Abstract
While it has been demonstrated that cytotoxic T lymphocytes (CTL) can be raised against tumor-associated self-antigens, achieving consistent and effective clinical responses has proven challenging. Superagonist altered peptide ligands (APL) can often elicit potent anti-tumor CTL responses where the native tumor-associated epitope fails. Current methods have identified a limited number of superagonist APLs, including the prototypic 27L mutant of MART-1. However, more comprehensive screening strategies would be desirable. In this study we utilize a novel genetic screen, involving recombinant technology and class I antigen cross-presentation, to search for supraoptimal superagonists of the 27L MART-1 mutant by surveying the effectiveness of virtually every single amino acid substitution mutant of 27L to activate human antigen-specific CTL clones recognizing the wild-type MART-126-35 epitope. We identify 3 novel mutant epitopes with superagonist properties that are functionally superior to 27L; however, the ability of a given analog to act as superagonist varies among patients and suggest that a given superagonist altered peptide ligand may be ideally suited to different patients. These findings endorse the use of comprehensive methods to establish panels of potential superagonist APLs to individualize tumor peptide vaccines among patients.
Keywords: Superagonists, Altered peptide ligands, Antigen cross-presentation, cytotoxic T cells
Introduction
It has been well established that cytotoxic T lymphocytes (1) have the potential to directly kill malignant cells, which express and display specific antigenic peptides in the context of specific class I MHC molecules (2, 3). These antigenic peptides, often referred to as CTL epitopes, are peptides of unique amino acid sequence, usually 9-11 amino acids in length. The tumor-associated antigenic peptide that is being targeted can be used as a peptide-based vaccine to promote the anti-tumor CTL response (4, 5). However, when the target peptide is derived from non-mutated differentiation antigens as is often the case (e.g. melanosomal proteins) , it can be insufficient to engender robust and sustained anti-tumor CTL responses (6, 7). This is a result of immune tolerance mechanisms that generally suppress or eliminate high avidity auto-reactive T cells (8). As a result of these mechanisms, the vast majority of tumor-specific CTL, specifically those that recognize non-mutated tumor-associated antigens, are eliminated in the thymus and in the periphery. What remains is a low frequency of tumor-specific CTL, and/or CTL that bear low avidity T cell receptors for the cognate tumor antigen (9-12).
It has been shown that one way to activate and mobilize these rare and low avidity tumor-specific CTL is with the use of superagonist altered peptide ligands (13, 14). These are mutant peptide ligands that deviate from the native peptide sequence by one or more amino acids, and which activate specific CTL clones more effectively than the native epitope. Generally, these alterations either allow the peptide to bind better to the restricting class I MHC molecule (13, 14) or interact more favorably with the TCR of a given tumor-specific CTL subset (1). Importantly, superagonist APLs have demonstrated favorable responses in clinical studies (15, 16).
To date, the study and utilization of APLs remains limited due to a lack of comprehensive methods for which to identify them. A common method to identify superagonist APLs involves comparing the amino acid sequence of the tumor-associated CTL epitope to the so-called consensus binding motif for the restricting class I MHC allotype (13, 14). Where the tumor-associated epitope deviates from the consensus sequence, the appropriate amino acids can be substituted, allowing the peptide to bind better to the class I MHC molecule. However, not all poorly stimulatory CTL epitopes deviate from the consensus motif, and thus render this approach less feasible. Some researchers take more random approaches. One of these involves substituting one or more specific amino acids into every position of the epitope; an example of this type of approach includes alanine scanning (17, 18). Another of these random approaches includes making every single amino acid substitution at one or two positions- positions either predicted to play a role in class I MHC secondary binding or to be directly involved in engaging the TCR (1, 19). While the above approaches have identified a number of superagonist APLs of clinically relevant antigens, they are severely limited in scope, and potentially overlook a large number of superagonist APLs.
In this study, we utilize a novel comprehensive genetic approach to identify superagonist APLs. Using saturation mutagenesis, we construct recombinant mutant peptide libraries that theoretically contain every single amino acid substitution of the cognate ligand. This approach takes advantage of the speed and simplicity of molecular biology, and exploits the unique ability of dendritic cells to cross-present extra-cellular antigens on class I MHC molecules.
Materials and Methods
Oligonucleotides, Peptids and DNA Sequence Analysis
The oligonucleotides utilized in this study were purchased from Operon Biotechnologies, Inc. They were designed to have a complimentary 5’ KpnI site and a complimentary 3’ PstI site. The sequences of the saturation mutagenesis sense strands of the MART-126-35 positional oligonucleotides were as follows (each sense strand has a corresponding mutant antisense strand):
NNN represents totally randomized codons. This can be likened to a slot machine with three positions and 4 possibilities at each position. Each pull of the machines lever should yield one of 64 possibilities. Thus, NNN represents any one of the sixty-four codons. In a given positional library consisting of 100 mutant oligonucleotide pairings, each codon has high likelihood of being represented.
Synthetic peptides were purchased from Sigma-Genosys @ >90% purity and re-suspended in DMSO (Sigma). Peptides were: ELAGIGILTV (A27L), GLAGIGILTV (E26G), SLAGIGILTV (E26S), ELAGIGIMTV (L33M).
DNA sequence analysis was carried out at ACGT, Inc.
Cloning, expression and purification of saturation mutagenesis constructs
Saturation mutagenesis oligonucleotides were cloned into the expression vector pQE40 (Qiagen). The plasmids were transformed into E. Coli (M15 pREP) and cultivated in Luria Broth. Expression of the mini-gene was induced using IPTG. Mini-gene products were expressed as fusion proteins containing 6x-histidine tags. Following recombinant protein induction, bacteria were lysed with 8M Urea, pH 8.0. Lysate was harvested and applied to Mg2+ coated paramagnetic beads (Talon beads, Dynal), which bind specifically to 6x-histidine.
For saturation mutagenesis libraries, bacterial clones were cultured individually in wells of 96-well plates.
