ) and cloned into mammalian expression vector pBudCE4.1 from Invitrogen (Carlsbad, CA). pCR3.1/ADAMTS-18 was kindly provided by Dr. Andrew Connolly (Stanford University, CA). Optimized ADAMTS-18-cDNA was synthesized by GenScript (Piscataway, NJ) and then cloned into pcDNA3.1. Protease inhibitors Complete Mini Cocktail and Complete Mini EDTA-free were purchased from Roche (Mannheim, Germany).
Peptide synthesized and mass spectrum assay
ADAMTS-18 peptide was digested with thrombin (5 U/ml) at room temperature for one hour with/without huridin (5 ug/ml). The digested samples were analyzed by mass spectrum assay at NYULMC protein core facility. Briefly, 10 mg/ml Alpha-Cyano-4-Hydroxycinnamic Acid (CHCA, Agilent Technologies) was used as the MALDI matrix. A total of 1.0 μL of sample in 50% acetonitrile and 0.1% TFA was mixed with equal volume of matrix solution, spotted on the MALDI plate, and air-dried. Samples for mass spectrometry determination were analyzed in positive, reflectron mode on a Bruker Autoflex (Billerica, MA). Samples for peptide sequence determination were analyzed in positive, reflectron mode on a Waters Q-Tof Ultima (Milford, MA).
In vitro translation
35S-methionine–labeled ADAMTS-18 was translated from pBudCE4.1/AD18, pBudCE4.1/AD18 mutants, pBluescript/AD18, pcDNA3.1/AD18, or pCR3.1/AD18 using an in vitro translation kit (TNT Coupled Reticulocyte Lysate Systems; Promega, Madison, WI) following the protocol provided by the manufacturer. Various protease inhibitors: Complete Mini Cocktail, Complete Mini(-EDTA) 100ug/ml or α2-macroglobulin (4mU-16mU) were added to the reactions. The translational products were resolved on SDS-PAGE gel. Film was developed by exposing to the dried gel at 4°C overnight.
Construct the catalytic domain mutant ADAMTS-18 expression vector
Mutant primers were synthesized by Sigma. The point mutations were introduced into the catalytic domain of ADAMTS-18 as previous described.[29; 30] Briefly, 10ng DNA fragments from the ADAMTS-18 plasmid were amplified using high fidelity Pfu from Stratagene (Santa Clara,CA) in a final volume of 100 u1 and subjected to 30 cycles of denaturation (1 min, 94 °C), annealing (2 min, 50 °C), and extension (3 min, 72 °C) using a DNA Thermal Cycler (Perkin Elmer, Waltham, MA). The products of the reaction were analyzed and purified through agarose gel electrophoresis. Purified DNA fragments from an initial set of reactions were used in a subsequent overlap extension reaction. Overlapping fragments were mixed and subjected to PCR amplification using the external primers. Full length ADAMTS-18 mutant fragment was purified from agarose gel using Qiagen PCR product purification kit and then cloned into pcDNA3.1. Sequences of all constructs were confirmed by sequencing.
Immunoblotting
Twenty-five micrograms of protein from a variety of tissue lysis from C57BL/6 mice embryo were separated by 7% SDS/PAGE gels and transferred to a PVDF membrane before immunoblotting with rabbit anti-ADAMTS-18 IgG (sc-68416, Santa Cruz Biotechnology, Inc.) overnight (1:1000 dilution). The membrane was then blotted with Biotin conjugated anti-Rabbit IgG (1:10,000) for 2 h, followed by horseradish peroxide (HRP)-conjugated Avidin (1:20,000) for half hour. The ADAMTS-18 band was detected with chemiluminescence substrate (Thermo Fisher Scientific, Rockford, IL).
