Abstract
The development of multispecies oral microbial communities involves complex intra- and interspecies interactions at various levels. The ability to adhere to the resident bacteria or the biofilm matrix and overcome community resistance are among the key factors that determine whether a bacterium can integrate into a community. In this study, we focus on community integration of Fusobacterium nucleatum, a prevalent Gram-negative oral bacterial species that is considered an important member of the oral community due to its ability to adhere to Gram-positive as well as Gram-negative species. This interaction with a variety of different species is thought to facilitate the establishment of multispecies oral microbial community. However, the majority of experiments thus far has focused on the physical adherence between two species as measured by in vitro co-aggregation assays, while the community-based effects on the integration of F. nucleatum into multispecies microbial community remains to be investigated. In this study, we demonstrated using an established in vitro mice oral microbiota (O-mix) that the viability of F. nucleatum was significantly reduced upon addition to the O-mix due to cell contact-dependent induction of hydrogen peroxide (H2O2) production by oral community. Interestingly, this inhibitory effect was significantly alleviated when F. nucleatum was allowed to adhere to its known interacting partner species (such as Streptococcus sanguinis) prior to addition. Furthermore, this aggregate formation-dependent protection was absent in the F. nucleatum mutant strain ΔFn1526 that is unable to bind to a number of Gram-positive species. More importantly, this protective effect was also observed during integration of F. nucleatum into a human salivary microbial community (S-mix). These results support the idea that by adhering to other oral microbes, such as streptococci, F. nucleatum is able to mask the surface components that are recognized by H2O2 producing oral community members. This evasion strategy prevents detection by antagonistic oral bacteria and allows integration into the developing oral microbial community.
Keywords: coaggregation, Fusobacterium nucleatum, microbial flora, oral cavity, community resistance
Introduction
Several hundred different taxa of bacteria contribute to the complex ecosystem in the oral cavity [1,2,3,4] and form highly structured, multispecies communities that are responsible for two major human oral diseases: caries (tooth decay) and gingivitis/periodontitis (gum diseases) [5,6,7]. The resident bacteria of the oral microbial community carry out a plethora of interactions at various levels to form sophisticated, multispecies biofilm structures, perform physiological functions and induce microbial pathogenesis [8,9,10,11]. The development of these oral microbial communities, especially the sequence of adherence to host tissue by the early bacterial colonizers as well as the subsequent recruitment of intermediate and late colonizers to the already attached microbes through coadhesion have been well investigated [8,10]. However, most of these studies have focused on in vivo observation of biofilm formation and its architecture or in vitro investigation of one-on-one adherence event [8,10,12,13], while detailed knowledge regarding how a bacterial species can integrate into an existing community remains to be examined.
During the development of highly structured microbial communities, the incoming bacteria must be able to attach to resident members or the extracellular matrix, and more importantly, overcome the invasion resistance in order to integrate into the community [14]. Recently, we demonstrated in vitro that the cultivable predominantly Gram-positive oral microbiota of mice (O-mix) developed invasion resistance and responded to the presence of the Gram-negative gut isolate Escherichia coli by producing hydrogen peroxide (H2O2). E. coli cells are more sensitive to this bactericidal agent than the oral isolates comprising the O-mix thus resulting in selective killing of the “intruder” [15]. Further analysis revealed that the lipopolysaccrides (LPS) of E. coli were the main determinant responsible for triggering the H2O2 production by oral communities [15,16].
The intriguing observation that the oral community has the collective ability to protect itself from Gram-negative species such as E. coli that are not typically part of the normal oral microbiota, naturally raises the question how Gram-negative oral species are able to integrate into the oral biofilm. We chose Fusobacterium nucleatum as a model oral Gram-negative microorganism, since it is ubiquitously found in the oral cavity of humans and other mammals, including mouse and dog [17,18]. While commonly regarded as a subgingival bacterium, F. nucleatum has been frequently isolated from supragingival and saliva samples [19,20,21] which harbor microbial population dominated by Gram-positive species [22,23,24]. F. nucleatum has been suggested to play an important role in biofilm community architecture due to its ability to adhere to a very large variety of different microorganisms [9,10], and detailed molecular studies have revealed the membrane components involved in co-adherence with other oral bacterial species [25]. However, many of the previous studies were performed in vitro at the dual species level and little is known about the effects exerted by other bacterial species on F. nucleatum during its integration into oral multispecies communities. Considering that our previous studies revealed LPS-induced hydrogen-peroxide production by Gram-positive oral community members in response to E. coli, another intriguing question is if F. nucleatum as a Gram-negative bacterium carrying LPS on its cell surface, would experience similar resistance when encountering the Gram-positive species dominated O-mix? In this study, using established in vitro systems, we investigated the integration of F. nucleatum into our established model system, the O-mix community that was isolated from mice [15,16], as well as a human saliva-derived microbial community (S-mix).
