Abstract
An algorithmically-derived abstraction network, called the partial-area taxonomy, for a SNOMED hierarchy has led to the identification of concepts considered complex. The designation “complex” is arrived at automatically on the basis of structural analyses of overlap among the constituent concept groups of the partial-area taxonomy. Such complex concepts, called overlapping concepts, constitute a tangled portion of a hierarchy and can be obstacles to users trying to gain an understanding of the hierarchy’s content. A new methodology for partitioning the entire collection of overlapping concepts into singly-rooted groups, that are more manageable to work with and comprehend, is presented. Different kinds of overlapping concepts with varying degrees of complexity are identified. This leads to an abstract model of the overlapping concepts called the disjoint partial-area taxonomy, which serves as a vehicle for enhanced, high-level display. The methodology is demonstrated with an application to SNOMED’s Specimen hierarchy. Overall, the resulting disjoint partial-area taxonomy offers a refined view of the hierarchy’s structural organization and conceptual content that can aid users, such as maintenance personnel, working with SNOMED. The utility of the disjoint partial-area taxonomy as the basis for a SNOMED auditing regimen is presented in a companion paper.
Keywords: SNOMED, Terminology, Partitioning, Abstraction network, Modeling, Taxonomy
1. Introduction
SNOMED CT [1] has proven to be an important resource to the healthcare and biomedical community since its origination in 2002. However, its expanding size (295,000 concepts designated as “current” in the July 2011 release) and inherent complexity may hinder its usability and further deployment. Advanced tools for the display of aspects of SNOMED’s conceptual content—facilitating orientation and comprehension—are needed.
In previous work, we have devised high-level abstraction networks based on analyses of a SNOMED hierarchy’s attribute relationships and their patterns of inheritance [2]. A hierarchy’s concepts were partitioned into groups, called areas, according to their specific attribute relationships. From this partition, an abstraction network, referred to as the area taxonomy, affording a summary view of the distribution of the attribute relationships was constructed. Further refinement of areas led to another abstraction network, the partial-area taxonomy, which conveyed information about sub-area hierarchical arrangements. In addition to their support for orientation to and comprehension of a SNOMED hierarchy, the two networks have served as the bases of our formulation of structural methodologies for auditing SNOMED hierarchies [2]. Importantly, we found that many concept errors manifested themselves as structural anomalies at the taxonomy level, and thus the taxonomies proved to be effective building blocks for automated auditing regimens.
In this work, we further extend the taxonomy paradigm to overcome some deficiencies in the framework in dealing with particularly complex portions of a SNOMED hierarchy. A recurring theme of our previous terminological analyses has been that complex concepts—characterized by various structural features—are often obstacles to orientation and comprehension efforts and usually are natural places to look for modeling errors. Of course, there are numerous ways, in different contexts, to qualify the notion of “complex.” We have typically used the idea that concepts are complex when they simultaneously belong to multiple groups along some given categorizing dimension. In the context of SNOMED auditing, concepts appearing in regions of the partial-area taxonomy characterized by the convergence of multiple ancestral inheritance paths were deemed to be complex and given auditing priority [3].
In this paper, we focus on another variety of complex concepts, where again a structural feature (relatively easily computed) is being used to determine “complex.” In this case, the structural feature is set overlap, and the concepts are those that reside in overlapping portions of two or more sub-area groupings called partial-areas. As it happens, the entire collection of these overlapping concepts may constitute a highly tangled subhierarchy. We wish to impose some order on such a subhierarchy to facilitate orientation and comprehension for various users. In particular, we present an automated methodology for partitioning the entire set of overlapping concepts to form what we refer to as a disjoint partial-area taxonomy, an abstraction network that captures the prevailing hierarchical configuration of the overlaps. Through this taxonomy, the user is presented with a view showing the gestalt of the overlaps, allowing for easier comprehension of their content.
One class of user, in particular, that can benefit from the refined, high-level display offered by the new abstraction network is the domain-expert auditor. In the current paper, we present the details of the abstraction network and its derivation. An enhanced auditing regimen based on this network and the overlapping concepts is expounded in a companion paper [4].
2. Background
2.1. SNOMED CT
SNOMED CT (Systematized Nomenclature of Medicine – Clinical Terms) [1], formed through the merger of SNOMED RT (Reference Terminology) and the UK’s CTV3 (Clinical Terms Version 3), is a description-logic-based (DL-based) medical terminology covering a wide range of clinical concepts, including diseases, clinical findings, procedures, specimens, substances, etc. In this paper, we are using the inferred (distributed) view of SNOMED CT as opposed to its stated view expressed in its native DL language [5].
Each SNOMED1 concept has a unique descriptive term, called its “fully specified name” (FSN), a preferred term, and, typically, a set of synonyms. Concepts are organized in 19 top-level, singly-rooted hierarchies to capture broad, clinically-related groupings, such as Clinical Finding, Substance, Body Structure, and so on. Concepts within one hierarchy are linked by IS-A (subsumption) relationships in such a way that each hierarchy forms a directed acyclic graph (DAG).
Concepts also have attribute relationships directed to other concepts. These attribute relationships (relationships, for short) serve in definitional capacities. For example, the concept Ear problem (in the Clinical Finding hierarchy) has the relationship finding site2 to the concept Ear structure (in the Body Structure hierarchy) specifying that Ear structure is the site of Ear problem. In SNOMED, each kind of relationship is defined to span from a source hierarchy to a target hierarchy (perhaps more than one). For example, five kinds of relationships, namely, specimen substance, specimen procedure, specimen source morphology, specimen source topography, and specimen source identity, are defined with the Specimen hierarchy as their source hierarchy.3 The respective target hierarchies of the first four are Substance, Procedure, Body Structure, and Body Structure. The relationship identity has four target hierarchies itself: Social Context, Physical Object, Qualifier Value, and Environment or Geographical Location.
2.2. Area and partial-area taxonomies
In previous work, we have carried out structural analyses of SNOMED hierarchies yielding two types of high-level abstraction networks: the area taxonomy and the partial-area taxonomy [2]. Each serves to capture the relationship distribution within a hierarchy from a high-level perspective. Both networks are derived based on the respective relationships exhibited by the concepts in the hierarchy. The latter network refines the former by including additional hierarchical grouping knowledge. In the following, we present the important details pertaining to these two networks.
The basis of the area taxonomy is a partition of the concepts into what we call areas according to their sets of (non-hierarchical) relationships. An area is defined as the complete set of concepts having exactly the same given set of relationships, irrespective of the targets of those relationships for the particular concepts. Formally, let {r1, r2, …, rn} be a set of relationships. The area defined with respect to this set of relationships is as follows, where relshps(C) is the entire set of relationships exhibited by the concept C:
| (1) |
Given the fact that an area is defined by its set of relationships, we denote it as “{r1, r2, …, rn}.” If a given combination of relationships does not exist (i.e., the set defined in (1) is empty), then the area is excluded from analysis and is deemed not to exist. As an example, in the context of the Specimen hierarchy, there are 380 concepts that have the relationships procedure and topography (and only those relationships). Examples include the concepts Ear swab sample, Specimen from thymus gland obtained by biopsy, and Cervical biopsy sample. Therefore, there is an area named {topography, procedure} in the Specimen hierarchy. Areas are by definition disjoint, i.e., each concept belongs to one and only one area, and they therefore provide a partition of the hierarchy.
The area taxonomy is a directed acyclic graph (DAG) constructed by making each area a node and then arranging them hierarchically—analogously to the underlying concepts—using what we refer to as child-of relationships as edges. The child-of’s are derived from the concepts’ IS-A links. Before we describe how this is done, we need to define the notion of a root of an area. Let area(C) be the area of concept C, and let parents(C) denote the set of parents of C in the IS-A hierarchy.
Definition
(Root of an Area): A concept O in area A is a root of A if ∀C ∈ parents(O), area(C) ≠ area(O).
That is, a root is a concept whose parents all reside in other areas, or, in other words, its set of relationships is different from all its parents’. A root is not unique; an area can have more than one of them. Let us note that a root is of considerable significance in the makeup of a SNOMED hierarchy because, from a top-down hierarchical perspective, it is the first concept with a given combination of relationships. In this sense, it constitutes a cornerstone in the successive build-up of knowledge that is a conceptual hierarchy.
A child-of link in the area taxonomy is derived as follows. Let A and B be two areas such that a root of A has a parent in B. Then there exists a child-of from A to B. Overall, the collection of area nodes and child-of edges forms a DAG.
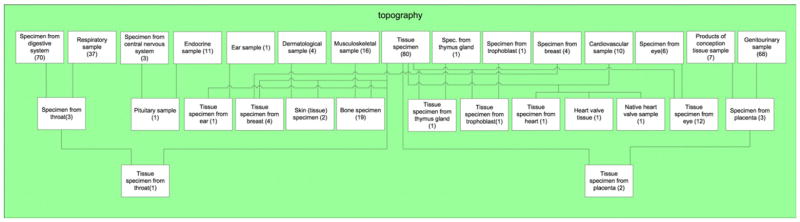
The area taxonomy of the 1056-concept Specimen hierarchy (July ’07 release) has a total of 24 areas distributed over five levels (Fig. 1). The boxes are the areas and the edges (directed upward) are the child-of’s. It should be noted that the top-level area 0̸ (“empty set”) comprises all concepts having no relationships at all. Thus, it has been given the name “0̸” to indicate an empty set of relationships. Note that the color-coded levels of the area taxonomy distinguish the numbers of relationships of the areas. The area 0̸ is on Level 0 and has zero relationships. The five green rectangles on Level 1 are the areas having exactly one relationship each.
Fig. 1.
Area taxonomy for SNOMED’s Specimen hierarchy (July 2007). The nodes (boxes) are the areas, and the lines are the child-of relationships.