Cell lines and Peripheral Blood Mononuclear Cells
Melanoma cell lines A375 and Mel 526, CTL clones and the TAP-deficient cell line T2 were maintained in RPMI 1640, containing 25mM HEPES, 2mM L-glutamine, 50U/ml penicillin, 50mg/ml streptomycin (Invitrogen Life Technologies) and 10% human serum from normal donors. Dendritic cells were prepared from adherent monocytes, isolated from the PBMC of HLA-A2+ healthy donors. IL-4 (500U/mL; R&D Systems, Minneapolis, MN) and GM-CSF (800U/mL; Amgen, Thousand Oaks, CA) were added to the monocytes to promote their differentiation into dendritic cells. MART-126-35-specific CTL clones were generated as described by Li et al (20). PBMC used in this study were obtained from HLA-A2+ melanoma patients.
Saturation Mutagenesis APL Screen
Following the isolation of the recombinant mini-gene APL products on Talon beads, the bead-bound products were “fed” to 1×105 immature dendritic cells in 96-well plates. Following a 4-hour incubation at 37°C, 1×105 of MART-126-35-specific CTL clones were added to DC/bead preparations. Following a 12-hour incubation at 37°C, the supernatant was harvested and assayed for the concentration of IFN-γ induced by the APL clones. Anti-IFN-γ antibodies (Endogen) used in the sandwich ELISA were used at 1μg/ml in PBS/0.1% BSA.
In Vitro PBMC stimulations with analog peptides and tetramer staining
On day 0, monocyte-derived dendritic cells were pulsed with 1 μM of each MART-126-35 analog peptide for 2 hours at 37°C. The DCs were washed and added to 5×106 HLA-A2+ PBMC from melanoma patients at a 1:20 ratio in 24-well plates. On day 2, 12.5 U/ml of IL-2, 5 ng/ml IL-7, 1 ng/ml IL-15, and 10 ng/ml of IL-21 were added to each culture. Cytokines were replenished every 2-3 days for 1-week. Following the 1-week primary stimulation, cultures were re-stimulated with 1×106 irradiated monocytes pulsed with 10 μM of the peptide used in the primary stimulation. IL-2, IL-7 and IL-15 were added to secondary stimulations on day 2. Cytokines were replenished every 2-3 days. 5×105 cells from each culture were stained with APC-labeled anti-CD8 antibody (Caltag Lab, Burlingame, CA) and PE-labeled MART-126-35 HLA-A2.1 tetramers (Immune Monitoring Lab, Fred Hutchinson Cancer Research Center, Seattle, WA). Stained cells were analyzed using FACScalibur flow cytometer and CellQuest (BD PharMingen) and analyzed using FlowJo software v8.5 (Tree Star, San Carlos, CA). Cells were stained with tetramers in 25 μl of 2% FCS/BSA for 1 hour at room temperature, followed by anti-CD8 antibody for 15 minutes at 4°C.
Generation of MART-126-35 polyclonal cell lines
Following in vitro peptide stimulation of PBMC from MelPt-B, MelPt-C, MelPt-D, MelPt-F and a healthy donor (Healthy-1) MART-126-35 tetramer and CD8 positive cells were sorted and isolated on BD FACSaria. Isolated cells were replicated using 30 ng/ml anti-CD3 antibody (OKT3) and IL-2 at 50 U/ml in the presence of irradiated feeder PBMC and LCL for 2 weeks. IL-2 was replenished every 2-3 days. Following the stimulation, cultures were stained for the generation of MART-126-35 tetramer and CD8 positive cell populations. The polyclonal cell lines were tested for lytic activity and TCR Vβ usage (MelPt-C only) (described below).
In vitro cytotoxicity assay
Target cells were labeled with 100 μCi of 51Cr and co-cultured with effector cells for 4 hours at 37°C plus 5% CO2. Targets were melanoma cell lines A375 (HLA-A2+/NY-ESO-1+) and Mel 526 (HLA-A2+/MART-1+), and T2 cells pulsed with 1 μM of MART-126-35 (positive control) or NY-ESO-1157-165 (negative control). Effector cells were MART-126-35-tetramer positive polyclonal cell lines generated with either A27L, E26S, or L33M peptides. Assays were performed in triplicate at a 50:1, 25:1 or 12.5:1 effector to target ratio. Released 51Cr was measured with a gamma scintillation counter and percent specific lysis was determined by using the formula: percent specific release = (experimental release-spontaneous release)/(maximum release-spontaneous release). Spontaneous release was <10% of the maximum release.
TCR Spectratype analysis
TCR Vβ spectratype analysis was carried out by the Immune Monitoring Laboratory at Fred Hutchinson Cancer Research Center. Briefly, cDNA was generated from 1×106 MART-126-35 tetramer staining polyclonal cell lines. Multiplex Vβ PCR primers were then used to amplify the variable regions of the complementarity-determining region 3 (CDR3) of the TCR β chain. Sequence analysis to determine the Vβ usage of the TCRs was conducted with GenScan.
Results
Mart-126-35 specific CTL clones can detect Enhanced CTL epitopes as reflected by IFN-γ expression
To identify superagonist APLs in this study, we utilize a novel genetic system. This system employs saturation mutagenesis of agonist peptide-encoding oligonucleotides, which when expressed in E. Coli will contain position specific single amino acid substitutions. The positional libraries are designed such that the codon of interest is totally randomized (NNN), resulting in a pool of oligonucleotides which contains every given codon sequence. This mutagenesis approach might be likened to a slot machine which contains three positions (a codon) and each position has the same 4 possibilities (A,C,G or T). When pulled, there is a 1 in 64 chance of getting any combination of 3. If pulled 100 times there is a high probability that every sequence will be represented (80% certainty, according to a Poisson distribution ). Here, the 100 pulls represent 100 bacterial colonies, each containing a different mutant agonist peptide-encoding oligonucleotide. When cloned and expressed, each amino acid should be represented in a library of 100, with 80% certainty, according to a Poisson distribution. A positional library can be generated for each position (amino acid) of the target peptide. The APL min-gene constructs are fused to a 6x-histidine tag, and can easily be separated from bacterial proteins on Co2+-coated paramagnetic beads (Dynal). APLs are screened for the ability to activate epitope-specific CTL clones following cross-presentation of the bead-bound ligand on class I MHC molecules by immature dendritic cells (DC).