Materials and Methods
Bacterial strains and growth conditions
Wild type strain F. nucleatum 23726 and its mutant derivatives lacking each one of the large fusobacterial outer membrane proteins including Fn0254, Fn0387, Fn1449 (Fap2), Fn1526 (RadD), Fn1554, Fn1893, Fn2047 or Fn2058 (Aim1) [25,26,27], were cultivated in Brain Heart Infusion (BHI) (Difco, Detroit, MI, USA) supplemented with hemin (5 µg/ml), vitamin K (0.5 µg/ml), sucrose (0.1%), mannose (0.1%) and glucose (0.1%), at 37 °C under anaerobic condition (nitrogen 85%, carbon dioxide 5%, hydrogen 10%). Thiamphenicol (MP Biomedicals, Irvine, CA, USA) at 5µg/mL was used for selection and maintenance of F. nucleatum strains possessing the catP determinant. Other oral isolates, including mice oral isolate Streptococcus sanguinis OI101and human salivary isolate S. sanguinis SI101 were also grown in supplemented BHI medium.
The cultivated mice oral microbiota were recovered from a lab stock that was described in a previous study [16]. Briefly, 50 µl of BHI-cultivated O-mix stock was inoculated into 5 ml of supplemented BHI broth. The cultures were incubated at 37°C under anaerobic conditions until the exponential growth phase was reached.
Cultivating human saliva-derived microbiota (S-mix)
The preparation and establishment of the BHI cultivable S-mix were performed under UCLA-IRB #09-08-068-02A as previously described [28]. Briefly, saliva samples were collected from 3 healthy subjects, ages 25 to 39 that were not being treated for any systemic disease or taking any prescription or non-prescription medications. Subjects were asked to refrain from any food or drink 2 hr prior to donating saliva and spit 2 ml of saliva directly into the saliva collection tube. Saliva samples were pooled together and centrifuged at 2,600 × g for 10 min to spin down large debris and eukaryotic cells. One ml of this pooled saliva sample was inoculated into 5 ml of BHI medium. The cultures were incubated under anaerobic condition at 37°C until turbid. Frozen stocks of cultured microbiota were prepared by adding glycerol to the samples to a final concentration of 25% and stored at −80° C as the stock of BHI-cultivable S-mix, which was used in current study. Isolation and identification of major bacterial species within the S-mix was performed as previously described [16].
Co-aggregation of F. nucleatum and S. sanguinis
Co-aggregation assays were performed in modified co-aggregation buffer (CAB) containing 150 mM NaCl, 1 mM Tris/HCl pH 8, 0.1 mM CaCl2, and 0.1 mM MgCl2 as previously described [29]. NaN3 was not included in CAB due to its bactericidal effect. Bacterial cells were collected in mid-exponential phase of growth, washed and re-suspended in reduced CAB to a final OD600nm of 1. Equal volumes of each bacterial test species were added to the reaction tube, vortex-mixed for 10 sec, and the suspensions were allowed to settle at room temperature under anaerobic condition for 10 min to allow the bacteria to attach to each other.
H2O2 production assay
F. nucleatum wild type and mutant strains, O-mix and the original mice oral isolate S. sanguinis-OI101 were grown to the mid-exponential growth phase, cells were harvested and resuspended in fresh BHI medium to a final OD600 of 1. The O-mix was mixed with different bacterial test strains or F. nucleatum co-aggregated with S. sanguinis at a 10:1 ratio before being subjected to the H2O2 production assay.
The H2O2 production assay was performed as previously described [30]. Briefly, spots of 5 µl of Horseradish peroxidase (1 mg/ml) (Thermo scientific, Rockford, IL, USA) were added onto a BHI agar plate containing 1 mg/ml leuco crystal violet (Acros Organics, New Jersey, USA). After the liquid was absorbed into the agar, 5 µl of the above mixtures were inoculated on top of the peroxidase containing spots. Plates were incubated overnight at 37°C under anaerobic condition and then inspected for the development of a purple color in and around the colony.