The partial-area taxonomy is designed in an effort to achieve hierarchical coherence in addition to the structural congruity of each of the areas. The network is a refinement of the area taxonomy. As noted, areas may have more than one root. In such a case, each root and its respective collection of descendants (within the area) can be seen as being a distinct unit of subject matter that has the same set of relationships as some other such units. Moreover, that unit of knowledge is hierarchically coherent due to the single root. All concepts in the unit are specializations of the single root. With this in mind, we define the notion of partial-area with respect to a root O. In the following, desc(X, Y) denotes the fact that concept X is a descendant of concept Y in the IS-A hierarchy.
| (2) |
That is, the partial-area is a subset of the area consisting of the root O and all its descendants in the area. The partial-area defined by (2) is named “O” after its constituent root, since it is the defining characteristic.
The partial-areas are drawn as separate nodes in the partial-area taxonomy embedded inside their area nodes, which are retained. The partial-area nodes within a given area are not connected via child-of links. However, partial-areas residing in different areas are connected by child-of’s in a manner analogous to that for areas. In the complete partial-area taxonomy, the child-of’s between areas are replaced by those between their partial-areas. However, we often use certain abbreviation conventions or abridgment to reduce clutter. See [2] for details.
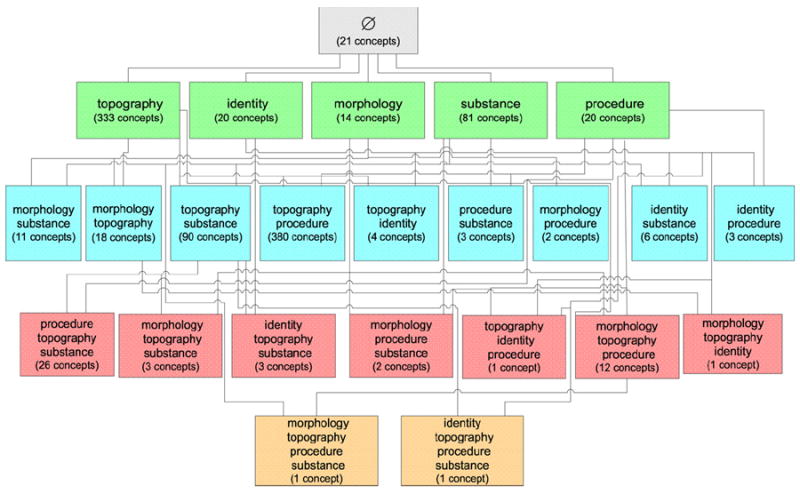
Fig. 2 shows the (abridged) partial-area taxonomy of the Specimen hierarchy. Each partial-area appears as a box inside its area node. In each partial-area node, the number in parentheses is the number of concepts it contains. For example, we see that the area {identity} (second lower green box from left) has two partial-areas, Device specimen and Specimen from patient, of 19 and two concepts, respectively. All 19 concepts of the partial-area Device specimen represent specimens from devices. The partial-area taxonomy has a total of 361 partial-areas. An example child-of can be seen on the left side of the figure extending from partial-area Effusion sample to Body substance sample. Many child-of’s have been omitted.
Fig. 2.
Partial-area taxonomy (abridged) for the Specimen hierarchy (July 2007). The main boxes are the areas, and the lines are the child-of relationships. The embedded boxes are the partial-areas.
Note that in some areas in Fig. 2, the numbers of concepts in the partial-areas do not add up to the total number of concepts in the area in Fig. 1. As a matter of fact, the area {substance} only contains 81 concepts, while the sum of concept numbers appearing in parentheses of its partial-areas is 136. This is due to overlaps among partial-areas, an issue we will deal with in detail in this paper.
In [6], the taxonomy framework was extended to hierarchies having no outgoing relationships by utilizing implicit converse relationships. The connection between auditing and complexity measures expressed with the taxonomy framework was explored in [7].
2.3. Terminology abstraction networks and automatic suspicious-concept identification
Due to the typically limited availability of resources for auditing, it is important to focus efforts on concepts or groups of concepts where those efforts are most likely to be needed. In this way, a better return, measured in the number of errors found, can be expected for a given amount of auditing work. Therefore, it is essential that research on terminology auditing develop techniques for automatically identifying concepts expected to have errors at a higher rate.
We have proposed and implemented SNOMED auditing regimens that make use of the two programmatically derived taxonomies introduced above [2,3]. We have shown that the taxonomies are extremely helpful in promoting more efficient and effective auditing. Different kinds of concept errors have been found to manifest themselves as anomalies at the taxonomy level, allowing for efficient discovery. The auditing methodology presented in the companion paper [4] is based on additional refinements to the partial-area taxonomy introduced herein.
Similar utilization of abstraction-network anomalies for the identification of suspicious concepts (i.e., those that are likely erroneous) has been done with other terminologies as well. Partial-area taxonomies were used for this purpose in the context of the National Cancer Institute thesaurus (NCIt) where partial-areas containing only a few concepts often held erroneous ones [8]. For the UMLS, inconsistencies between hierarchical relationships involving concepts and the corresponding relationships involving their semantic types in the Semantic Network have been used as an effective means of identifying suspicious concepts, particularly those having incorrect relationships and type assignments [9-12]. The Refined Semantic Network [13] for the UMLS supports this effort as well through the use of its “intersection semantic types” when they contain a minimal number of concepts [14,15].
3. Methods
The partial-area taxonomy has proven to be a useful vehicle for comprehending the overall structure of a SNOMED hierarchy, locating potential errors within it, and identifying modeling aspects that can be improved [2,3]. However, the taxonomy does lack a characteristic we call semantic uniformity that we have found useful in the realms of both comprehension and auditing. This deficiency is due to the potential overlap between partial-areas that was alluded to above. Let us look at an example to demonstrate this. The area {identity} has two roots, Device specimen and Specimen from patient (see Fig. 2). Device specimen and its 18 descendants (including Blood bag specimen) form one partial-area. Specimen from patient and its child Blood bag specimen, from patient form another. Blood bag specimen, from patient also happens to be a child of Blood bag specimen. Thus, Blood bag specimen, from patient is in two partial-areas: Device specimen and Specimen from patient. This situation is illustrated in Fig. 3. We say that these two partial-areas Device specimen and Specimen from patient “overlap” and call the concept Blood bag specimen, from patient an “overlapping concept.” The entire set of overlapping concepts is denoted V.
Fig. 3.
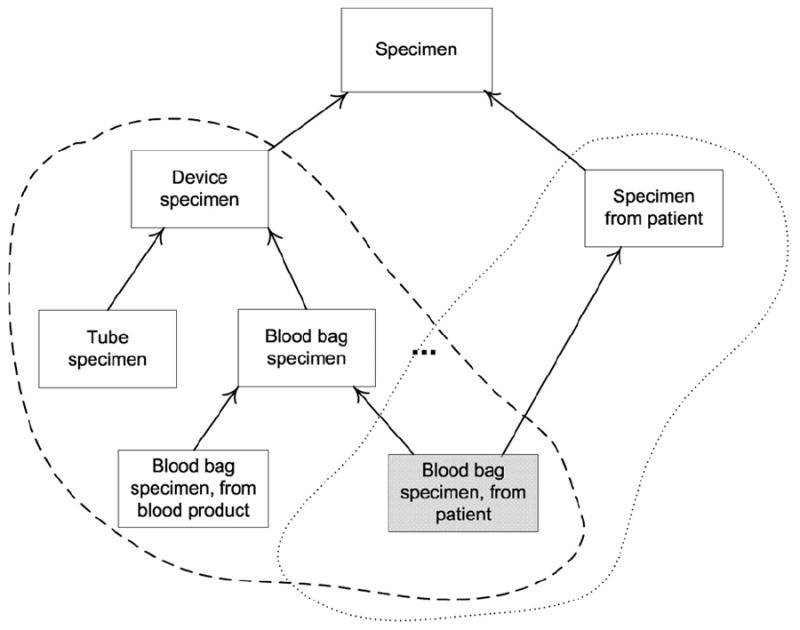
The overlapping concept Blood bag specimen, from patient resides in two partial-areas, Device specimen and Specimen from patient, demarcated by the dashed bubbles.
This raises two important issues. First, the entire collection of partial-areas does not form a partition of the hierarchy. This is in contrast to the collection of areas which does. Second, when two partial-areas overlap, some concepts in a partial-area, like the concept Blood bag specimen, elaborate only the semantics of one root (i.e., Device specimen) while the overlapping concepts in that same partial-area, in this case, the concept Blood bag specimen, from patient, elaborate the semantics of two roots (i.e., Device specimen and Specimen from patient). The situation gets worse when we have three overlapping partial-areas, say, R1, R2, and R3. In this situation, some concepts in R1 are elaborating the semantics of the root R1, while others may be elaborating the semantics of the two roots R1 and R2, and others are elaborating the semantics of all three roots R1, R2, and R3. In this sense, the partial-area R1 is not semantically uniform with respect to its root.
This deficiency of the partial-area taxonomy actually presents us with opportunities that we avail ourselves of in this paper and a companion paper [4]. One is the fact that the overlapping concepts lend themselves nicely to auditing scrutiny. Such concepts elaborate the semantics of two or more significant root concepts in the hierarchy and thus warrant the designation “complex concept,” which underpins an auditing methodology that we are introducing in [4].
The second opportunity pertains to the refinement of the partial-area taxonomy. We will extend its theoretical underpinning and refine it to further facilitate comprehending the terminology as well as the job of an auditor. In particular, we wish to partition the overlapping concepts systematically such that each resulting group of concepts is singly-rooted. The single root of each such group will provide a uniform semantics for the whole group. This is important because, as we will see, the overlapping concepts can collectively constitute quite a tangled hierarchy. The partition paves the way for the formation of an enhanced partial-area taxonomy that provides a view of the prevailing hierarchical configuration of the overlapping concepts. This will aid the subject-domain-expert editor and user in seeing the gestalt of the partial-area overlaps and more easily comprehending their content. Furthermore, such enhanced comprehension will enable an auditor to recognize any troublesome aspects. In this context, a new auditing methodology is proposed in the companion paper [4].