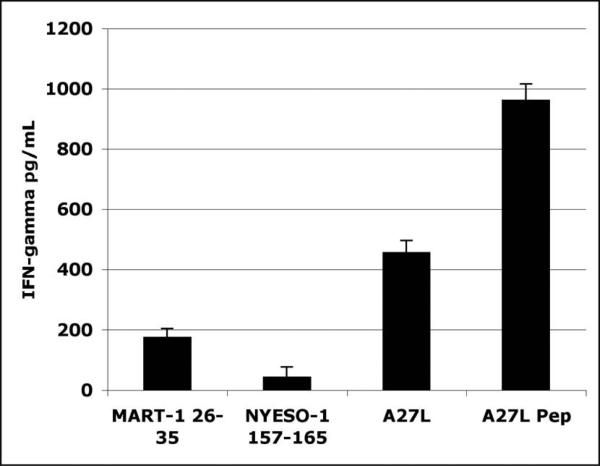
To validate this system and to verify that it was sensitive enough to detect our model tumor-associated HLA-A2 restricted antigenic peptide, MART-126-35, as well as an APL superagonist epitope of MART-126-35, called MART-126-35A27L (henceforward referred to as A27L), oligonucleotides encoding the appropriate peptide sequences were cloned, expressed and assayed for the ability to activate antigen specific CTL clones as described in materials and methods. The CTL clone used in this assay, called M26-H1, is specific for MART-126-35, and expresses IFN-γ in response to HLA-A2/ MART-126-35 complexes. Here, the IFN-γ response elicited by the recombinant unmodified MART-126-35 cross-presented construct is significantly higher than that elicited by the HLA-A2 restricted negative control, NYESO-1157-165 (Figure 1). Further, the IFN-γ response elicited by the recombinant superagonist APL, A27L, was more than 2-fold higher than that elicited by the recombinant wild type construct. Yet, the activation of M26-H1 by the unmodified MART-126-35 construct was clearly distinguishable from that elicited by the HLA-A2 restricted negative control construct, NYESO1157-165. These results suggest that the HLA-A2 cross-presented recombinant ligands are sufficient to elicit detectable antigen-specific responses from CTL clones, and also that superagonist APLs can be distinguished based on an increase in IFN-γ expression, relative to the wild type CTL ligand.
Figure 1. Native and superagonist CTL determinants can be distinguised in bead-based cross-presentation assay.
Oligonucleotides encoding MART-126-35, NY-ESO-1157-165, or MART-126-35A27L were cloned into and expressed by pQE40 expression vectors in 5ml bacterial cultures. The mini-gene products were isolated and “fed” to immature dendritic cells as described in materials and methods. MART-126-35-specific CTL clones were used to detect the presence of the cross-presented mini-gene products. Induced IFN-γ expression was determined by standard sandwich ELISA. A27L synthetic peptide at 1μM was used a positive control. Experiment was performed in triplicate.
Saturation Mutagenesis can effectively generate random amino acids in the parental antigenic peptide from which enhanced agonist APLs can be identified
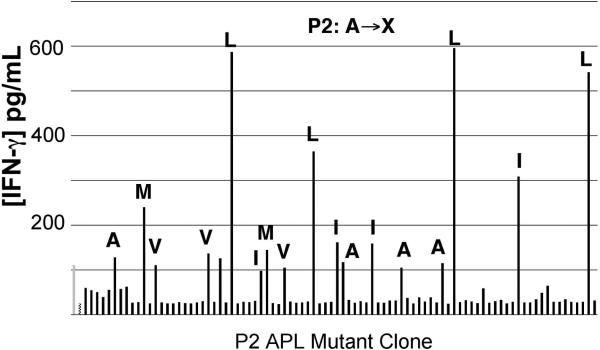
The saturation mutagenesis APL library screen depends on 200 μl bacterial expression cultures in 96-well plates. Figure 1 clearly shows that cross-presented recombinant ligands can be detected by antigen-specific CTL. However, in that experiment recombinant proteins were produced at high concentrations in 5 ml cultures. To determine whether the recombinant protein produced in these significantly smaller cultures would be sufficient to reflect detectable and varying degrees of activation, we constructed a position 2 (P2) library of MART-126-35 (EXAGIGILTV). By screening this library, in addition to determining if 200 μl cultures produce sufficient concentrations of recombinant protein we would be able to determine whether previously identified superagonist APLs, including A27L, could be identified from among 88 unique mutant APL clones. The P2 library screen (Figure 2), using the CTL clone M26-H1, clearly shows that the wild type recombinant ligand MART-126-35 (in green on the chart), elicits significantly more IFN-γ than the negative control (in red on the chart). Furthermore, the APL clones from the library that contained leucine residues at P2 (A27L), elicited significantly more IFN-γ expression in comparison to the wild type ligand. Amino acid content was determined from replicated glycerol stock of the P2 bacterial library. Interestingly, APL clones containing methionine residues at P2 also elicited greater IFN-γ expression than wild type MART-126-35, although not as great as that elicited by the leucine containing APLs, A27L. Like A27L, A27M has also been described as a superagonist APL of MART-126-35. Thus, 200 μl bacterial cultures produce sufficient concentrations of the recombinant ligands to be detected in this screen. Also, superagonist APLs can be identified in a library of at least 88 unique APL clones.
Figure 2. Previously described superagonists identified in MART-126-35 Position 2 saturation mutagenesis APL screen.
88 P2 saturation mutagenesis clones were screened as described in Materials and Methods. MART-126-35 control construct is depicted in light gray (first bar on left), while the NY-ESO-1157-165 negative control construct is depicted in checkered pattern (second bar from left). DNA sequence analysis was performed for APL clones eliciting comparable IFN-γ expression as the native construct. The amino acid is indicated above clones that were sequenced for codon content.