Quantitative measurement of H2O2 concentration in bacterial culture
The H2O2 concentration in bacterial co-cultures of O-mix or S-mix with F. nucleatum (pre-aggregated with and without S. sanguinis) was measured as previously described [31]. Briefly, 100 µl of bacterial co-culture was subjected to centrifugation at 14,000 ×g for 3 min. Twenty µl of supernatant or a 1:2 serial dilution of supernatant with PBS was mixed with 20 µl of horseradish peroxidase (0.5 mg/ml) and 50 µl buffer solution (0.1 mg/ml of 3,3',5,5'-tetramethylbenzidine, 0.05 M phosphate citrate buffer at pH 5.0). The mixture was incubated at room temperature for 1 min, and 100 µl of 2M H2SO4 was added to stop the reaction. Color intensity was measured at 420 nm using micro-plate reader.
H2O2 susceptibility tests
The MICs (minimal inhibitory concentrations) for H2O2 were determined for F. nucleatum and major bacterial species within O-mix by the following dilution method: a two-fold dilution series was prepared in BHI for H2O2 concentrations between 0 and 5 mM. One hundred µl of each dilution was added into individual wells of a 96 well plate. Exponentially growing cultures of each isolate were harvested, resuspended, diluted to 106 cfu/ml in fresh BHI medium, and 100 µl were seeded into the wells containing different concentrations of H2O2. Plates were incubated at 37°C overnight. MIC was defined as the lowest concentration that inhibited visible growth of bacteria.
F. nucleatum cell membrane preparation
The F. nucleatum cell membrane was prepared as previously described [25]. Briefly, exponentially growing F. nucleatum cells were harvested, the cell pellet was washed twice with PBS and resuspended in lysis buffer (50 mM Tris/HCl pH 8.0, 10 mM β-mercaptoethanol, 1 mM EDTA, and 50 mM NaCl). Cells were passed three times through a French Press at 20,000 psi. Cell debris was removed by centrifugation (10,000 × g at 4°C for 10 min). Membranes were pelleted by ultracentrifugation at 150,000 × g for 30 min at 4°C, re-suspended in buffer and stored at −80°C. Membrane protein concentrations were determined with the BCA protein assay kit (Pierce, Rockford, IL).
Pronase treated membrane was prepared by adding pronase (with a final concentration of 1 mg/ml) to the F. nucleatum membrane fraction obtained from ~1010 cells. The sample was incubated at 37°C for 3 hrs, followed by centrifugation at 150,000 × g for 30 min at 4°C. The pellet was resuspended in buffer and stored at −80°C for later use.
Heat and pronase treatments of F. nucleatum cells
Heat and pronase treatments of F. nucleatum cells were performed as previously described [25]. Briefly, exponentially growing F. nucleatum cells were harvested, washed two times, resuspended in PBS to an OD600nm of 1 prior to the following treatments: 1) Heat-treatment: cells were incubated at 90°C for 15 min; 2) Pronase treatment: Pronase was added to the cell suspension to a final concentration of 1 mg/ml, and the sample was incubated at 37°C for 3 hr. After the above treatments, cells were spun down, washed three times and resuspended in fresh PBS to a final OD600nm of 1.
Two-chamber assay
O-mix, S. sanguinis, F. nucleatum wild type and its mutant derivative lacking radD were grown to the exponential growth phase, harvested and resuspended in fresh supplemented BHI medium to OD600nm of 1. The two-chamber assay was performed as previously described [15]. Briefly, 2 ml of the O-mix was inoculated into the bottom chamber of a 12-well plate containing a 0.4-µm PET membrane insert (Millipore, Billerica, MA), while 0.7 ml of wild-type F. nucleatum was added to the top chamber. Alternatively, the O-mix, together with either F. nucleatum cell membranes, heat-killed, pronase treated or untreated wild type F. nucleatum cells (in a 5:1 O-mix to F. nucleatum ratio), or a coaggregated F. nucleatum and S. sanguinis mixture were added to the bottom chamber, while wild-type F. nucleatum was added to the top chamber. Induction of killing under each condition was monitored by performing viable counts on the wild type F. nucleatum in the upper chamber. Specifically, culture samples were taken periodically, vortexed for 1 min, subjected to serial dilution and plated onto selective plates. Plates were incubated for 4 days at 37°C under anaerobic condition before colonies were counted.
Statistical Analysis
Significance of differences between average values was analyzed by Student's t-tests.