In the remainder of this section, we first discuss further the issue of the complexity of overlapping concepts. After that, we devise a singly-rooted partitioning scheme for the overlapping concepts of an area. This begins with the definition of what we call overlapping roots. From the partition, we are able to define a new refined abstraction network for the concepts of a SNOMED hierarchy. This refined abstraction network will better support comprehension of a SNOMED hierarchy by maintenance personnel, including editors and auditors, by providing a disjoint partition of the hierarchy’s concepts—the overlapping concepts, among them—into semantically uniform groups. It will also form the basis for an enhanced auditing regimen for the overlapping concepts of such a hierarchy [4].
3.1. Overlapping concepts are complex concepts
To further motivate the focus on overlapping concepts and see their inherent complexity, let us look at some examples. In the area {substance} (Fig. 2), the three direct children, Body substance sample, Fluid sample, and Drug specimen, of the top-level concept Specimen induce three partial-areas, respectively. Fig. 4 shows the three root concepts, along with two of their descendants (shaded). The partial-areas are demarcated with dashed bubbles, where the different border styles denote the different partial-areas. Body fluid sample, being a child of both Body substance sample and Fluid sample, resides in the intersection of the two partial-areas. It inherits the relationship substance directed to Body fluid in the Substance hierarchy from both its parents.
Fig. 4.
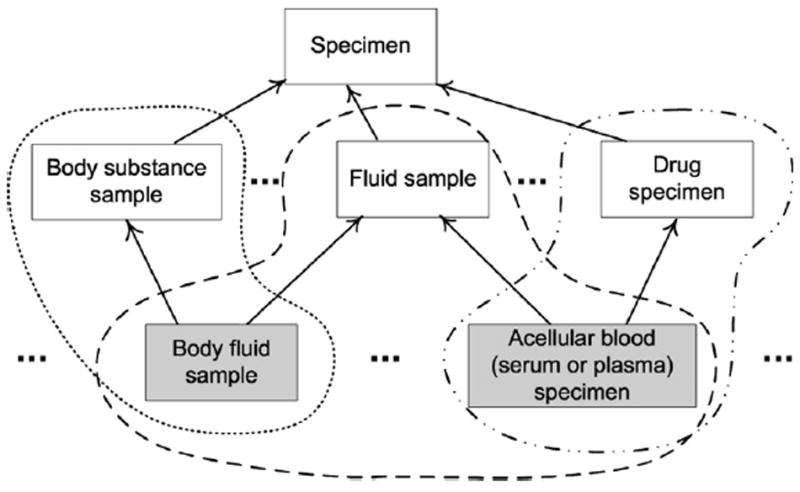
The overlapping concepts Body fluid sample and Acellular blood (serum or plasma) specimen (shaded) in the area {substance}.
The other shaded concept in Fig. 4, Acellular blood (serum or plasma) specimen, sits in the intersection of the partial-areas Fluid sample and Drug specimen. Thus, it elaborates the semantics of both parents, and inherits the relationship substance and the accompanying targets. Different from the previous example, Acellular blood (serum or plasma) specimen has two occurrences of the substance relationship, one pointing at Liquid substance and the other pointing at Blood component, a descendant of Drug or medicament, the target of the relationship substance of Drug specimen.
Overall, the area {substance} (Fig. 2) contains ten partial-areas and has quite a few overlapping concepts. This can be gathered from the fact that the sum of the numbers of concepts in its partial-areas (136) is much higher than the actual number of concepts in the area (81). The increased complexity of overlapping concepts is a consequence of the fact that they represent combination specializations deriving from multiple root concepts. For example, Body fluid sample and all its descendants residing in {substance} are overlapping concepts belonging to the partial-areas Body substance sample and Fluid sample. All these concepts that are both body substance and fluid examples, e.g., Amniotic fluid specimen and Lymph sample, are inherently more complex than concepts that are solely fluid samples, e.g., Water specimen, or only body substance samples, e.g., Calculus specimen. They each elaborate the semantics of a dual specialization.
The amount of overlapping, and attendant complexity, may increase as we traverse downward along the IS-A hierarchy. In {substance}, we find 15 concepts belonging to exactly two partial-areas, and 20 concepts belonging to three partial-areas. From this, we get its actual number of concepts: 136 − (2 − 1) · 15 − (3 − 1) · 20 = 81.
Differing degrees of complexity are seen for the overlapping concepts in Fig. 5, which contains a small fragment of the Specimen hierarchy consisting of nine concepts from the area {substance} (along with the hierarchy’s root). The three bubbles with different border styles enclose three partial-areas. Their roots are children of Specimen. All concepts below the roots (in colors) are overlapping concepts. The first of these are the yellow concepts Body fluid sample and Acellular blood (serum or plasma) specimen. As we traverse downward along the IS-A hierarchy, we find examples of overlapping concepts that are even more complex. For example, one of the children of the overlapping concept Body fluid sample, Blood specimen (in orange), has another parent Drug specimen that is the root of its partial-area. In this case, Blood specimen is the specialization of three roots and thus resides in the intersection of three separate partial-areas. But from the complexity point of view, it is a child of one overlapping concept and one root of a partial-area. Hence, it is more complex than the two yellow overlapping concepts that are children of roots of partial-areas. Other—more complex—cases can be seen with the green concepts, Serum specimen and Serum specimen from blood product, each having two parents that are overlapping concepts themselves. Let us note that a move down the hierarchy does not necessarily imply an increase in complexity. This is illustrated by Amniotic fluid sample, whose only parent is Body fluid sample. Being singly parented, it does not lie at a significant knowledge convergence point and is thus considered no more complex than Body fluid sample from a structural standpoint.
Fig. 5.
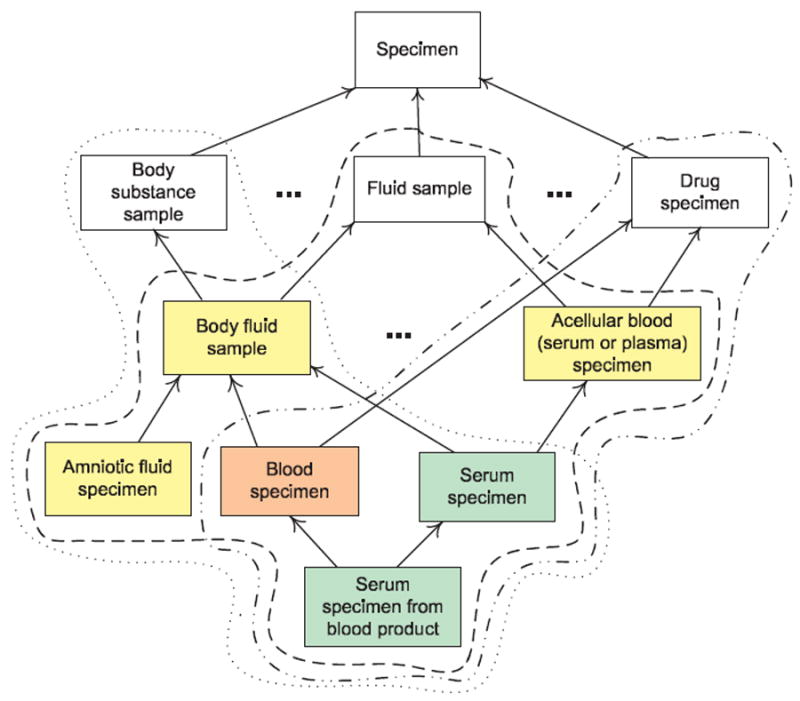
Differing degrees of complexity for overlapping concepts in the area {substance}. The green overlapping concepts are more complex than the orange overlapping concept which is more complex than the yellow overlapping concepts.
3.2. Foundations of the partition: overlapping roots
As discussed, the portion of an area consisting of the overlapping concepts may constitute a highly tangled hierarchy. Our goal is to impose some order on it by partitioning it in such a way as to obtain a collection of concept groups exhibiting semantic uniformity by satisfying single-rootedness and no overlaps. Thus, our first task is to identify those overlapping concepts that will serve as the roots of the concept groups. We will call them overlapping roots. Just like the root of a partial-area, an overlapping root will capture the overarching semantics of its group of overlapping concepts. The grouping process that we introduce proceeds in a deeply nested (recursive) fashion.
We will actually define two kinds of overlapping roots: those at the true “tops” of the overlapping portions of the partial-areas and those residing beneath them—perhaps quite deep in the overlap. Let us first define the fundamental kind of overlapping root called a base overlapping root, where, again, V is the entire set of overlapping concepts.
Definition
(Base Overlapping Root): A concept L ∈ V is a base overlapping root if ∀C ∈ parents(L), C ∉ V.
We have shown examples of overlapping concepts in Fig. 5. Among them, for instance, Body fluid sample is a base overlapping root because both of its parents, Body substance sample and Fluid sample, are non-overlapping concepts. They are, in fact, partial-area roots. Another example is Acellular blood (serum or plasma) specimen with the non-overlapping parents Fluid sample and Drug specimen.
In the progressive build-up of knowledge that is a concept hierarchy, the significance of a base overlapping root is that it lies at the confluence of multiple independent lines of knowledge—originating from the roots of the area. In this sense, such a concept can be seen as denoting a change of conceptual context within the hierarchy as one moves downward. The roots of a partial-area are significant in terms of unique sets of relationships. The base overlapping roots do not differ from their partial-area roots in regard to their relationships (they have the same relationships, in fact), but each one does represent a new combination in the downward direction of individual knowledge artifacts, each of which was first expressed by some partial-area root.