Putative enhanced CTL epitopes of Mart-126-35 A27L are identified in APL library screens
On the basis of previous experiments demonstrating that superagonist APLs can be uncovered using the saturation mutagenesis screen, remaining positional libraries of MART-126-35, (with the exception of P10, which already contains an anchor residue that conforms to the HLA-A2 C-terminal consensus binding motif) were screened using similar methods. Because a potent superagonist APL of MART-126-35 has already been identified in A27L, we chose to use A27L as the basis for our mutational strategy. That is, we chose to fix leucine in position 2, while mutating other positions independently. We reasoned that this would allow us to identify superagonist APLs more effective than A27L. The APL libraries were screened with two different high avidity MART-126-35-specific CTL clones (screening results are presented for M26-H1, only). A high avidity TCR is defined here as having the ability to recognize tumor cells that express both MART-1 and HLA-A2 class I molecules. The vast majority of the MART-126-35 derivative mutant peptide clones screened from each of the positional libraries were not as effective as A27L at activating the MART-126-35-specific CTL clone (Figure 3). However, several clones from the P1, P3 (not shown) and P8 libraries appeared to work similarly as well as the A27L recombinant construct. It should be noted that the initial screen was conducted by screening 2 unique APL library clones simultaneously in a single well. While this approach allows us to screen twice as many APL clones, the potency of any agonist APL in the pool is potentially underestimated in the initial screen. Agonist candidates were selected and re-screened based on their ability to elicit more or comparable levels of IFN-γ from M26-H1 in the initial screen (Figure 3b). When tested independently, both of the clones from the P3 libraries elicited less IFN-γ expression from the MART-126-35-specific CTL clone, relative to A27L (data not shown). When re-screened independently, it was apparent that only one of the two mutant peptide clones from the P1 and P8 wells was responsible for the increased IFN-γ expression. The DNA encoding these putative MART-126-35 agonist peptides was prepared from the duplicated bacterial glycerol stocks. We found that the enhancing mutations for the P1 putative agonists contained either glycine (E26G) or serine (E26S) residues at P1 instead of the naturally occurring glutamate residue. The P8 putative agonist contained a methionine residue (L33M) at position 8 rather than the naturally occurring leucine residue. No additional putative agonists were identified from the library screens using the second CTL clone, M26-H2 (data not shown).
Figure 3. 8 positional libraries of A27L were screened using the saturation mutagenesis technique.
88 mutant clones were screened for each of 8 positional libraries of A27L- P1, P3 (not shown), P4, P5 (not shown), P6, P7, P8 and P9- as described in materials and methods (3a). 2 clones are screened simultaneously for each library. Activation was assessed by IFN-γ expression. IFN-γ was measured by sandwich ELISA. Positive control (A27L) is illustrated in light gray (far left bar) while the negative control (NYESO-1157-165) is illustrated in checkered (second from left). APL clonal wells indicated with an arrow were de-convoluted and each mutant APL re-screened separately. The table in 3b shows the IFN-γ activity elicited by individual clones, relative to the activity elicited by A27L. The clones that were initially assayed together are indicated by shading. A bold number indicates the APL clone which is most responsible for the activation of the screening CTL clone. DNA sequence analysis was used to determine the amino acid encoded.
MART-126-35 Agonist Peptides Display a differential capacity to activate different MART-126-35-specific CTL Clones
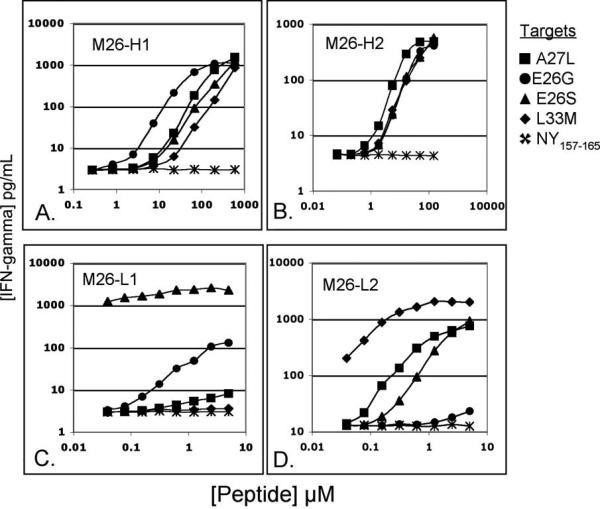
To analyze the putative superagonist APLs on a molar basis, individual peptides were synthesized at greater than 90% purity. To determine whether these APLs would be similarly recognized by unique MART-126-35-specific CTL clones, the APLs were tested against 4 clones bearing unique T cell receptors (TCR). These included 2 high avidity CTL clones (M26-H1 and M26-H2) and 2 low avidity CTL clones (M26-L1 and M26-L2) (Figure 4). We define low-avidity TCR as having the ability to respond HLA-A2 positive peptide-pulsed target cells but not to cells displaying naturally processed and presented determinants from HLA-A2/MART-1 positive tumors. It has been suggested that low-aviditiy T cells have the potential to mediate antigen-specific cell and tissue destruction (10,12).
Figure 4. APLs identified in saturation mutagenesis screen activate unique MART-126-35-specific CTL clones differently.
Two unique high avidity MART-126-35-specific CTL clones, M26-H1 (A) and M26-H2 (B), and two unique low avidity MART-126-35-specific CTL clones, M26-L1(C) and M26-L2 (D), were assayed against the agonist peptides A27L (square), E26G (circle), E26S (triangle), L33M (diamond) and NY-ESO-1157-165 (x). Peptides were titrated on T2 target cell. IFN-γ expression was measured by standard ELISA. Assay was performed in triplicate.
Figure 4a shows that each of the newly identified agonist peptides is similarly effective in activating M26-H1- the high-avidity CTL clone used in the initial screen (figure 3)- as compared to MART-126-35 superagonist peptide, A27L. A similar pattern of activation is found when the identified agonist peptides are used to stimulate the CTL clone M26-H2. In contrast to the above results, the low-avidity MART-126-35-specific CTL clones yielded widely divergent results in response to different agonist peptides. For example, while the CTL clone M26-L1 recognizes the peptide E26S more than 100-fold better than A27L (based on half-maximal activation), the CTL clone M26-L2 recognizes A27L better than it does E26S. Similarly, while L33M is scarcely recognized by the CTL clone M26-L1, it is the most effective agonist for activating M26-L2. Thus, these analogs might be considered “conditional” agonists, as they do not elicit generalized patterns of activation among unique antigen-specific clonotypes.