Results
O-mix killed F. nucleatum by generating H2O2 upon physical contact
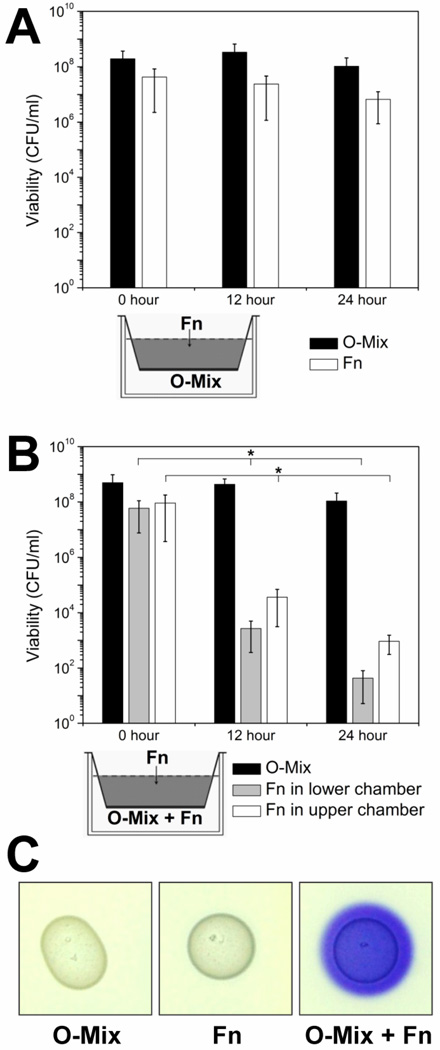
Since the ability of F. nucleatum to interact with a variety of Gram-negative as well as Gram-positive oral microbes has been demonstrated in vitro predominantly on a one-on-one level[10], little is known regarding the mechanisms involved in its integration into an existing oral community. Previous studies have shown that a biofilm community comprised of cultivable mice oral microbial microbiota (O-mix), which is primarily comprised of Gram-positive bacterial species that are closely related to human oral species (Suppl. Table 1) has the ability to prevent community integration of Gram-negative “intruders” such as E. coli [16]. Intrigued by this observation, we employed the same two-chamber assay developed for that study, to investigate the process of F. nucleatum integration into a preexisting oral community. F. nucleatum persisted well when it was physically separated by a membrane from the O-mix cells while sharing the same culture medium (Fig. 1A). However, when the lower chamber contained both O-mix and F. nucleatum cells, a reduction of more than three orders of magnitude in the viable counts was observed for F. nucleatum cells in both chambers at 12 hours (Fig. 1B), and no viable F. nucleatum cells were detected in both chambers after 48 hr incubation (data not shown). This scenario resembled our previous study in which the same O-mix produced H2O2 upon direct contact to inhibit the growth and prevent the integration of an E. coli gut isolate [15]. To investigate if the same mechanism was responsible for the reduction in F. nucleatum viable counts, we tested H2O2 production of the O-mix in physical contact with F. nucleatum. Similar to E. coli, physical contact with F. nucleatum induced H2O2 production in the O-mix as indicated by the intense purple color, while O-mix alone didn’t produce significant amount of H2O2 (Fig. 1C). The H2O2 susceptibility test showed a MIC of 0.15 mM for F. nucleatum, while most of the oral isolates tested exhibited a higher resistance to H2O2 up to 1.5 mM (Suppl. Table 2).
Figure 1. Contact-dependent killing of F. nucleatum by the O-mix.
(A) Two-chamber assay in which F. nucleatum (Fn) was inoculated into the upper chamber and physically separated from the O-mix in the lower chamber by a 0.4 µm pore size membrane (as shown in the small diagram). Viability of F. nucleatum (open bars) and O-mix (solid bars) was tracked every 12 hours by determining viable counts.
(B) Two-chamber assay in which F. nucleatum was added to the upper chamber, while O-mix, together with F. nucleatum (at a 10:1 ratio) was inoculated into the lower chamber. Viability was monitored for O-mix and F. nucleatum in both chambers (black solid bars represent total viable counts of O-mix, grey bars and open bars represent viable count of F. nucleatum in the lower and upper chamber, respectively). Two independent experiments were performed in three replicates each, and average values ± SD are shown. The asterisk indicates that the values of 12 and 24 hour were significantly lower than the one for 0 hour (Student’s t-test p value < 0.05).
(C) F. nucleatum triggers the release of H2O2 in O-mix cells. O-mix and wild type F. nucleatum were spotted individually or mixed together on BHI agar plates containing leuco crystal violet and Horseradish peroxidase, purple color development was monitored after overnight incubation. Three replicates were performed and a representative result is shown.