With the definition of base overlapping root now in place, we can define the general notion of overlapping root in a recursive manner as follows.
Definition
(Overlapping Root): A concept L ∈ V is an overlapping root if either (1) it is a base overlapping root; or there exist concepts C1 and C2 (C1 ≠ C2) such that desc(L, C1), desc(L, C2), and either (2) C1 is an overlapping root and C2 is a partial-area root or (3) both C1 and C2 are overlapping roots. For both Cases (2) and (3), the hierarchical paths from L to C1 and from L to C2 do not contain other (intermediate) overlapping roots.
Let us note that the qualifying pair of ancestors (C1, C2) is not necessarily unique. That is, more than one pair of ancestors might satisfy the requirements. The definition of overlapping root is well illustrated in Fig. 5. The yellow concepts, Body fluid sample and Acellular blood (serum or plasma) specimen, are base overlapping roots (Case (1)). The orange concept Blood specimen follows Case (2) since one parent, Body fluid sample, is an overlapping root and the other, Drug specimen, is a partial-area root. Finally, the green concepts Serum specimen and Serum specimen from blood product are overlapping roots according to Case (3) since each is a child of two overlapping roots.
Case (1) denotes the fact that base overlapping roots, defined above, form the foundation upon which other overlapping roots are defined. Cases (2) and (3) of the definition (the recurrences) designate certain points in the hierarchy below the level of the base overlapping concepts as being significant convergences of knowledge and thus warranting new grouping structures. A concept satisfying Case (2) or Case (3) in particular is called a derived overlapping root.
In Fig. 5, Blood specimen is a derived overlapping root according to Case (2). Its two qualifying ancestors are its parents Body fluid sample, a base overlapping root, and Drug specimen, a partial-area root. Serum specimen and Serum specimen from blood product are also derived overlapping roots. The two parents of Serum specimen are base overlapping roots. On the other hand, the two parents of Serum specimen from blood product are both derived overlapping roots.
The excerpt of the Specimen hierarchy’s area {substance} in Fig. 6a—some of which we saw already in Fig. 5—shows six of its overlapping roots, highlighted with multi-coloring. (All lines in the figure are IS-As.) This coloring scheme allows for easy identification of an overlapping root’s respective partial-area root ancestors.4 The three partial-area roots are the single-colored concepts on the top level of the figure. So, for example, Body fluid sample, colored orange and blue on the second level, is an overlapping root that is a descendant of Body substance sample (orange) and Fluid sample (blue). In fact, it happens to be a child of both and is thus a base overlapping root. In Level 2, we find another base overlapping root Acellular blood (serum or plasma) specimen, colored blue and yellow, as well as the non-overlapping concept Stool specimen, a descendant of only one partial-area root Body substance sample. Fecal fluid sample, colored orange and blue on Level 3, is also a base overlapping root due to the fact that its two parents are non-overlapping concepts. The derived overlapping roots begin to appear on that level, too. They are the two concepts Blood specimen and Serum specimen, both colored orange, blue, and yellow. Blood specimen is a child of one base overlapping root, Body fluid sample, in Level 2 and one partial-area root, Drug specimen (see Case (2) of the definition). Serum specimen is a child of the two base overlapping roots in Level 2 (Case (3)). Note that both have descendants that are not overlapping roots (e.g., Mixed venous blood specimen). On Level 4, we find the last derived overlapping root Serum specimen from blood product.
Fig. 6.
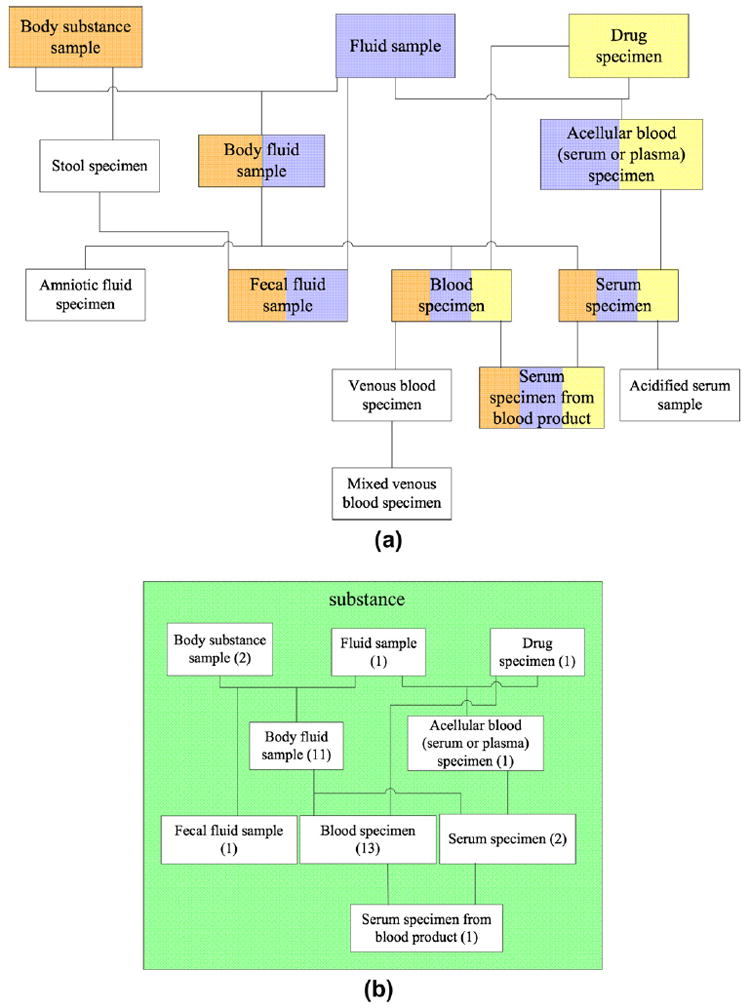
(a) Some overlapping roots (shown as multi-colored boxes) from the area {substance} in the Specimen hierarchy; (b) corresponding excerpt of the d-partial-area taxonomy representation of {substance}, where the embedded boxes are d-partial-areas.
It should be noted that overlapping concepts having a single parent cannot be overlapping roots. Again, the purpose of this designation is to highlight knowledge convergence points for which multiple parents are necessary. As an example, the concept Acidified serum sample has as its only parent the derived overlapping root Serum specimen and is thus not an overlapping root (see Fig. 6a). Similarly, the derived overlapping root Blood specimen has 12 descendants, such as Whole blood sample, Arterial blood specimen, and Cord blood specimen, none of which are overlapping roots. (Note that these descendants are not shown in the excerpt in Fig. 6a. They will be shown in the full figure in Section 4.) As these examples demonstrate, there are overlapping concepts that are not overlapping roots, even though their parents are derived overlapping roots.
3.3. Disjoint partial-areas
With the definition of overlapping root in place, we can now proceed to establish a partition of an entire area whose partial-areas overlap. Moreover, each of the concept groups collectively forming the partition will be singly-rooted. We will refer to such concept groups as disjoint partial-areas (d-partial-areas, for short). The initial set of d-partial-areas is derived by removing those portions of the original partial-areas that constitute overlaps, leaving only non-overlapping concepts. For example, the d-partial-area Body substance sample contains one additional concept Stool specimen beyond its root. It is obtained from the original partial-area of the same name having 47 total concepts by removing the overlapping roots Fecal fluid sample and Body fluid sample along with the latter’s descendants (see Fig. 6a). Clearly, such d-partial-areas are all disjoint with respect to each other and also with respect to the entire set of overlapping concepts. And they are each singly-rooted.
The remainder of the d-partial-areas are created in the context of the set of overlapping concepts based on the overlapping roots. In fact, each overlapping root will be the root of its own newly derived d-partial-area. Intuitively, such a d-partial-area is the portion of the area residing “between” an overlapping root, say, CR and the descendants of CR that are also overlapping roots. For example, consider the overlapping root Body fluid sample. The concepts that are removed in order to form its d-partial-area are the overlapping root child Blood specimen along with all its respective descendants and the other overlapping root child Serum specimen with its two children (see Fig. 6a). The concepts that are left in the d-partial-area rooted at Body fluid sample are, besides itself, its seven children (e.g., Amniotic fluid specimen) and its grandchildren which are children of the child Cerebrospinal fluid sample. (Note that only one such child is shown in Fig. 6a, as it is an excerpt. All ten descendants appear in the full figure in Section 4.)
More formally, let CR be an overlapping root. Then it is designated as the root of its own d-partial-area with the name “CR.” Furthermore, let C be an overlapping concept—but not an overlapping root—which is a descendant of CR such that there are no other overlapping roots on the paths between C and CR. Then C is a member of the d-partial-area CR. For example, consider the overlapping root Blood specimen and its descendant Mixed venous blood specimen in Fig. 6a. Since the intermediate concept Venous blood specimen on the only path from Mixed venous blood specimen to Blood specimen is not an overlapping root, Mixed venous blood specimen belongs to the d-partial-area Blood specimen. It is possible to prove that CR is unique for any given C, and hence C’s membership in a d-partial-area is well-defined. Moreover, it is possible to prove that for each overlapping concept C there is always such a CR.
3.4. Disjoint partial-area taxonomy
From the d-partial-areas, we can form an abstraction network that enhances the previous partial-area taxonomy framework [2] and highlights the structural subtleties of the overlapping portions of the partial-areas. This new network is called the disjoint partial-area taxonomy (d-partial-area taxonomy, for short). Those d-partial-areas derived directly from the existing partial-areas—and consisting only of non-overlapping concepts—hold the same place as their predecessors in the d-partial-area taxonomy. Moreover, partial-areas originally having no overlapping concepts retain their places as nodes and are also designated d-partial-areas in the new network. The child-of relationships emanating from these d-partial-areas and extending into other areas are derived as done previously for the partial-areas.