Results
Primary thrombin cleavage site is between Arg775 and Ser776
Previously, we have reported that thrombin is able to cleave ADAMTS-18 and release a C-terminal fragment.[22] To identify the thrombin cleavage site, we have synthesized a ADAMTS-18 peptide containing the predicted putative thrombin cleavage site (Figure 1a,b). Mass spectrum assay of ADAMTS-18 peptide cleaved by thrombin reveals two peptide peaks (MW1561 and MW1447), which reflect molecular weight of half the size of the synthesized peptide (Figure 1c). Mass spectrum sequencing confirms that MW1561 is the N terminal end of the peptide suggesting that the thrombin cleavage site is between Arg775 and Ser776. Hirudin completely inhibited the generation of MW1561 and MW1447 peak suggesting the specificity of thrombin cleavage (Figure 1c).
Figure 1.
a. Schematic of ADAMTS-18 showing putative thrombin cleavage site; b. Synthesized ADAMTS-18 peptide with predict thrombin cleavage site; c. Mass spectrometry assay to confirm thrombin cleavage site.
In vitro translation of ADAMTS-18 shows a short form
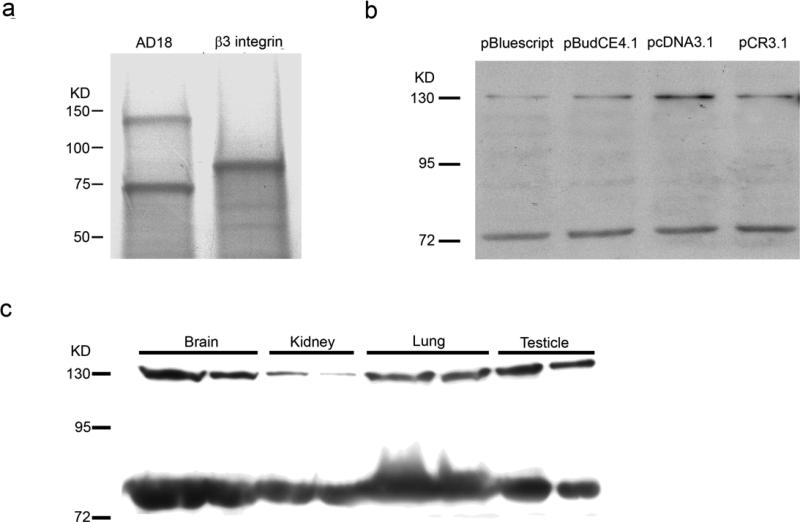
35S-methione-labeled ADAMTS-18 was generated by in vitro translation using the expression vector pBudCE4.1/ADAMTS-18. Since a non-specific band sometimes also presents in an in vitro translation assay, pBudCE4.1/β3 integrin was used as a control. As shown in Figure 2a, pBudCE4.1/AD18 has two bands (135 kd and 75 kd). There is only one 85kd band in control. In addition, since 75 kd band was also found in translational product using other clone vectors: pBluescript/ADAMTS-18, pCR3.1/ADAMTS-18, our data suggests that the 75 kd band is indeed from ADMTS-18 cDNA (Figure 2b).
Figure 2.
a. In vitro translation using pBudCE4.1/ ADAMTS-18 and pBudCE4.1/β3 integrin ; b. In vitro translation using pBluescript/ADAMTS-18, pBudCE4.1/ADAMTS-18, pcDNA3.1/ADAMTS-18 (genetic codon optimized cDNA), and pCR3.1/ADAMTS-18; c. Western blot to detect ADAMTS-18 in brain, kidney, lung, and testicle from C57BL/6 mouse embryo.
Genetic codon optimization has no effect on production of short form ADAMTS-18
ADAMTS-18 contains a significant amount of rare genetic codons. These rare codons may lead to the premature translational stop.[31] To test whether the formation of the short is due to such a rare genetic codon, we re-synthesized the full lenghth cDNA of ADAMTS-18 with optimized genetic code and cloned into pcDNA3.1. Again, the optimization of ADAMTS-18 has no effect on the short form ADAMTS-18 formation (Figure 2b).