F. nucleatum membrane associated protein(s) are involved in triggering H2O2 production in the O-mix
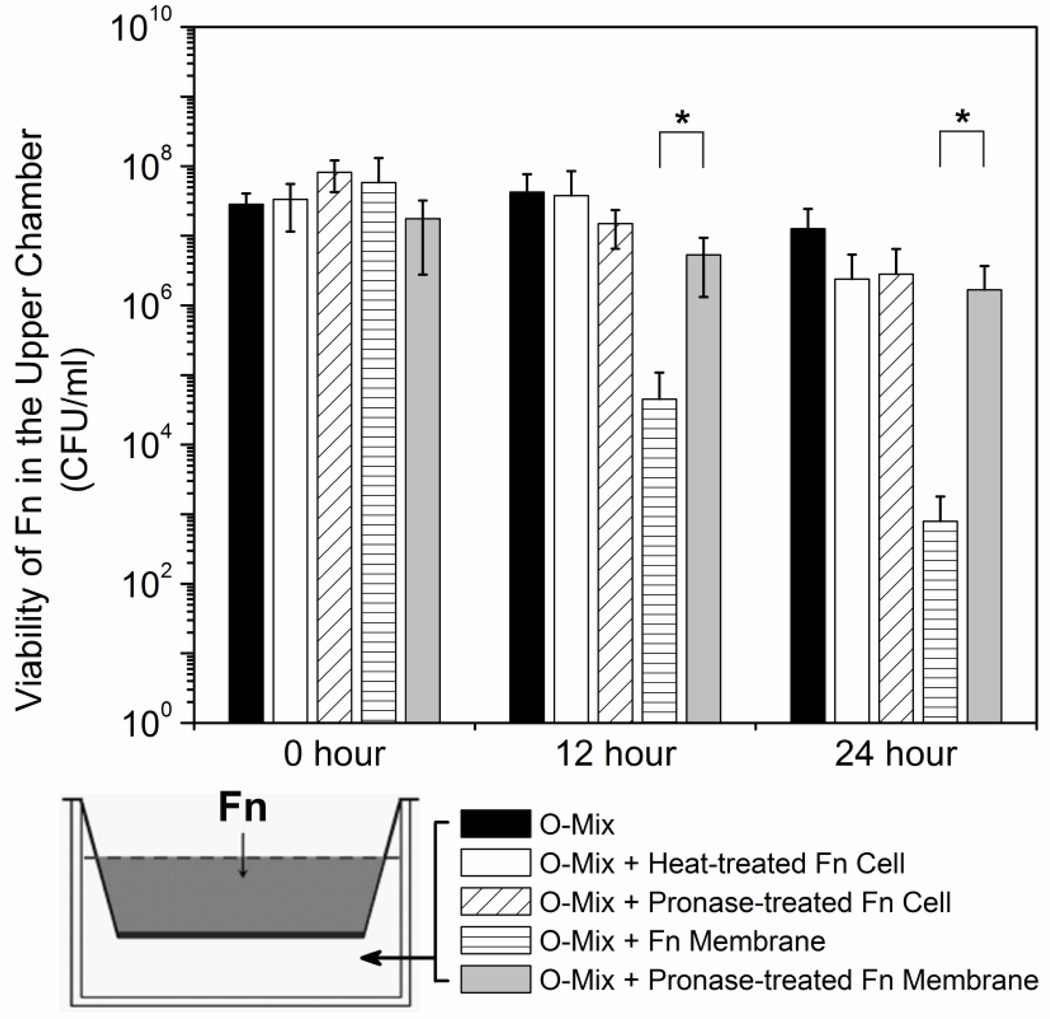
The results of the two-chamber and H2O2 production assays indicated that the O-mix cells generate H2O2 upon physical contact with F. nucleatum cells. The following experiments were performed to examine if the triggering component(s) are located on the F. nucleatum cell surface and to distinguish if they are surface proteins, exopolysacchrides (EPS) or LPS: heat-killed or pronase-treated F. nucleatum cells were added to the lower chamber of the two-chamber setup together with O-mix cells, and the viability of wild type F. nucleatum in the upper chamber was monitored periodically. Addition of heat-killed or pronase treated whole cells of F. nucleatum did not result in any significant killing of the F. nucleatum cells in the upper chamber (Fig. 2), indicating that heat-sensitive protein(s) were the triggering component(s). Untreated F. nucleatum cell membrane fractions induced killing to a level similar to whole cells; while pronase treatment greatly reduced this phenomenon (Fig. 2). This was consistent with our hypothesis that the triggering components are likely F. nucleatum cell surface proteins. In an initial effort to identify these proteins, we tested the eight F. nucleatum mutant strains defective in individual large outer membrane protein that we constructed previously [25]. However, none of them displayed a significant reduction in the ability to elicit killing (Data not shown).