The d-partial-areas comprising overlapping concepts are also elevated to the status of nodes in the d-partial-area taxonomy. Each is displayed as a box with its name (i.e., its unique overlapping root) inside and its number of concepts in parentheses. Child-of links are defined for these new nodes in a similar manner to those for areas and partial-areas, but here the overlapping roots play a role. Let A and B be two d-partial-areas, such that the concept A (the overlapping root of the former) has a parent in the latter. Then there exists a child-of from the d-partial-area A to the d-partial-area B. A portion of the d-partial-area taxonomy for the area {substance} derived from the excerpt of its hierarchy shown in Fig. 6a can be seen in Fig. 6b. For example, there is a child-of from the d-partial-area Fecal fluid sample to the d-partial-area Body substance sample since, in Fig. 6a, there is an IS-A from the concept Fecal fluid sample to the concept Stool specimen which resides in Body substance sample. As can be seen in Fig. 6b, the d-partial-area nodes, like the partial-area nodes, are embedded in their respective area, which in this case is {substance}, colored green following Fig. 1.
3.5. Enhanced abstraction of the complex overlapping concepts in disjoint partial-areas
The described taxonomies provide abstraction-level views of the content of a SNOMED hierarchy. For example, the area taxonomy (Fig. 1) shows that there are 81 concepts having exactly the one relationship substance. The partial-area taxonomy (Fig. 2) also conveys the overarching semantics of these concepts. There are 44 fluid samples, 23 drug specimens, 47 body substance samples, and 13 food specimens. Those four large groups constitute most of the concepts representing specimens with only one relationship to the Substance hierarchy of SNOMED. There are some other small groups, including Gaseous material specimen (3), Microbial isolate specimen (2), and Plant specimen (1). Reviewing this information, the user gets a summary of the content of this area. In contrast, the area {morphology} has just one partial-area Lesion sample of 14 concepts. (This consolidated view was obtained following the auditing of the 2004 release of the Specimen hierarchy supported by the taxonomies [2]. The area {morphology} had six partial-areas in the earlier version, but the auditing found that all fall under Lesion sample.)
When users want to view concepts with both substance and morphology relationships, they can utilize the area {morphology, substance} in the second level having 11 concepts. This area is a child of both {substance} and {morphology} (Fig. 1). As it happens, the area has 11 partial-areas of one concept each, e.g., Effusion sample and Cyst fluid sample (Fig. 2). As shown in [2,3], this view provided by the partial-area taxonomy was very helpful in exposing errors in the Specimen hierarchy.
The partial-area taxonomy view is particularly useful when the different partial-areas of an area are disjoint, but it is somewhat deficient when the partial-areas overlap. As was discussed above, those overlapping parts of a partial-area contain concepts that are semantically more complex than concepts of non-overlapping parts of the same partial-area. Furthermore, the unit of a partial-area with an overlap is not semantically uniform. Hence, the difficulty of comprehending such concepts is magnified. For example, out of the 23 drug-specimen concepts in the partial-area of that name in the area {substance}, 21 are also fluid samples, while 20 are also body substance samples. Furthermore, 12 concepts are both fluid samples and body substance samples. Hence, the knowledge conveyed by the partial-areas of the area {substance} (Fig. 2) is hiding a more complex situation. They provide a relatively superficial perspective where a more refined view is needed. Furthermore, as shown in Fig. 1, the area {substance} contains only 81 concepts, where overlapping concepts appear in multiple counts of the sizes of the partial-areas in Fig. 2.
The desired refined view of an area with overlapping partial-areas is provided by the d-partial-area taxonomy introduced above. In Fig. 6b, we see that the overlap of the three partial-areas just discussed is concentrated under two d-partial-areas: Body fluid sample of 11 concepts, capturing an overlap of Body substance sample and Fluid sample; and Acellular blood (serum or plasma) specimen of one concept, capturing an overlap of Drug specimen and Fluid sample. In the d-partial-area taxonomy, the children of these two d-partial-areas, Blood specimen of 13 concepts and Serum specimen of two concepts, denote the overlaps of the three partial-areas. In turn, a deeper level of overlap is indicated by the grandchild d-partial-area Serum specimen from blood product of one concept. As we see, the names (overlapping roots) of the d-partial-areas communicate more precise knowledge of the content of the overlapping concepts. The full d-partial-area taxonomy for that portion of the area {substance} from which Fig. 6b was extracted will appear in Section 4 below. More such knowledge was excluded from Fig. 6a and 6b for the sake of brevity and clarity.
Importantly, each d-partial-area of the overlapping concepts consists of a semantically uniform group, where its name, e.g., Blood specimen, characterizes the concepts of the group very well. Hence, the d-partial-area taxonomy is a vehicle for more readily comprehending the nature of the overlapping concepts. In another example corresponding to Fig. 3, the d-partial-area taxonomy will have a minimal overlap of just one concept, Blood bag specimen, from patient, between the two partial-areas of Fig. 3, Device specimen and Specimen from patient. This overlap appears as one d-partial-area, Blood bag specimen, from patient, containing only that concept. Note that in the d-partial-area taxonomy, this d-partial-area is the child of the two semantically uniform d-partial-areas Device specimen (18) and Specimen from patient (1), which are now uniform due to the removal of the overlapping concept (Fig. 7). Thus, the d-partial-area taxonomy reveals both the uniform semantics of the overlapping subgroup and the precise size of its extent (by the number appearing alongside the name) as well as the uniform semantics of the d-partial-areas obtained by the removal of the overlapping concepts from the partial-areas of the partial-area taxonomy. This enhanced view afforded by the d-partial-area taxonomy supports a better auditing regimen for the complex overlapping concepts, which is demonstrated in the companion paper [4].
Fig. 7.
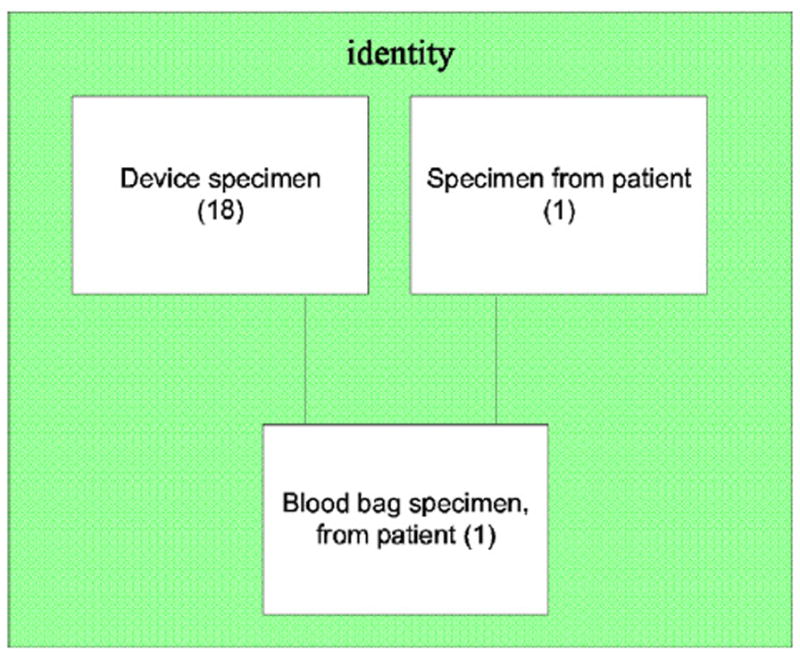
The d-partial-areas Device specimen, Specimen from patient, and Blood bag specimen, from patient of the area {identity}.
There are two issues regarding the display of the d-partial-area taxonomy. One is the arrangement of d-partial-areas within an area. In the partial-area taxonomy (e.g., Fig. 2), no child-of hierarchical relationships exist between partial-areas of the same area because each is based on and contains a root of the area. When we display one partial-area below another (see, e.g., the area {substance} in Fig. 2), no hierarchical arrangement is implied. It is just a layout expediency.
In the d-partial-area taxonomy, there are child-of’s between d-partial-areas in a given area. In fact, any d-partial-area rooted at an overlapping root (be it base or derived) has multiple child-of’s to other d-partial-areas of the same area. To reflect the hierarchical nature of these child-of’s, we try to position the d-partial-areas such that they are below their respective parents, and the child-of’s are in an upward direction.
As a result, there is a contrast between the detailed display of an area of many overlapping concepts, such as {substance} in Fig. 6b, and an area without overlapping concepts, such as {morphology}. The d-partial-area taxonomy contains both kinds of areas. Thus, there is a disparity in the display of these two kinds of areas in regard to their nature and level of detail. As we will discuss below, the three taxonomies are best used in concert in a kind of multiscale display.
4. Results
The July 2007 release of the Specimen hierarchy of SNOMED consists of 1056 active concepts, of which 162 are overlapping. We have used the July 2007 release in this paper because in the companion paper [4], we report on the application of a systematic auditing regimen to both the July 2007 and 2009 releases. The partial-area taxonomy and the d-partial-area taxonomy for July 2009, whose contents were affected by the audit of the July 2007 release, will appear in [4]. Most of the overlapping concepts reside in Level l areas, i.e., those having one relationship. In fact, roughly one third (155 out of 468) of the Level 1 concepts are overlapping, and these are found primarily in {topography} and {substance}. Overlapping concepts also appear in the partial-areas of areas with two relationships, but in far fewer numbers. In fact, there are only seven of them. Six are in {topography, procedure}, and the other is in {topography, morphology}. The statistics of the overlapping concepts in Levels 1 and 2 are given in Table 1. For each area, we list its total number of concepts C (Column 2), number of overlapping concepts V (Column 3), the percentage of overlapping concepts (Column 4), the number of d-partial-areas with overlapping roots D (Column 5), and the average number of overlapping concepts per d-partial-area: V/D (Column 6). For example, {substance} has 81 concepts and 35 of them are overlapping (43%). It also has nine overlapping roots which head d-partial-areas, with about four concepts per each such d-partial-area, on average.