MART-126-35 APLs demonstrate patient specific enhanced generation of MART-126-35 CTL populations from the PBMC of melanoma patient donors
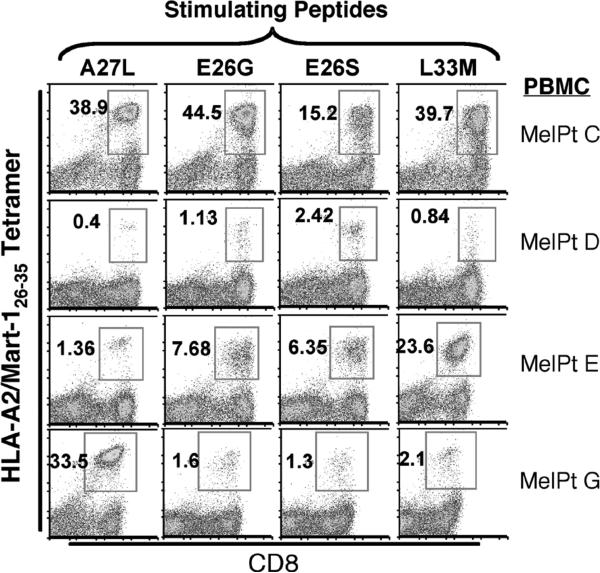
To determine how well the identified agonist APLs could prospectively generate MART-126-35-specific CTL populations from melanoma patient peripheral blood mononuclear cell (PBMC) preparations, the APLs were used to stimulate 8 different patient PBMC samples under standard in vitro conditions (Table 1). One week following the second in vitro stimulation, cultures were stained with the wild-type MART-126-35/HLA-A2 tetramer. Similar to the observations made using different MART-126-35-specific CTL clones, none of the peptide ligands were universally effective in generating MART-126-35-specific CTL populations from all patient PBMC samples (Figure 6). Any given APL was more or less effective in generating antigen-specific CTL from any given patient PBMC sample. For example, while the agonist peptide E26S is the least effective at generating MART-126-35-specific CD8 positive populations from the PBMC of MelPt-C (3-fold < A27L), it is the most effective APL for generating such T cell populations from MelPt-D (5-fold > A27L). Similarly, whereas the agonist peptide L33M is 14-fold more effective than A27L in generating of MART-126-35-specific CD8 positive populations from the PBMC of MelPt-E, it is 14-fold less effective than A27L in generating MART-126-35-specific CD8 populations from the PBMC of MelPt-G. These findings demonstrate that any one CTL ligand may or may not be effective at generating antigen-specific CTL populations from the PBMC of any given patient; and suggest the importance of establishing a panel of potential superagonist APLs.
Table 1.
| MART-126-35 Library | Sense strand of MART-126-35 positional Saturation Mutagenesis Oligonucleotides |
|---|---|
| P1 | CATCGAGGGAAGGNNNCTCGCCGGAATCGGCATTCTGACCGTTTAATGAATTCTGCA |
| P2 | CATCGAGGGAAGGGAGNNNGCCGGAATCGGCATTCTGACCGTTTAATGAATTCTGCA |
| P3 | CATCGAGGGAAGGCAGCTCNNNGGAATCGGCATTCTGACCGTTTAATGAATTCTGCA |
| P4 | CATCGAGGGAAGGCAGCTCGCCNNNATCGGCATTCTGACCGTTTAATGAATTCTGCA |
| P5 | CATCGAGGGAAGGCAGCTCGCCGGANNNGGCATTCTGACCGTTTAATGAATTCTGCA |
| P6 | CATCGAGGGAAGGCAGCTCGCCGGAATCNNNATTCTGACCGTTTAATGAATTCTGCA |
| P7 | CATCGAGGGAAGGCAGCTCGCCGGAATCGGCNNNCTGACCGTTTAATGAATTCTGCA |
| P8 | CATCGAGGGAAGGCAGCTCGCCGGAATCGGCATTNNNACCGTTTAATGAATTCTGCA |
| P9 | CATCGAGGGAAGGCAGCTCGCCGGAATCGGCATTCTGNNNGTTTAATGAATTCTGCA |
*N represents any one of the 4 chemical bases.
CD8 positive MART-126-35-specific polyclonal cell lines generated with the identified MART-126-35 Agonist APLs can kill HLA-A2+ tumors expressing endogenous MART-1
In recent vaccination studies it has been demonstrated that the use of altered peptide ligands poses the risk of generating antigen-specific T cells which display relatively low anti-tumor functional avidity (7, 28). To determine whether the MART-126-35-specific CTL that were generated with these novel MART-126-35 agonist peptides were of sufficient functional avidity to kill HLA-A2/MART-1 positive tumor targets, polyclonal lines of CD8 positive MART-126-35 tetramer-staining cells were established from the PBMC of MelPt-B, MelPt-C, MelPt-D, MelPt-F or a healthy donor (Healthy1), stimulated with either A27L, E26S or L33M agonist peptides. These cell lines were screened for reactivity to unmodified MART-126-35 peptide pulsed HLA-A2 positive targets and to HLA-A2/MART-1 positive tumor targets at varying effector to target ratios in a standard chromium release assay (Table II). The results illustrate that the CTL populations that were generated from each PBMC source with either of the altered peptide ligands can kill targets that display wild-type MART-126-35 in the context of HLA-A2, and recognize the epitope with sufficient affinity to kill tumors expressing MART-1.
Table II.
Identified MART-126-35 APLs exhibit differential capacities to generate MART-126-35-specific CTL populations from the PBMC of different melanoma patient donors.
| %M26 Tetramer Positive | ||||
|---|---|---|---|---|
| Patient | A27L | E26G | E26S | L33M |
| MelPt-A | 3.14 (1) | 1.68 (0.53) | 3.36 (1.07) | 0.98 (0.31) |
| MelPt-B | 2.97 (1) | 1.31 (0.44) | 4.3 (1.45) | 7.7 (2.6) |
| MelPt-C | 40.6 (1) | 45.6 (1.12) | 15.6 (0.38) | 41.1 (1.02) |
| MelPt-D | 0.65 (1) | 1.73 (2.66) | 3.43 (5.27) | 2.07 (3.1) |
| MelPt-E | 1.77 (1) | 8.42 (4.75) | 6.88 (3.88) | 24.2 (13.67) |
| MelPt-F | 5.45 (1) | 3.35 (0.61) | 3.72 (0.68) | 3.07 (0.56) |
| MelPt-G | 33.4 (1) | 1.89 (.06) | 1.75 (.05) | 2.37 (.07) |
| MelPt-H | 1.24 (1) | 2.03 (1.63) | 1.31 (1.06) | 2.77 (2.2) |
APLs were used to stimulate PBMC cultures in vitro. Following a 1-week secondary stimulation cells were stained with FITC-labeled anti-CD8 antibodies and APC-labeled HLA-A2/ MART-126-35 tetramers and analyzed by flow cytometry. Percent tetramer positive indicated. Fold difference relative to A27L is indicated in parentheses. Differences of more than two-fold are indicated in bold.