Short form of ADAMTS-18 exists in brain, kidney, lung, and testicle from C57BL/6 embryo
To confirm whether the short form of ADAMTS-18 is present in vivo, we examined ADAMTS-18 expression in normal tissues by western blot. Protein samples from tissues of mouse embryo: brain, kidney, lung, and testicle were resolved on SDS-PAGE gel and transferred to PVDF membrane to detect ADAMTS-18. Our result shows that both two forms of ADAMTS-18 present in these tissues (Figure 2c).
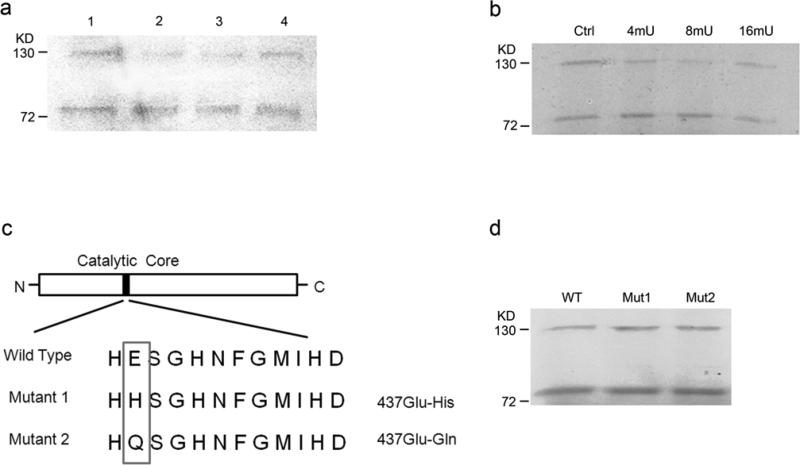
Protease inhibitors and mutations in catalytic domain have no effect on 75 kd ADAMTS-18 formation
We next explored the potential mechanism for the generation of the short form ADAMTS-18. To test whether the 75 kd form is due to protease cleavage, in vitro translation was done in the presence of a variety of protease inhibitor. Protease inhibitors have no significant effect on the generation of the short form (Figure 3a,b). Since ADAMTS-18 is the closest homology to ADAMTS-16, which has been shown to be able to cleave α2-macroglobulin, we decided to test whether the short form is due to the autocatalytic activity of ADAMTS-18.[20; 32] Point mutations were introduced into the catalytic domain based on previous reports (Figure 3c).[33; 34; 35; 36] These mutations in ADAMTS-18 catalytic domain had no effect on the generation of the 75 kd form in in vitro translation assays (Figure 3d). Thus, it appears another mechanism may be involved in the generation of the 75 kd ADAMTS-18.
Figure 3.
a. In vitro translation of pBudCE4.1/ADAMTS-18 with protease inhibitors: Control (lane 1); Complete Mini EDTA free 100ug/ml (lane 2); Complete Mini Cocktail 100ug/ml (lane 3); α2-macroglobulin 4mU/ml (lane 4); b. In vitro translation of pBudCE4.1/ADAMTS-18 with α2-macroglobulin: Control (lane 1); 4mU(lane 2); 8mU(lane 3); 16mU(lane4); c. Schematic of point mutations introduced into ADAMTS-18 catalytic domain; d. In vitro translation of pBudCE4.1/ADAMTS-18 wildtype and mutants.
Discussion
Alternative forms have been reported in many ADAM members as well as ADAMTS members. Different isoforms resulting from alternative splicing have been shown in ADAM-8,-9,-10,-11,-12,-15,-19,-22,-28,-29,-30, and -33 or ADAMTS-4, ADAMTS-6.[23] Isoforms could also result from post-translational modulations from a processing of the metalloproteinase domain itself or by other metalloproteinases.[23] The different forms of ADAMTS could function differently in vivo. It has been shown that there are two distinct active forms of ADAMTS-1 generated by two consecutive steps to release p87 and p65 forms. The full length of ADAMTS-1 and ADAMTS-1 fragments possess pro- or anti-metastatic activity, respectively.[37; 38] Thus, it is of particular interest to identify whether a ADAMTS member has an isoform and what its function may be.