Figure 2.
Induction of “diffusible killing factor” (H2O2) by F. nucleatum cell fractions. Live wild type F. nucleatum (Fn) cells were added to the upper chamber of the two chamber system, while O-mix alone, or O-mix, together with heat-treated, pronase-treated F. nucleatum cells, F. nulceatum membrane fraction or pronase-treated membranes were inoculated to the lower chamber. Viability of wild type F. nulceatum in the upper chamber in each setup (represented by the different bars) was monitored every 12 hours. Two independent experiments were performed in duplicate each, and average values ± SD are shown. The asterisk indicates significant differences between two values (Student’s t-test p value < 0.05).
Adherence of wild type F. nucleatum to S. sanguinis decreased induction of H2O2 production by the O-mix
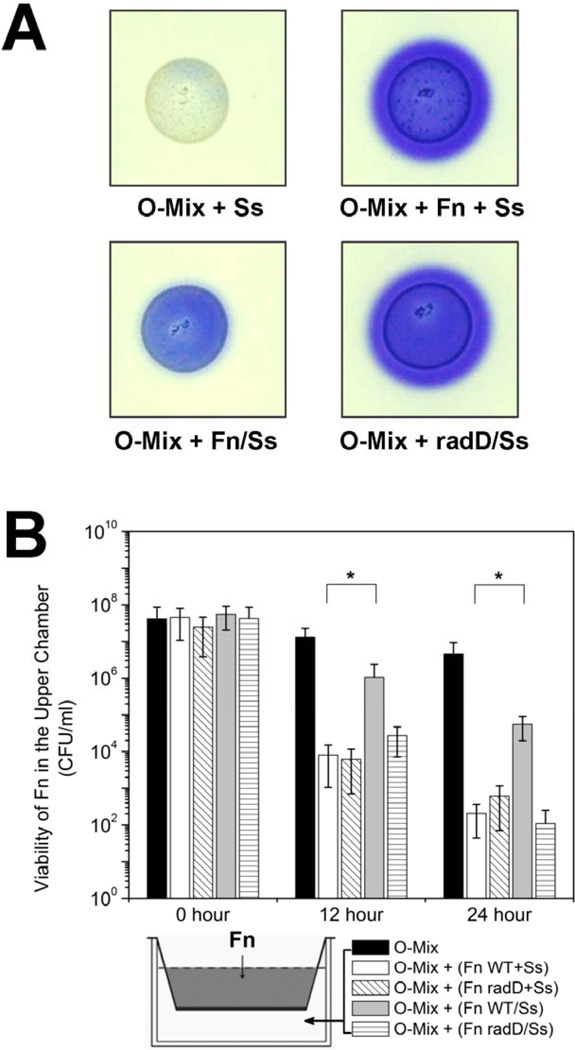
The result in the previous section showed that the O-mix killed wild type F. nucleatum by producing H2O2, which raised an interesting question: how does F. nucleatum manage to integrate and persist in supra-gingival biofilms which are dominated by Gram-positive bacteria? One of the remarkable features of F. nucleatum is its ability to adhere and form aggregates with a number of oral bacteria and thus plays pivotal roles in facilitating the development of oral community [9,10]. We hypothesized that, in addition to facilitating biofilm development, co-adherance with other oral bacteria, such as streptococci, could “mask” the Gram-negative cell surface of F. nucleatum and allow its integration into the O-mix. To test this hypothesis, we allowed wild type F. nucleatum to adhere to one of its known partner species, S. sanguinis prior to addition to the O-mix. Formation of aggregates between the two species was confirmed by phase-contrast microscopy (data not shown). The greatly reduced purple color development in the H2O2 production assay (Fig. 3A) supported our hypothesis that adherence to S. sanguinis significantly decreased induction of this killing agent in the O-mix by F. nucleatum. This finding was corroborated by two-chamber assay data, since F. nucleatum cells in the upper well displayed better survival when F. nucleatum aggregates with S. sanguinis (Fig. 3B) instead of F. nucleatum cells alone (Fig. 1B) were added to the lower chamber together with the O-mix. Meanwhile, the addition of S. sanguinis alone to the lower chamber together with the O-mix did not induce significant killing of F. nucleatum in the upper chamber (data not shown).