To determine whether unique or shared MART-126-35-specific CTL clonotypes were generated with each of the peptide ligands (A27L, E26S and L33M), spectratype analysis was performed on CTL lines derived from MelPt-C PBMC to determine their Vβ TCR usage. We found that the agonist peptides A27L, E26S and L33M generated CTL populations that primarily (>90%) utilized TCR Vβ24, Vβ8 and Vβ3, respectively. This suggests that the different analog peptides preferentially generate specific TCR utilizing CTL subsets. Taken together, these results demonstrate the ability of the identified APLs to elicit MART-126-35-specific CTL responses that are capable of directly killing MART-1 expressing tumors, and suggest that unique MART-126-35-specific TCR subpopulations are being preferentially generated by the different MART-126-35 analog peptides.
Discussion
In this report we utilize a novel genetic technique to screen for potential superagonist APLs of a clinically relevant tumor-associated antigen, MART-1. Rather than screening a limited subset of possible agonists, this technique allowed us to screen virtually every single amino acid mutant of MART-126-35 in a rapid and cost-effective manner. A comprehensive approach to identifying APLs is perhaps the most ideal, given the tremendous ramifications that even subtle amino acid substitutions can induce on specific CTL responses (21-23). Borbulevych et al. has demonstrated somewhat of disconnect between the overall structure of a given CTL epitope and the CTL response that it elicits (21). In a 2007 study, the authors demonstrated that structurally dissimilar analog peptides of MART-127-35 could more effectively induce MART-127-35-specific CTL responses than analog peptides of MART-127-35 with very similar HLA-A2 associated conformations. This presents a challenge for the ability to predict superagonist APLs, and highlights the necessity of utilizing thoroughly comprehensive screening techniques.
Other approaches include the use of a positional scanning synthetic combinatorial library (PS-SCL) which represents a powerful technique involves a totally comprehensive method for screening peptide libraries, and allows for the identification of undefined CTL epitopes as well as superagonist ligands (24, 25). In contrast to the genetic screen described here, which identifies single amino acid substitution APLs, PS-SCL screens peptides that are randomized at every position except one. While PS-SCL scanning has been shown capable of identifying superagonist analogs (25, 26), the need to deduce the sequence of the putative superagonists, and the subsequent testing of large numbers of potentially nonproductive altered peptide ligands which are non-cross-reactive for the native eptiope, present additional hurdles.
In this study, from a screen of 9 positional APL libraries we identify three novel agonists of MART-126-35. Because our goal was to identify APLs of MART-126-35 more effective than the most effective APL described to date, MART-126-35 A27L, we chose to use A27L as the basis of our screen, and fixed leucine into the second position (P2) of all APLs screened. Two of the identified agonist peptides contained P1 substitutions of glutamate for glycine and serine. Previous studies have demonstrated that glutamate at P1 of MART-126-35 is not ideal for the most effective binding of the peptide to HLA-A2 (14, 18). In 1998 Valmori et al. demonstrated that substituting an alanine residue for the glutamate residue at P1 of MART-126-35 resulted in significantly higher affinity of the peptide for HLA-A2. Importantly, as compared to the native sequence, the alanine-containing agonist peptide was recognized significantly better by MART-126-35-specific CTL clones. The same group demonstrated that a substitution of glutamate at P1 of MART-126-35 with tyrosine or phenylalanine residues also increased both the binding of these agonists to HLA-A2 and their recognition by MART-126-35-specific CTL clones. Given these findings, it is not surprising that we would identify enhancing mutations at P1. However, in the studies just described, it was not made clear what effect the P1 mutants would have when leucine simultaneously occupies the N-terminal anchor position.
In this report we identified an agonist of MART-126-35 which contains a methionine substitution at position P8 for leucine (L33M). Preliminary data suggests that this agonist binds better to HLA-A2 relative to A27L, but like the other agonists, is not recognized by all of the MART-126-35-specific CTL clones used in this study. The crystal structures of MART-126-35 A27L generated by Sliz et al. and Borbulevych et al. (21, 27) do not suggest that L33 is closely associated with binding to HLA-A2.
The finding that different agonist peptides could be recognized by different CTL clones suggested to us that any given agonist APL may be more or less effective for different patients. We confirmed this hypothesis by performing in vitro stimulations with the identified agonist peptides on the PBMC of multiple melanoma patients. We found that while a given agonist APL might have a high stimulatory capacity for one patient it could be relatively ineffective for another patient. This would suggest that different clones are being mobilized with different agonist peptides. Indeed we went on to show that, at least for one patient, unique MART-126-35-specific CTL clones are being generated with different agonist peptides. Importantly, all of these clones were of sufficient affinity to recognize endogenously processed and presented epitopes of MART-1 and kill MART-1+, HLA-A2+ tumors. This is not the first demonstration that different agonists can activate different antigen-specific CTL populations. Recently, Hou et al. demonstrated that the a superagonist epitope of carcinoembryonic antigen (CEA), called Cap1-6D, generated unique CEA-specific CTL clones in comparison to Cap1, the native epitope (7). In that report, however, the superagonist proved to generate CTL of lower “functional avidity,” which were significantly less effective at killing CEA positive tumor targets. Similarly, in a recent vaccination study by Speiser et al. (28) utilizing unmodified MART-126-35 or the superagonist MART-126-35A27L in conjuction with the potent adjuvant CpG, the authors found that while the superagonist MART-126-35A27L routinely generated a higher percentage of antigen-specific CTL, the CTL that were generated displayed considerably lower anti-tumor functional avidity than the antigen-specific CTL which were generated using the unmodified antigenic peptide. In 1998 Rivoltini et al. (16) demonstrated that a superagonist of MART-127-35, called 1L, could generate unique MART-1-specific CTL clonotypes relative to the native peptide. In that study the authors showed that 1L could more effectively elicit MART-1-specific CTL responses from the PBMC of 5 different melanoma patients.