Table 1.
Statistics of overlapping concepts at Levels 1 and 2.
| Area | C | V | V/C (%) | D | Avg = V/D |
|---|---|---|---|---|---|
| Substance | 81 | 35 | 43 | 9 | 3.9 |
| Topography | 333 | 116 | 35 | 52 | 2.2 |
| Procedure | 20 | 3 | 15 | 3 | 1.0 |
| Identity | 20 | 1 | 5 | 1 | 1.0 |
| Topography, procedure | 380 | 6 | 2 | 6 | 1.0 |
| Topography, morphology | 18 | 1 | 6 | 1 | 1.0 |
| Total | 852 | 162 | 19 | 72 | 2.3 |
C = # concepts; V = # overlapping concepts; D = # overlapping roots.
Most overlapping concepts in the area {topography} are found in intersections with the partial-area Tissue specimen which contains 126 concepts. We have tabulated these results separately in Table 2. For example, the partial-area Specimen from eye has 18 concepts. Its intersection with Tissue specimen has 12 of them (67%).
Table 2.
Intersections involving partial-area Tissue specimen.
| Second partial-area | C | V | V/C (%) |
|---|---|---|---|
| Specimen from eye | 18 | 12 | 67 |
| Ear sample | 2 | 1 | 50 |
| Specimen from breast | 8 | 4 | 50 |
| Cardiovascular sample | 13 | 3 | 23 |
| Products of conception tissue sample | 12 | 3 | 8 |
| Genitourinary sample | 73 | 22 | 27 |
| Dermatological sample | 6 | 2 | 33 |
| Spec. from digestive system | 74 | 30 | 39 |
| Musculoskeletal sample | 35 | 22 | 63 |
| Respiratory sample | 41 | 7 | 16 |
| Endocrine sample | 12 | 3 | 25 |
| Specimen from central nervous system | 4 | 1 | 25 |
| Spec. from thymus gland | 2 | 1 | 50 |
| Specimen from trophoblast | 2 | 1 | 50 |
| Total | 302 | 112 | 35 |
C = # concepts; V = # overlapping concepts.
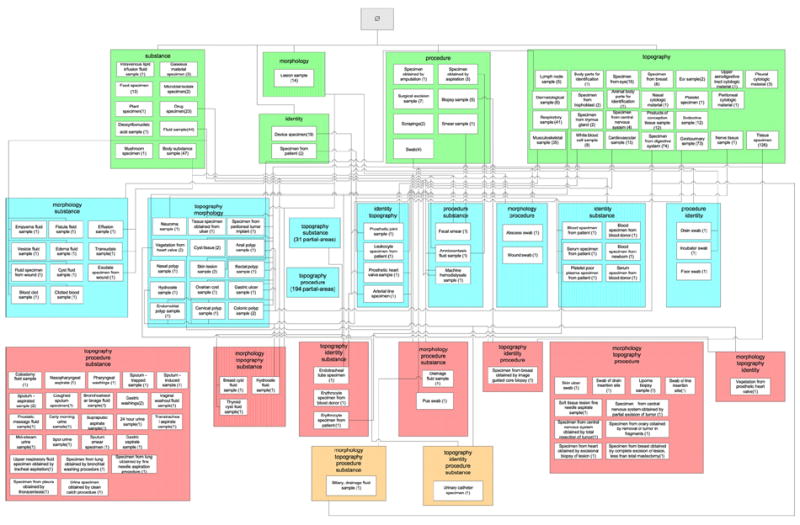
The full complement of nine overlapping roots from the area {substance} can be seen as the multi-colored boxes in the excerpt in Fig. 8. The figure follows the color conventions of Fig. 6a. The top four concepts are the area’s roots. Among the overlapping roots, five are base overlapping roots and four are derived overlapping roots. The remaining white concepts are overlapping concepts that elaborate the semantics of the overlapping roots of their respective d-partial-areas.
Fig. 8.
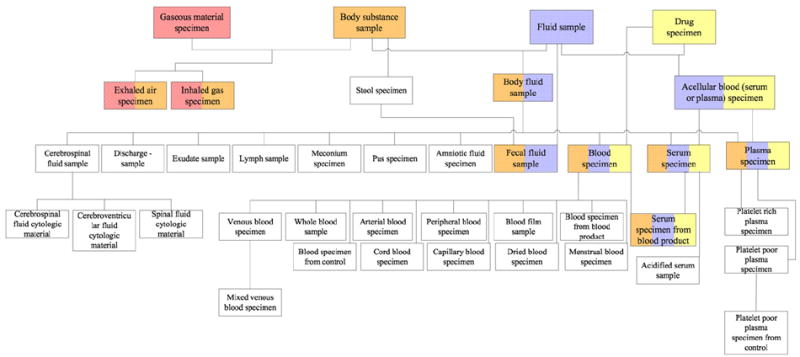
The nine overlapping roots from the area {substance} are shown as multi-colored boxes among other concepts.
The portion of the d-partial-area taxonomy for the area {substance} corresponding to the concept diagram in Fig. 8 is shown in Fig. 9. It presents a precise abstraction of the configuration of the overlapping concepts within {substance}. Note that the numbers of concepts listed for the top-level d-partial-areas are actually the numbers of non-overlapping concepts appearing in the original partial-areas from which these d-partial-areas are derived. For example, Drug specimen (2) has the two non-overlapping concepts from the partial-area of the same name, containing a total of 23 concepts, in Fig. 2. They are the area root Drug specimen plus a non-overlapping child not shown in Fig. 8. The entire content of the partial-area Drug specimen is distributed among the d-partial-area Drug specimen and all its descendants. This can be seen by summing up the numbers of concepts in those d-partial-areas: 2 + 1 + 13 + 2 + 4 + 1 = 23. The same holds true for the other top-level d-partial-areas and their respective descendants in the figure.
Fig. 9.
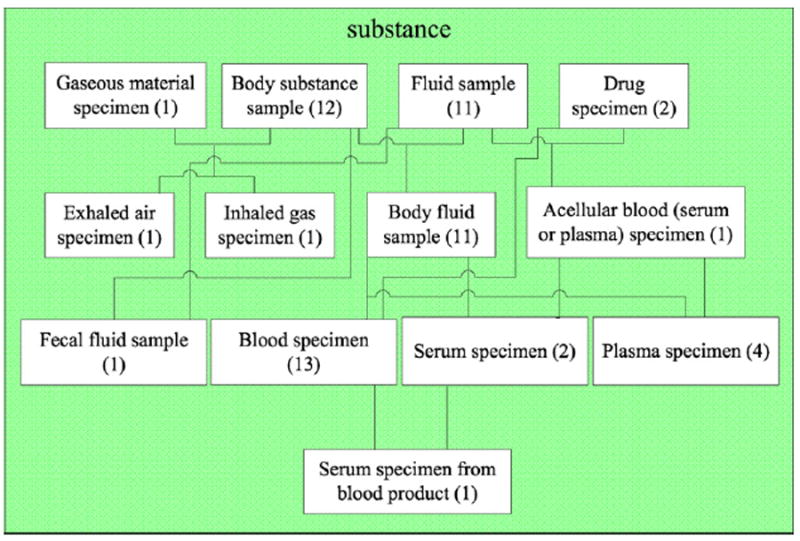
The d-partial-area taxonomy excerpt consisting of 13 d-partial-areas corresponding to the concept network appearing in Fig. 8.
The complete node for {substance} in the d-partial-area taxonomy is shown in Fig. 10, which differs from Fig. 9 only in the inclusion of the six additional d-partial-areas derived from the corresponding six partial-areas (Fig. 2) that do not contain any overlapping concepts. The isolation of these d-partial-areas from the others conveys the absence of overlaps. Overall, this network can be used, for example, as a vehicle for comprehending the details of the kinds of overlapping concepts and their numbers in the underlying SNOMED hierarchy.
Fig. 10.
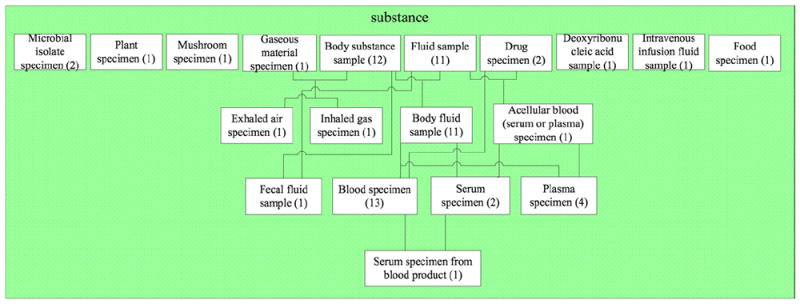
The d-partial-area taxonomy node for the area {substance} containing 19 embedded d-partial-areas. The numbers in parentheses indicate the numbers of concepts in the respective d-partial-areas.
Fig. 11 provides a larger excerpt of the portion of the d-partial-area taxonomy appearing within the area {topography}, highlighting the extensive overlapping among its partial-areas. As shown in Table 1, this area has 116 overlapping concepts distributed among 52 d-partial-areas. Most of the overlapping concepts have Tissue specimen as one of their partial-areas, as listed in Table 2. In the top level of Fig. 11, we see the 15 d-partial-areas obtained by removing all overlapping concepts from the original partial-areas. On the next level down, we find 13 d-partial-areas having base overlapping roots. Two d-partial-areas with derived overlapping roots appear on the bottom level. Many other d-partial-areas with few concepts have been omitted. Again, it should be noted that the intersection of two partial-areas may contain several overlapping roots. For example, the intersection of Tissue specimen and Cardiovascular sample has three overlapping roots, as shown in the figure: Tissue specimen from heart, Heart valve tissue, and Native heart valve sample.