In this study, we have identified the thrombin cleavage site and confirmed the presence of a short form of ADAMTS-18 in vivo. Although the potential function of the short form is not clear, our result has revealed a potential mechanism may involve in the regulation of ADAMTS-18 activity. The changes of ADAMTS-18 activity could be of significant biological implication in vivo. In fact, previous publications have shown that ADAMTS-18 expression level is associated with the tumor metastasis.[39] The expression profile of ADAMTS-18 has been studied in gastric, colorectal and pancreatic cancers and the a down regulation of ADAMTS-18 is associated with DNA methylation of its promoter.[40] The methylation levels in ADAMTS-18 gene promoter region has been studied in tissue from patients samples:100 gastric cancers, 100 colorectal cancers, 70 pancreatic cancers, and equal number of adjacent normal tissues. Methylation levels in all three types of cancers were significantly higher compared to normal tissues. This is the first study to estimate the prevalence of ADAMTS-18 methylation based on large amount of tumor samples, showing that epigenetic regulation of ADAMTS-18 expression level is associated with carcinogenesis.[40] Further studies are needed for better understand how ADAMTS-18 activity is regulated and what the biological effect could result.
Since the protease inhibitors used in this study do not inhibit the formation of short form, the potential mechanism underlying the generation of short form ADAMTS-18 is not clear. It is possible that the protease inhibitors do not completely inhibit all proteases activity given the fact that ADAMTS-16 the closest homology of ADATMTS18 could cleave α2-Macroglobulin.[20] Other possibilities could be either there is a cryptic translation start site or a translation stop site. It has been shown that RNA secondary structure can affect the translation.[41] Using computer program Mfold (Mfold Web Server, The RNA Institute, Albany, SUNY), we have found a similar mRNA secondary structure with mRNA sequences from wildtype ADAMTS-18 or codon optimized ADAMTS-18. In addition, it has been reported that the 5’ untranslated region of ADAMTS-6 may be involved in translational control.[42] Thus, the role of mRNA secondary structure in generation of ADAMTS-18 short form cannot be excluded. Further study will help us understand better how ADAMTS-18 short form is generated in vivo.
Previously, we have shown that thrombin cleaves ADAMTS-18 and the cleavage site is not clear.
In present study, we have identified that the thrombin cleavage site is between Arg775 and Ser776.
Our data also reveals that a short form ADAMTS-18 presents in a variety of C57BL/6 tissues.
Acknowledgement
The mass spectrum assay was done with the help of Mr. Steven Blais in Protein Analysis Facility of Skirball Institute of Biomolecular Medicine of NYUMC. We thank Mr.Michael Nardi for reviewing the manuscript and his suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem, United States. 2009:31493–7. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tortorella MD, Malfait F, Barve RA, Shieh HS, Malfait AM. A review of the ADAMTS family, pharmaceutical targets of the future. Curr Pharm Des. 2009;15:2359–74. doi: 10.2174/138161209788682433. [DOI] [PubMed] [Google Scholar]
- 3.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J, England. 2005:15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wight TN. The ADAMTS proteases, extracellular matrix, and vascular disease: waking the sleeping giant(s)! Arterioscler Thromb Vasc Biol, United States. 2005:12–4. doi: 10.1161/01.ATV.0000150043.43083.aa. [DOI] [PubMed] [Google Scholar]