Figure 3.
Induction of H2O2 production by F. nucleatum cells with and without adherence to partner species. (A) H2O2 production assay: O-mix samples mixed with either S. sanguinis (Ss), F. nucleatum (Fn) plus S. sanguinis (separately (+), after aggregate formation (/)), or the mutant derivative of F. nucleatum lacking RadD (radD) coincubated with S. sanguinis under coaggregation conditions (/) were spotted on to BHI agar plate containing leuco crystal violet and Horseradish peroxidase. Purple color development was monitored after overnight incubation. Three replicate experiments were performed and a representative result is shown.
(B) Two-chamber assay in which wild type F. nucleatum (Fn) was inoculated into the upper chamber, while O-mix alone, together with wild type F. nucleatum and S. sanguinis (Ss) (separately or after aggregate formation), or the F. nulceatum radD mutant and S. sanguinis (separately or after incubation under aggregation conditions) was inoculated into the lower chamber. Viability of wild type F. nucleatum in the upper chamber of the different setups was monitored. Two independent experiments were performed in three replicates each, and average values ± SD are shown. ‘/’ indicates two species were treated with co-aggregation buffer prior to addition; ‘+’ indicates two species were added at the same time but without prior co-aggregation. The asterisk indicates significant differences between two values (Student’s t-test p value < 0.05).
Furthermore, when F. nucleatum and S. sanguinis were included in the O-mix together without being allowed to adhere to each other prior to addition, H2O2 was still produced at levels similar to the presence of F. nucleatum alone (Fig. 3A, 1C), and the viability of F. nucleatum cells in the upper chamber was significantly reduced (about 3 orders of magnitude) within 12 hour of co-cultivation in the two-chamber assay (Fig. 3B).
The F. nucleatum outer membrane protein RadD is required for the protection mediated by its adherence to partner species
RadD, one of the large fusobacterial outer membrane proteins was previously identified as the arginine-inhibitable adhesin that mediates the adherence of F. nucleatum to early colonizers, including S. sanguinis [25]. To further investigate the protective effect of aggregate formation with S. sanguinis, we tested if a F. nucleatum mutant strain lacking radD would induce H2O2 production in the oral microbiota, since cells carrying this mutation are unable to adhere to a number of early colonizing partner species even under co-aggregation condition [25]. The F. nucleatum radD mutant strain was mixed with S. sanguinis in co-aggregation buffer prior to addition to the O-mix. H2O2 production was monitored and the levels observed were similar to those measured upon the addition of wild type F. nucleatum cells alone (Fig. 3A). These results correlated with the two-chamber assay in which the mixture of the radD mutant and S. sanguinis treated under co-aggregation conditions prior to addition to the O-mix in the lower chamber resulted in a drastic reduction in viable counts of wild type F. nucleatum in the upper chamber (Fig. 3B). Microscopic observation confirmed that the radD mutant failed to form aggregates with S. sanguinis even upon co-incubation in co-aggregation buffer (data not shown).