Fig. 11.
An excerpt of the d-partial-area taxonomy for the area {topography} consisting of 30 d-partial-areas.
To illustrate the general applicability of our abstraction approach, we have applied it to all seven of the SNOMED hierarchies that have outgoing lateral relationships. (The other 12 hierarchies have no such relationships, rendering our methodology inapplicable to them.) The results are listed in Table 3. For each of the seven hierarchies, the table gives its total number of concepts, the number of overlapping concepts and their percentage, and the number of overlapping roots. For example, the Pharmaceutical Product hierarchy has a total of 17,410 concepts, of which 1047 are overlapping (6.1%). The number of overlapping roots is 949. Let us note that in Pharmaceutical Product almost all the overlapping concepts are overlapping roots (1047 compared to 949). As it happens, the hierarchies Event and Body Structure have no overlapping concepts whatsoever.
Table 3.
Concept distributions in seven SNOMED hierarchies.
| Hierarchy | C | V | V/C (%) | D | Cmult | Cmult/C (%) | Vmult |
|---|---|---|---|---|---|---|---|
| Event | 3661 | 0 | 0 | 0 | 86 | 2.4 | 0 |
| Situation | 3237 | 86 | 2.7 | 67 | 387 | 12.0 | 67 |
| Pharmaceutical product | 17,140 | 1047 | 6.1 | 949 | 7721 | 45.1 | 963 |
| Procedure | 52,687 | 7878 | 15.0 | 3374 | 27,031 | 51.3 | 5846 |
| Specimen | 1330 | 191 | 14.4 | 80 | 788 | 59.3 | 130 |
| Body structure | 31,155 | 0 | 0 | 0 | 13,418 | 43.1 | 0 |
| Clinical finding | 98,414 | 13,943 | 14.2 | 3127 | 44,544 | 45.3 | 9841 |
| Total | 207,624 | 23,145 | 11.2 | 7597 | 93,975 | 45.3 | 16,847 |
C = # concepts; V = # overlapping concepts; D = # overlapping roots; Cmult = # concepts having multiple parents; Vmult = # overlapping concepts having multiple parents.
As a point of comparison, we also list in Table 3 the number of concepts having multiple parents (and their percentage), along with the number of overlapping concepts having that characteristic. These numbers will be discussed further below. The Pharmaceutical Product hierarchy has 7721 concepts (45%) with multiple parents, of which only 963 (5.6%) are overlapping. As can be seen, there are only 14 (=963 − 949) non-root overlapping concepts having multiple parents. Note that 84 overlapping concepts have only one parent.
5. Discussion
5.1. Taxonomy support for presentation of terminology content
The value of a terminological knowledge-base depends on the accuracy and reliability of its constituent knowledge. This is true from the perspective of both ad hoc users and developers of software systems, such as EHR software and decision-support systems, that are dependent on that knowledge. Moreover, the ability to visualize and assess the knowledge’s underlying structural organization is a critical factor contributing to terminology usability, deployment, and maintenance. The area and partial-area taxonomy abstraction networks have been shown to support maintenance efforts for SNOMED [2] and the NCIt [8]. However, in this paper, we have discussed some deficiencies in these abstraction networks regarding complex portions of the terminology involving what we call overlapping concepts. The d-partial-area taxonomy that we have introduced extends the area taxonomy paradigm to more properly present the overlapping concepts by highlighting semantically uniform groups and their sizes. For example, Fig. 9 highlights the groups Blood specimen (13), Serum specimen (2), and Plasma specimen (4), which were originally hidden but tacitly accounted for multiple times in Body substance sample (47), Fluid sample (44), and Drug specimen (23) in Fig. 2.
In Fig. 11, showing the area {topography}, we find only two d-partial-areas with derived overlapping roots. We find more than twice that number, with many concepts in their d-partial-areas, in the excerpt of {substance} in Fig. 9. What we see in {topography} is extensive overlapping with many base overlapping roots but not as complex a pattern as is found in {substance}. An interesting finding revealed by Fig. 11 is that Products of conception tissue sample, the second d-partial-area from the right in Level 1, represents a modeling error. Its root should not actually have been a root but rather an overlapping concept of Tissue specimen and Genitourinary sample.
In the present paper, we studied the complexity of overlapping concepts, finding what we call “overlapping roots” that represent the convergence of multiple hierarchical paths originating at the roots of an area (see Section 3.1). A variety we call “base overlapping root” is less complex than the “derived overlapping root.” Within the latter, we have identified different kinds according to Cases (2) and (3) of the definition (see Section 3.2). The organizational subtleties of the various kinds of overlapping concepts are abstracted in the d-partial-area taxonomy that we have introduced in this paper. The network breaks down the highly tangled group of overlapping concepts of an area into subsets in a manner that summarizes their hierarchical configuration and supports orientation into their nature. This phenomenon is demonstrated, for example, in Fig. 10, where nine d-partial-areas (rooted at derived overlapping roots) on Levels 2 and 3 expose the very complex modeling of the 35 overlapping concepts in a clear and unambiguous way, while all this knowledge is hidden “under the hood” in the partial-area taxonomy of Fig. 2. The refined view helps in assessing the correctness of the modeling of this highly complex portion of the SNOMED hierarchy.
5.2. Taxonomy support for auditing
We have proposed and implemented SNOMED auditing regimens that make use of the programmatically derived area and partial-area taxonomies [2,3] and have shown that they promote more efficient and effective auditing. In the companion paper [4] to this paper, we present an auditing methodology based on the additional refinements appearing in the d-partial-area taxonomy, thus demonstrating its usefulness and significance in SNOMED quality assurance.
In [16], we presented an initial study regarding the complexity of overlapping concepts of multiple partial-areas and their likelihood of being erroneous. The observations in [16] are in line with the theme that complex concepts in their many varieties are often more probably in error. The auditing approach used there just called for a review of all the overlapping concepts.
For the sake of modularity and readability, we have deferred the presentation of a systematic auditing regimen based on the d-partial-area taxonomy to the companion paper [4]. The new methodology is more detailed and sophisticated than the initial one employed in [16], making use of the inter-relationships among the overlapping concepts and their groupings into d-partial-areas. In fact, the partition of the overlapping concepts into disjoint sets exhibiting semantic uniformity suggests a regimen of “group-based auditing,” a theme which we have successfully employed in other related contexts [3,9,10,17].
5.3. Further applicability of the methodology
While our abstraction methodology was formulated in the context of SNOMED, its applicability extends to other DL-based terminologies such as the NCIt. Moreover, terminologies such as Kaiser-Permanente’s CMT [18] and the VA’s ERT [19], that have been derived in part from SNOMED, may prove to be fertile grounds for additional applications. By 2015, SNOMED is slated to become a standard for problem-list encoding in EHRs under the HITECH initiative [20]. It is thus reasonable to assume that further derivatives from SNOMED will emerge. SNOMED’s design, in fact, anticipates the need for extensions and subsets in order to craft terminological artifacts that are tuned to the needs of individual hospitals and other organizations. Its “reference set specification” [21] serves the purpose of extracting components of SNOMED tailored to particular organizational preferences and use-cases.
5.4. Abstraction networks
In reviewing abstraction networks of terminologies, we distinguish between extrinsic and intrinsic networks. In the former, the nodes of the network represent categories obtained from knowledge outside the terminology itself. The Semantic Network [22] of the UMLS [23] is such an example. The nodes of the Semantic Network, called semantic types, represent known, broad categories in the biomedical field. A modified abstraction network for the UMLS is the Refined Semantic Network (RSN) [13,24]. A defining feature of the RSN is the fact that the extents of its types (i.e., the sets of concepts assigned respective types) are semantically uniform and disjoint, offering a clearer abstraction view.
In an intrinsic abstraction network, the categories are derived directly from the specific concepts of the terminology. An example of such a network is the partial-area taxonomy, which has been described above. In addition to its derivation from SNOMED hierarchies, it has been used in the context of the NCIt [8]. The d-partial-area taxonomy is also clearly intrinsic. Another example of an intrinsic network is the object-oriented schema [25,26] that has been derived from the MED [27]. As with the taxonomies, concepts that exhibit more relationships than their parents serve as roots of and to name the classes in the schema. In terms of size, the schema is relatively small in its number of nodes compared to the partial-area taxonomy.
5.5. Limitations and future work
The area and partial-area taxonomies are available only for DL-based terminologies. Abstraction of terminologies is very delicate, and no one model of abstraction networks is expected to fit all terminologies. However, more research is needed to explore abstraction networks for other families of terminologies and terminological systems, e.g., the ones mentioned in Section 5.4. The benefits obtained from abstraction networks in regard to auditing should motivate more research in this direction.
A limitation of the taxonomy approach is that it depends on the existing relationships defined for a hierarchy of SNOMED. Hence, the methodology of this paper is not applicable to a SNOMED hierarchy without any outgoing relationships at all. An initial effort to handle such a hierarchy based on converse relationships appeared in [6]. Moreover, the d-partial-area taxonomy is only pertinent when there are overlapping partial-areas within the partial-area taxonomy. Otherwise, the two taxonomies are identical.
In general, an abstraction network should represent a significant reduction in size (i.e., number of nodes) vis-à-vis its underlying concept network. For the Specimen hierarchy, the area taxonomy provides a 0.023 reduction factor (24 areas versus 1056 concepts). The partial-area taxonomy has a reduction factor of 0.34 (361 partial-areas versus 1056 concepts). The d-partial-area taxonomy only has a reduction factor of 0.41 (433 d-partial-areas versus 1056 concepts). Note that the higher reduction factor for the d-partial-area taxonomy is the justifiable price paid for the enhanced view obtained by the inclusion of the d-partial-areas that abstract the more complex overlapping concepts. There is no impact on the representation of those partial-areas experiencing no overlap in the partial-area taxonomy. Let us also note that the relatively high reduction factor for the partial-area taxonomy is a result of a large number of partial-areas containing just one concept each (so-called “singletons”). As was shown in [3], such partial-areas tend to signal errors. It is interesting to see if the number of such partial-areas will decrease as a result of auditing them. An initial promising result is brought up in [7]. Further research into this issue is required.