Our attempts to directly compare the functional avidity of MART-126-35-specific CTL generated via unmodified MART-126-35 peptide with CTL generated via the analogs described here, have been hampered by the inability to generate CD8+/ MART-126-35-tetramer positive T cell populations using the unmodified peptide (data not shown).While we do not directly address the functional avidity of the CTL generated in this study via agonist peptides relative to those generated with the unmodified peptide, we show that these newly identified agonist MART-126-35 peptides elicit different antigen-specific CTL responses from patient to patient, and that the generated CTL populations are capable of effectively killing tumors. Thus these agonist APLs might be considered “conditional” superagonist ligands. It is probable that by using several unique MART-126-35-specific CTL clones in the initial screen that we can identify other potential superagonist peptides. Given that naïve MART-1 specific CTL populations are uniquely abundant and highly diverse in healthy individuals, we will plan to determine how well our findings translate to other antigens which generally have significantly lower specific precursor CTL populations.
We believe the findings of this study suggest the promise of identifying panels of potential tumor-associated superagonist peptides, any of which may be most effective at generating potent anti-tumor CTL responses from a given patient. Further, we suggest that the novel genetic APL screen described here is a rapid, robust and inexpensive approach for identifying superagonist APLs.
Figure 5. APLs generate different CTL responses from the PBMC of different melanoma patients.
Identified APLs were used to stimulate the PBMC of different melanoma patients in vitro. Following a 1-week primary and 1-week secondary peptide stimulation, cultures were stained with FITC-labeled anti-CD8 antibody and APC-labeled HLA-A2/MART-126-35 tetramer and analyzed by flow cytometry. Data is representative of at least 3 different experiments.
Table III.
MART-126-35 and tumor specific lysis by CTL generated with MART-126-35 peptide analogs
| Percentage Specific Lysis from polyclonal CTL lines generated with the indicated peptide | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MART-126-35A27L | MART-126-35E26S | MART-126-35L33M | |||||||||||
| Patient | E/T | T2a | T2+M26b | A375 | Mel526 | T2 | T2+M26 | A375 | Mel526 | T2 | T2+M26 | A375 | Mel526 |
| MelPt-B | 50 25 12.5 |
* | 0c 0 2 |
40 31 21 |
6 4 8 |
58 45 30 |
2 2 4 |
63 52 41 |
13 12 9 |
80 70 58 |
|||
| MelPt-C | 50 25 12.5 |
9 10 9 |
73 52 42 |
3 5 2 |
35 30 25 |
2 7 1 |
87 66 54 |
8 5 2 |
41 32 25 |
9 12 11 |
91 84 76 |
0 2 5 |
58 52 45 |
| MelPt-D | 50 25 12.5 |
0 2 5 |
50 42 35 |
10 10 9 |
32 30 25 |
3 8 1 |
87 65 54 |
8 7 6 |
41 32 25 |
14 11 8 |
90 84 75 |
2 4 6 |
57 54 45 |
| MelPt-F | 50 25 12.5 |
6 3 2 |
92 81 73 |
5 5 5 |
65 50 43 |
23 21 22 |
70 72 65 |
2 3 5 |
44 42 38 |
||||
| Helthy1 | 50 25 12.5 |
2 4 1 |
52 42 31 |
3 7 6 |
58 42 35 |
0 0 0 |
34 26 18 |
9 5 8 |
28 15 10 |
2 6 5 |
57 49 38 |
13 13 10 |
65 57 44 |
Where values are not shown, polyclonal cell lines were not established for that condition.
T2 is a TAP-deficient cell line that expresses peptide-unbound HLA-A2 molecules unless pulsed extracellularly. Here T2, has been pulsed with NYESO-1157-165 unless indicated otherwise.
M26 is an abbreviation for the unmodified MART-126-35 peptide.
Numbers represent the percentage specific lysis obtained from each target. T375 is a HLA-A2 positive/MART-1 negative
Acknowledgments
The genetic approach utilized in this study to identify altered peptide ligands of previously identified T cell determinants was initially developed in the laboratory of Professor John G. Frelinger of the University of Rochester, School of Medicine and Dentistry. The approach was adapted here to be used with human dendritic cells and human T cell lines. We thank Dr. Frelinger for his support.
This work was supported by a grant from the National Institutes of Health and the National Cancer Institute (grant number 3 P50 CA083636)
Abbreviations used in this paper
- APL
altered peptide ligand
- CTL
cytotoxic T lymphocytes
- DC
dendritic cell
- PS-SCL
positional scanning synthetic combinatorial libraries
- CEA
carcinoembryonic antigen
REFERENCES
- 1.Zaremba S, Barzaga E, Zhu M, Soares N, Tsang KY, Schlom J. Identification of an enhancer agonist cytotoxic T lymphocyte peptide from human carcinoembryonic antigen. Cancer Res. 1997;57:4570–4577. [PubMed] [Google Scholar]
- 2.Boon T, van der Bruggen P. Human tumor antigens recognized by T lymphocytes. J Exp Med. 1996;183:725–729. doi: 10.1084/jem.183.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knuth A, Wolfel T, Klehmann E, Boon T, Meyer zum Buschenfelde KH. Cytolytic T-cell clones against an autologous human melanoma: specificity study and definition of three antigens by immunoselection. Proc Natl Acad Sci U S A. 1989;86:2804–2808. doi: 10.1073/pnas.86.8.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinoza-Delgado I. Cancer vaccines. Oncologist. 2002;7(Suppl 3):20–33. doi: 10.1634/theoncologist.7-suppl_3-20. [DOI] [PubMed] [Google Scholar]
- 5.Overwijk WW, Restifo NP. Creating therapeutic cancer vaccines: notes from the battlefield. Trends Immunol. 2001;22:5–7. doi: 10.1016/s1471-4906(00)01793-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Hou Y, Kavanagh B, Fong L. Distinct CD8+ T Cell Repertoires Primed with Agonist and Native Peptides Derived from a Tumor-Associated Antigen. J Immunol. 2008;180:1526–1534. doi: 10.4049/jimmunol.180.3.1526. [DOI] [PubMed] [Google Scholar]
- 8.Kazansky DB. Intrathymic selection: new insight into tumor immunology. Adv Exp Med Biol. 2007;601:133–144. doi: 10.1007/978-0-387-72005-0_14. [DOI] [PubMed] [Google Scholar]