- 5.Colige A, Beschin A, Samyn B, Goebels Y, Van Beeumen J, Nusgens BV, Lapiere CM. Characterization and partial amino acid sequencing of a 107-kDa procollagen I N-proteinase purified by affinity chromatography on immobilized type XIV collagen. J Biol Chem. 1995;270:16724–30. doi: 10.1074/jbc.270.28.16724. [DOI] [PubMed] [Google Scholar]
- 6.Colige A, Li SW, Sieron AL, Nusgens BV, Prockop DJ, Lapiere CM. cDNA cloning and expression of bovine procollagen I N-proteinase: a new member of the superfamily of zinc-metalloproteinases with binding sites for cells and other matrix components. Proc Natl Acad Sci U S A. 1997;94:2374–9. doi: 10.1073/pnas.94.6.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colige A, Vandenberghe I, Thiry M, Lambert CA, Van Beeumen J, Li SW, Prockop DJ, Lapiere CM, Nusgens BV. Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J Biol Chem. 2002;277:5756–66. doi: 10.1074/jbc.M105601200. [DOI] [PubMed] [Google Scholar]
- 8.Fernandes RJ, Hirohata S, Engle JM, Colige A, Cohn DH, Eyre DR, Apte SS. Procollagen II amino propeptide processing by ADAMTS-3. Insights on dermatosparaxis. J Biol Chem. 2001;276:31502–9. doi: 10.1074/jbc.M103466200. [DOI] [PubMed] [Google Scholar]
- 9.Cal S, Arguelles JM, Fernandez PL, Lopez-Otin C. Identification, characterization, and intracellular processing of ADAM-TS12, a novel human disintegrin with a complex structural organization involving multiple thrombospondin-1 repeats. J Biol Chem, United States. 2001:17932–40. doi: 10.1074/jbc.M100534200. [DOI] [PubMed] [Google Scholar]
- 10.Georgiadis KE, Hirohata S, Seldin MF, Apte SS. ADAM-TS8, a novel metalloprotease of the ADAM-TS family located on mouse chromosome 9 and human chromosome 11. Genomics. 1999;62:312–5. doi: 10.1006/geno.1999.6014. [DOI] [PubMed] [Google Scholar]
- 11.Clark ME, Kelner GS, Turbeville LA, Boyer A, Arden KC, Maki RA. ADAMTS9, a novel member of the ADAM-TS/ metallospondin gene family. Genomics. 2000;67:343–50. doi: 10.1006/geno.2000.6246. [DOI] [PubMed] [Google Scholar]
- 12.Robker RL, Russell DL, Espey LL, Lydon JP, O'Malley BW, Richards JS. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc Natl Acad Sci U S A. 2000;97:4689–94. doi: 10.1073/pnas.080073497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuno K, Okada Y, Kawashima H, Nakamura H, Miyasaka M, Ohno H, Matsushima K. ADAMTS-1 cleaves a cartilage proteoglycan, aggrecan. FEBS Lett. 2000;478:241–5. doi: 10.1016/s0014-5793(00)01854-8. [DOI] [PubMed] [Google Scholar]
- 14.Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, Link JR, Ellis DM, Tortorella MD, Pratta MA, Hollis JM, Wynn R, Duke JL, George HJ, Hillman MC, Jr., Murphy K, Wiswall BH, Copeland RA, Decicco CP, Bruckner R, Nagase H, Itoh Y, Newton RC, Magolda RL, Trzaskos JM, Burn TC, et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J Biol Chem. 1999;274:23443–50. doi: 10.1074/jbc.274.33.23443. [DOI] [PubMed] [Google Scholar]
- 15.Somerville RP, Longpre JM, Jungers KA, Engle JM, Ross M, Evanko S, Wight TN, Leduc R, Apte SS. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J Biol Chem. 2003;278:9503–13. doi: 10.1074/jbc.M211009200. [DOI] [PubMed] [Google Scholar]
- 16.Jin H, Wang X, Ying J, Wong AH, Li H, Lee KY, Srivastava G, Chan AT, Yeo W, Ma BB, Putti TC, Lung ML, Shen ZY, Xu LY, Langford C, Tao Q. Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas. Oncogene, England. 2007:7490–8. doi: 10.1038/sj.onc.1210559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagase H, Brew K. Designing TIMP (tissue inhibitor of metalloproteinases) variants that are selective metalloproteinase inhibitors. Biochem Soc Symp. 2003:201–12. doi: 10.1042/bss0700201. [DOI] [PubMed] [Google Scholar]