The reduction factors aside, the three taxonomies complement each other in terms of granularity of display, with a zooming effect achieved as one moves successively through them starting from the area taxonomy. When used together in this manner, they provide a multi-scale display. The area taxonomy offers a global view of the hierarchy’s layout and the partial-area taxonomy provides a more semantically focused view of the areas, whereas the real benefits of the d-partial-area taxonomy are seen at the local level—on the scale of an individual area—where it helps to reveal the complexity of the configuration of the overlapping concepts.
One might question whether there are simpler ways to identify “complex” concepts rather than having to go through the abstraction analysis presented in this paper. For example, one might choose to consider the easily identified concepts having multiple parents as being complex. Let us note that the overlapping concepts are not simply a subset of the multi-parent concepts. Only a root overlapping concept must have, by definition, more than one parent. As we see in Table 1, only 72 out of 162 overlapping concepts, in the Specimen hierarchy of July 2007, are overlapping roots. The other overlapping concepts have mostly a single parent. See also Fig. 8 where only the nine overlapping roots are multiparented. Similar statistics are seen in Table 3. In the seven hierarchies for which our analysis is applicable, we find a total of 93,975 multi-parented concepts (45.3%). In that same context, there are a total of 23,145 overlapping concepts, with 16,847 being multiparented.
6. Conclusion
SNOMED CT is one of the leading terminologies being used in a variety of applications worldwide. However, it contains hundreds of thousands of concepts and has an inherent complexity that could hinder its further adoption as well as its ongoing maintenance. A new abstraction network, called the disjoint partial-area taxonomy, has been introduced to provide a better high-level view of portions of a SNOMED hierarchy containing concepts of a particularly complex nature. It refines our previous abstraction network, the partial-area taxonomy, for SNOMED. The new network focuses on the location and number of such complex concepts and highlights their modeling and local neighborhoods. Overall, users are provided with a summary account of the “lay of the land” that can facilitate orientation to and assessment of SNOMED’s content. Our methodology was demonstrated by applying it to SNOMED’s Specimen hierarchy. In a companion paper [4], we present a systematic auditing regimen based on the disjoint partial-area taxonomy, demonstrating its utility to terminology maintenance personnel.
Acknowledgments
This work was partially supported by the NLM under Grant R-01-LM008912-01A1.
Footnotes
From now on, we will drop the “CT”.
Concept names are written in italics with the first letter capitalized; relationships are in italics.
For brevity, we will use just the last word of each relationship name in the remainder of the paper.
Let us note that the coloring used in this concept-level diagram has nothing to do with the coloring used previously in the taxonomy diagrams, where areas were colored to highlight their respective levels.
Contributor Information
Yue Wang, Email: yw44@njit.edu.
Michael Halper, Email: mikehhalper@yahoo.com.
Duo Wei, Email: Duo.Wei@stockton.edu.
Yehoshua Perl, Email: yehoshua.perl@gmail.com.
James Geller, Email: james.geller@gmail.com.
References
- 1.IHTSDO: SNOMED CT. [30.03.11]; < http://www.ihtsdo.org/snomed-ct>.
- 2.Wang Y, Halper M, Min H, Perl Y, Chen Y, Spackman KA. Structural methodologies for auditing SNOMED. J Biomed Inform. 2007;40(5):561–81. doi: 10.1016/j.jbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Halper M, Wang Y, Min H, Chen Y, Hripcsak G, Perl Y, et al. Analysis of error concentrations in SNOMED. In: Teich JM, Suermondt J, Hripcsak G, editors. Proceedings of 2007 AMIA annual symposium. Chicago, IL: 2007. pp. 314–8. [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Halper M, Wei D, Gu H, Perl Y, Xu J, et al. Auditing complex concepts of SNOMED using a refined hierarchical abstraction network. J Biomed Inform. doi: 10.1016/j.jbi.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.IHTSDO. SNOMED CT abstract logical models and representational forms (draft document) 2008 January; [Google Scholar]
- 6.Wei D, Halper M, Elhanan G, Chen Y, Perl Y, Geller J, et al. Auditing SNOMED relationships using a converse abstraction network. Proceedings of 2009 AMIA annual symposium; San Francisco, CA. 2009. pp. 685–9. [PMC free article] [PubMed] [Google Scholar]
- 7.Wei D, Wang Y, Perl Y, Xu J, Halper M, Spackman KA. Complexity measures to track the evolution of a SNOMED hierarchy. In: Suermondt J, Evans RS, Ohno-Machado L, editors. Proceedings of 2008 AMIA annual symposium. Washington, DC: 2008. pp. 778–82. [PMC free article] [PubMed] [Google Scholar]
- 8.Min H, Perl Y, Chen Y, Halper M, Geller J, Wang Y. Auditing as part of the terminology design life cycle. JAMIA. 2006;13(6):676–90. doi: 10.1197/jamia.M2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Gu H, Perl Y, Geller J, Halper M. Structural group auditing of a UMLS semantic type’s extent. J Biomed Inform. 2009;42(1):41–52. doi: 10.1016/j.jbi.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Gu H, Perl Y, Geller J. Structural group-based auditing of missing hierarchical relationships in UMLS. J Biomed Inform. 2009;42(3):452–67. doi: 10.1016/j.jbi.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geller J, Morrey CP, Xu J, Halper M, Elhanan G, Perl Y, et al. Comparing inconsistent relationship configurations indicating UMLS errors. Proceedings of 2009 AMIA annual symposium; San Francisco, CA. 2009. pp. 193–7. [PMC free article] [PubMed] [Google Scholar]
- 12.Cimino JJ, Min H, Perl Y. Consistency across the hierarchies of the UMLS Semantic Network and Metathesaurus. J Biomed Inform. 2003;36(6):450–61. doi: 10.1016/j.jbi.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Geller J, Gu H, Perl Y, Halper M. Semantic refinement and error correction in large terminological knowledge bases. Data Knowledge Eng. 2003;45(1):1–32. [Google Scholar]
- 14.Gu H, Perl Y, Elhanan G, Min H, Zhang L, Peng Y. Auditing concept categorizations in the UMLS. Artif Intell Med. 2004;31(1):29–44. doi: 10.1016/j.artmed.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Gu H, Hripcsak G, Chen Y, Morrey CP, Elhanan G, Cimino JJ, et al. Evaluation of a UMLS auditing process of semantic type assignments. In: Teich JM, Suermondt J, Hripcsak G, editors. Proceedings of 2007 AMIA annual symposium. Chicago, IL: 2007. pp. 294–8. [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Wei D, Xu J, Elhanan G, Perl Y, Halper M, et al. Auditing complex concepts in overlapping subsets of SNOMED. In: Suermondt J, Evans RS, Ohno-Machado L, editors. Proceedings of 2008 AMIA annual symposium. Washington, DC: 2008. pp. 273–7. [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Gu H, Perl Y, Halper M, Xu J. Expanding the extent of a UMLS semantic type via group neighborhood auditing. JAMIA. 2009;16(5):746–57. doi: 10.1197/jamia.M2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolin RH, Mattison JE, Cohn S, et al. Kaiser Permanente’s convergent medical terminology. In: Fieschi M, Coiera E, Li Y-C, editors. Proceedings of Medinfo 2004. San Francisco, CA: 2004. pp. 346–50. [PubMed] [Google Scholar]
- 19.Lincoln MJ, Brown SH, Nguyen V, Cromwell T, et al. US department of veterans affairs enterprise reference terminology strategic overview. In: Fieschi M, et al., editors. Proceedings of Medinfo2004. San Francisco, CA: 2004. pp. 391–5. [PubMed] [Google Scholar]
- 20.Department of Health and Human Services. Health information technology: initial set of standards, implementation specifications, and certification criteria for electronic health record technology. Final Rule 45 CFR Part 170. 2010 July 28; [PubMed] [Google Scholar]
- 21.SNOMED Clinical Terms reference sets: technical specification. Technical specification, College of American Pathologists: Jul, 2006. [Google Scholar]
- 22. [10.04.11];The UMLS Semantic Network. < http://semanticnetwork.nlm.nih.gov>.
- 23. [10.04.11];Unified Medical Language System (UMLS) – Home. < http://www.nlm.nih.gov/research/umls>.
- 24.Gu H, Perl Y, Geller J, Halper M, Liu L, Cimino JJ. Representing the UMLS as an OODB: modeling issues and advantages. JAMIA. 2000;7(1):66–80. doi: 10.1136/jamia.2000.0070066.. selected for reprint in: Haux R, Kulikowski C, editors. Yearbook of Medical Informatics: Digital Libraries and Medicine (International Medical Informatics Association) Schattauer, Stuttgart; Germany: 2001. pp. 271–285.
- 25.Gu H, Halper M, Geller J, Perl Y. Benefits of an object-oriented database representation for controlled medical terminologies. JAMIA. 1999;6(4):283–303. doi: 10.1136/jamia.1999.0060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Halper M, Geller J, Perl Y. Controlled vocabularies in OODBs: modeling issues and implementation. Distrib Parallel Datab. 1999;7(1):37–65. [Google Scholar]
- 27.Cimino JJ, Clayton PD, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology. JAMIA. 1994;1(1):35–50. doi: 10.1136/jamia.1994.95236135. [DOI] [PMC free article] [PubMed] [Google Scholar]