- 9.McMahan RH, Slansky JE. Mobilizing the low-avidity T cell repertoire to kill tumors. Semin Cancer Biol. 2007;17:317–329. doi: 10.1016/j.semcancer.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan DJ, Kreuwel HT, Fleck S, Levitsky HI, Pardoll DM, Sherman LA. Activation of low avidity CTL specific for a self epitope results in tumor rejection but not autoimmunity. J Immunol. 1998;160:643–651. [PubMed] [Google Scholar]
- 11.Ohashi PS, Oehen S, Buerki K, Pircher H, Ohashi CT, Odermatt B, Malissen B, Zinkernagel RM, Hengartner H. Ablation of “tolerance” and induction of diabetes by virus infection in viral antigen transgenic mice. Cell. 1991;65:305–317. doi: 10.1016/0092-8674(91)90164-t. [DOI] [PubMed] [Google Scholar]
- 12.Zehn D, Bevan MJ. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity. 2006;25:261–270. doi: 10.1016/j.immuni.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JL, Dunbar PR, Gileadi U, Jager E, Gnjatic S, Nagata Y, Stockert E, Panicali DL, Chen YT, Knuth A, Old LJ, Cerundolo V. Identification of NY-ESO-1 peptide analogues capable of improved stimulation of tumor-reactive CTL. J Immunol. 2000;165:948–955. doi: 10.4049/jimmunol.165.2.948. [DOI] [PubMed] [Google Scholar]
- 14.Valmori D, Fonteneau JF, Lizana CM, Gervois N, Lienard D, Rimoldi D, Jongeneel V, Jotereau F, Cerottini JC, Romero P. Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J Immunol. 1998;160:1750–1758. [PubMed] [Google Scholar]
- 15.Fong L, Hou Y, Rivas A, Benike C, Yuen A, Fisher GA, Davis MM, Engleman EG. Altered peptide ligand vaccination with Flt3 ligand expanded dendritic cells for tumor immunotherapy. Proc Natl Acad Sci U S A. 2001;98:8809–8814. doi: 10.1073/pnas.141226398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivoltini L, Kawakami Y, Sakaguchi K, Southwood S, Sette A, Robbins PF, Marincola FM, Salgaller ML, Yannelli JR, Appella E, et al. Induction of tumor-reactive CTL from peripheral blood and tumor-infiltrating lymphocytes of melanoma patients by in vitro stimulation with an immunodominant peptide of the human melanoma antigen MART-1. J Immunol. 1995;154:2257–2265. [PubMed] [Google Scholar]
- 17.Singh RA, Zang YC, Shrivastava A, Hong J, Wang GT, Li S, Tejada-Simon MV, Kozovska M, Rivera VM, Zhang JZ. Th1 and Th2 deviation of myelin-autoreactive T cells by altered peptide ligands is associated with reciprocal regulation of Lck, Fyn, and ZAP-70. J Immunol. 1999;163:6393–6402. [PubMed] [Google Scholar]
- 18.Valmori D, Gervois N, Rimoldi D, Fonteneau JF, Bonelo A, Lienard D, Rivoltini L, Jotereau F, Cerottini JC, Romero P. Diversity of the fine specificity displayed by HLA-A*0201-restricted CTL specific for the immunodominant Melan-A/MART-1 antigenic peptide. J Immunol. 1998;161:6956–6962. [PubMed] [Google Scholar]
- 19.McCue D, Ryan KR, Wraith DC, Anderton SM. Activation thresholds determine susceptibility to peptide-induced tolerance in a heterogeneous myelin-reactive T cell repertoire. J Neuroimmunol. 2004;156:96–106. doi: 10.1016/j.jneuroim.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Bleakley M, Yee C. IL-21 influences the frequency, phenotype, and affinity of the antigen-specific CD8 T cell response. J Immunol. 2005;175:2261–2269. doi: 10.4049/jimmunol.175.4.2261. [DOI] [PubMed] [Google Scholar]
- 21.Borbulevych OY, Insaidoo FK, Baxter TK, Powell DJ, Jr., Johnson LA, Restifo NP, Baker BM. Structures of MART-126/27-35 Peptide/HLA-A2 complexes reveal a remarkable disconnect between antigen structural homology and T cell recognition. J Mol Biol. 2007;372:1123–1136. doi: 10.1016/j.jmb.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding YH, Baker BM, Garboczi DN, Biddison WE, Wiley DC. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999;11:45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- 23.Kersh GJ, Miley MJ, Nelson CA, Grakoui A, Horvath S, Donermeyer DL, Kappler J, Allen PM, Fremont DH. Structural and functional consequences of altering a peptide MHC anchor residue. J Immunol. 2001;166:3345–3354. doi: 10.4049/jimmunol.166.5.3345. [DOI] [PubMed] [Google Scholar]
- 24.Nino-Vasquez JJ, Allicotti G, Borras E, Wilson DB, Valmori D, Simon R, Martin R, Pinilla C. A powerful combination: the use of positional scanning libraries and biometrical analysis to identify cross-reactive T cell epitopes. Mol Immunol. 2004;40:1063–1074. doi: 10.1016/j.molimm.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Rubio-Godoy V, Pinilla C, Dutoit V, Borras E, Simon R, Zhao Y, Cerottini JC, Romero P, Houghten R, Valmori D. Toward synthetic combinatorial peptide libraries in positional scanning format (PS-SCL)-based identification of CD8+ Tumor-reactive T-Cell Ligands: a comparative analysis of PS-SCL recognition by a single tumor-reactive CD8+ cytolytic T-lymphocyte clone. Cancer Res. 2002;62:2058–2063. [PubMed] [Google Scholar]
- 26.Venturini S, Allicotti G, Zhao Y, Simon R, Burton DR, Pinilla C, Poignard P. Identification of peptides from human pathogens able to cross-activate an HIV-1-gag-specific CD4+ T cell clone. Eur J Immunol. 2006;36:27–36. doi: 10.1002/eji.200425767. [DOI] [PubMed] [Google Scholar]
- 27.Sliz P, Michielin O, Cerottini JC, Luescher I, Romero P, Karplus M, Wiley DC. Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes. J Immunol. 2001;167:3276–3284. doi: 10.4049/jimmunol.167.6.3276. [DOI] [PubMed] [Google Scholar]
- 28.Speiser S, Baumgaertner P, Voelter V, Devevre E, Barbey C, Rufer N, Romero P. Unmodified self antigen triggers human CD8 T cells with stronger tumor reactivity than altered antigen. Proc Natl Acad Sci U S A. 2008;105:3849–3854. doi: 10.1073/pnas.0800080105. [DOI] [PMC free article] [PubMed] [Google Scholar]