- 18.Devel L, Czarny B, Beau F, Georgiadis D, Stura E, Dive V. Biochimie. 2010 Elsevier Masson SAS; France: 2010. Third generation of matrix metalloprotease inhibitors: Gain in selectivity by targeting the depth of the S1' cavity; pp. 1501–8. [DOI] [PubMed] [Google Scholar]
- 19.Surridge AK, Rodgers UR, Swingler TE, Davidson RK, Kevorkian L, Norton R, Waters JG, Goldring MB, Parker AE, Clark IM. Characterization and regulation of ADAMTS-16. Matrix Biol, Netherlands. 2009:416–24. doi: 10.1016/j.matbio.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao S, De Geyter C, Kossowska K, Zhang H. FSH stimulates the expression of the ADAMTS-16 protease in mature human ovarian follicles. Mol Hum Reprod. 2007;13:465–71. doi: 10.1093/molehr/gam037. [DOI] [PubMed] [Google Scholar]
- 21.Dunn JR, Reed JE, du Plessis DG, Shaw EJ, Reeves P, Gee AL, Warnke P, Walker C. Expression of ADAMTS-8, a secreted protease with antiangiogenic properties, is downregulated in brain tumours. Br J Cancer. 2006;94:1186–93. doi: 10.1038/sj.bjc.6603006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Nardi MA, Li YS, Zhang W, Pan R, Dang S, Yee H, Quartermain D, Jonas S, Karpatkin S. C-terminal ADAMTS-18 fragment induces oxidative platelet fragmentation, dissolves platelet aggregates, and protects against carotid artery occlusion and cerebral stroke. Blood, United States. 2009:6051–60. doi: 10.1182/blood-2008-07-170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocks N, Paulissen G, El Hour M, Quesada F, Crahay C, Gueders M, Foidart JM, Noel A, Cataldo D. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie. 2008;90:369–79. doi: 10.1016/j.biochi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Liu YJ, Xu Y, Yu Q. Full-length ADAMTS-1 and the ADAMTS-1 fragments display pro- and antimetastatic activity, respectively. Oncogene, England. 2006:2452–67. doi: 10.1038/sj.onc.1209287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao G, Plaas A, Thompson VP, Jin S, Zuo F, Sandy JD. ADAMTS4 (aggrecanase-1) activation on the cell surface involves C-terminal cleavage by glycosylphosphatidyl inositol- anchored membrane type 4-matrix metalloproteinase and binding of the activated proteinase to chondroitin sulfate and heparan sulfate on syndecan-1. J Biol Chem, United States. 2004:10042–51. doi: 10.1074/jbc.M312100200. [DOI] [PubMed] [Google Scholar]
- 26.Gao G, Westling J, Thompson VP, Howell TD, Gottschall PE, Sandy JD. Activation of the proteolytic activity of ADAMTS4 (aggrecanase-1) by C-terminal truncation. J Biol Chem, United States. 2002:11034–41. doi: 10.1074/jbc.M107443200. [DOI] [PubMed] [Google Scholar]
- 27.Cross AK, Haddock G, Stock CJ, Allan S, Surr J, Bunning RA, Buttle DJ, Woodroofe MN. ADAMTS-1 and -4 are up-regulated following transient middle cerebral artery occlusion in the rat and their expression is modulated by TNF in cultured astrocytes. Brain Res, Netherlands. 2006:19–30. doi: 10.1016/j.brainres.2006.02.136. [DOI] [PubMed] [Google Scholar]
- 28.Wang K, Vishwanath P, Eichler GS, Al-Sebaei MO, Edgar CM, Einhorn TA, Smith TF, Gerstenfeld LC. Analysis of fracture healing by large-scale transcriptional profile identified temporal relationships between metalloproteinase and ADAMTS mRNA expression. Matrix Biol. 2006;25:271–81. doi: 10.1016/j.matbio.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–9. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Nardi MA, Wu J, Pan R, Zhang W, Karpatkin S. Platelet fragmentation requires a specific structural conformation of human monoclonal antibody against beta3 integrin. J Biol Chem. 2008;283:3224–30. doi: 10.1074/jbc.M705902200. [DOI] [PubMed] [Google Scholar]
- 31.Parmley JL, Huynen MA. Clustering of codons with rare cognate tRNAs in human genes suggests an extra level of expression regulation. PLoS Genet. 2009;5:e1000548. doi: 10.1371/journal.pgen.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cal S, Obaya AJ, Llamazares M, Garabaya C, Quesada V, Lopez-Otin C. Cloning, expression analysis, and structural characterization of seven novel human ADAMTSs, a family of metalloproteinases with disintegrin and thrombospondin-1 domains. Gene, Netherlands. 2002:49–62. doi: 10.1016/s0378-1119(01)00861-7. [DOI] [PubMed] [Google Scholar]
- 33.Arza B, De Maeyer M, Felez J, Collen D, Lijnen HR. Critical role of glutamic acid 202 in the enzymatic activity of stromelysin-1 (MMP-3). Eur J Biochem. 2001;268:826–31. doi: 10.1046/j.1432-1327.2001.01943.x. [DOI] [PubMed] [Google Scholar]
- 34.Kang T, Park HI, Suh Y, Zhao YG, Tschesche H, Sang QX. Autolytic processing at Glu586-Ser587 within the cysteine-rich domain of human adamalysin 19/disintegrin metalloproteinase 19 is necessary for its proteolytic activity. J Biol Chem. 2002;277:48514–22. doi: 10.1074/jbc.M208961200. [DOI] [PubMed] [Google Scholar]
- 35.Vazeux G, Wang J, Corvol P, Llorens-Cortes C. Identification of glutamate residues essential for catalytic activity and zinc coordination in aminopeptidase A. J Biol Chem. 1996;271:9069–74. doi: 10.1074/jbc.271.15.9069. [DOI] [PubMed] [Google Scholar]
- 36.Windsor LJ, Bodden MK, Birkedal-Hansen B, Engler JA, Birkedal-Hansen H. Mutational analysis of residues in and around the active site of human fibroblast-type collagenase. J Biol Chem. 1994;269:26201–7. [PubMed] [Google Scholar]
- 37.Rodriguez-Manzaneque JC, Milchanowski AB, Dufour EK, Leduc R, Iruela-Arispe ML. Characterization of METH-1/ADAMTS1 processing reveals two distinct active forms. J Biol Chem. 2000;275:33471–9. doi: 10.1074/jbc.M002599200. [DOI] [PubMed] [Google Scholar]
- 38.Liu YJ, Xu Y, Yu Q. Full-length ADAMTS-1 and the ADAMTS-1 fragments display pro- and antimetastatic activity, respectively. Oncogene. 2006;25:2452–67. doi: 10.1038/sj.onc.1209287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei X, Prickett TD, Viloria CG, Molinolo A, Lin JC, Cardenas-Navia I, Cruz P, Rosenberg SA, Davies MA, Gershenwald JE, Lopez-Otin C, Samuels Y. Mutational and functional analysis reveals ADAMTS18 metalloproteinase as a novel driver in melanoma. Mol Cancer Res. 2010:1513–25. doi: 10.1158/1541-7786.MCR-10-0262. Aacr., United States, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Zhang W, Shao Y, Zhang C, Wu Q, Yang H, Wan X, Zhang J, Guan M, Wan J, Yu B. High-resolution melting analysis of ADAMTS18 methylation levels in gastric, colorectal and pancreatic cancers. Med Oncol. 2010;27:998–1004. doi: 10.1007/s12032-009-9323-8. [DOI] [PubMed] [Google Scholar]
- 41.Goldman E. Translation Control by RNA. In: Encyclopedia of Life Sciences (ELS) John Wiley&Sons,Ltd; Chichester: 2008. pp. 1–13. [Google Scholar]
- 42.Bevitt DJ, Li Z, Lindrop JL, Barker MD, Clarke MP, McKie N. Analysis of full length ADAMTS6 transcript reveals alternative splicing and a role for the 5' untranslated region in translational control. Gene. 2005;359:99–110. doi: 10.1016/j.gene.2005.06.011. [DOI] [PubMed] [Google Scholar]