Adherence to S. sanguinis renders F. nuleatum less sensitive to killing by H2O2
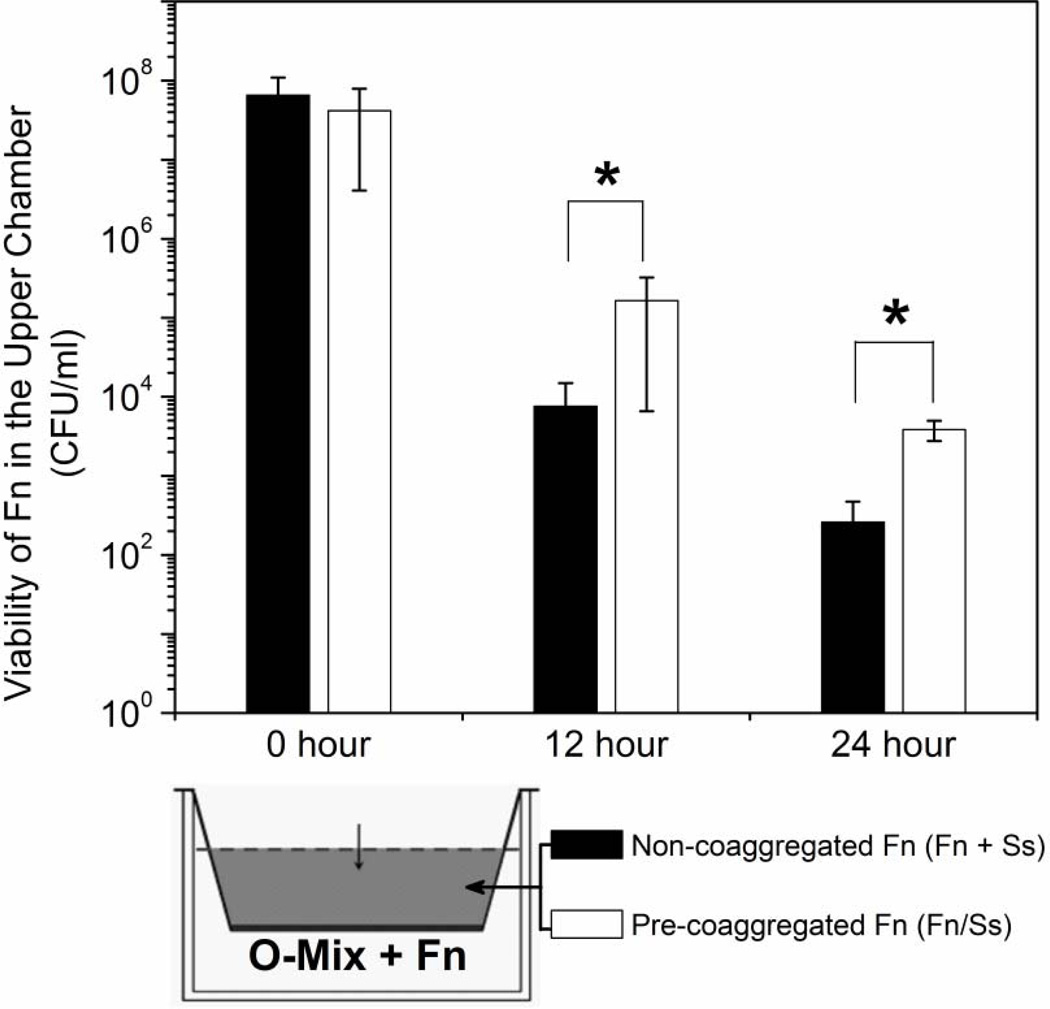
F. nucleatum adherence to S. sanguinis significantly reduced induction of H2O2 production in the O-mix (Fig. 3A). Next, we investigated if adherence to its interacting partner S. sanguinis can alter the sensitivity of F. nucleatum to the killing molecules generated by the O-mix using the two-chamber assay. F. nucleatum and S. sanguinis cells were added separately but at the same time or after aggregate formation to the upper chamber, while F. nucleatum alone was mixed with O-mix in the lower chamber to induce H2O2 production. Samples in the upper chamber were taken periodically for quantitative measurement of H2O2 levels and monitoring of F. nucleatum viability. While there was no significant difference in H2O2 concentration (~ 0.35 mM) between the two setups (data not shown), adherence of F. nucleatum to S. sanguinis prior to addition to the upper well resulted in an increased survival rate compared to F. nucleatum cells that were not allowed to form aggregates with its partner species (Fig. 4).
Figure 4.
Survival of F. nucleatum in aggregates with S. sanguinis and non-aggregated cells. The two chamber assay in which aggregated (Fn/Ss) and non-aggregated (Fn+Ss) F. nucleatum and S. sanguinis cells were added to the upper chamber, while F. nucleatum cells alone were co-cultured with O-mix in the lower chamber. Samples in the upper chamber were taken periodically to monitor F. nucleatum viability. Two independent experiments were performed in three replicates each, and average values ± SD are shown. The asterisk indicates significant differences between two values (Student’s T-test P value < 0.05).
Adherence to S. sanguinis resulted in better survival of F. nucleatum within a human saliva-derived microbial community (S-mix)
The above data suggested that aggregate formation with partner species such as S. sanguinis through adherence enables F. nucleatum to evade detection and killing by the O-mix community isolated from mice. To confirm that this mechanism is involved in protecting F. nucleatum cells and facilitate their integration into the human oral microbial community, we cultivated an in vitro S-mix from human saliva samples. Similar to the O-mix that was used for the majority of experiments performed in this study, the S-mix is mainly comprised of Gram-positive bacteria with a significant overlap in species (Suppl. Table 1) including Streptococci and Lactobacilli and the presence of F. nucleatum triggered a higher level of H2O2 production in S-mix as well (Suppl. Table 1).
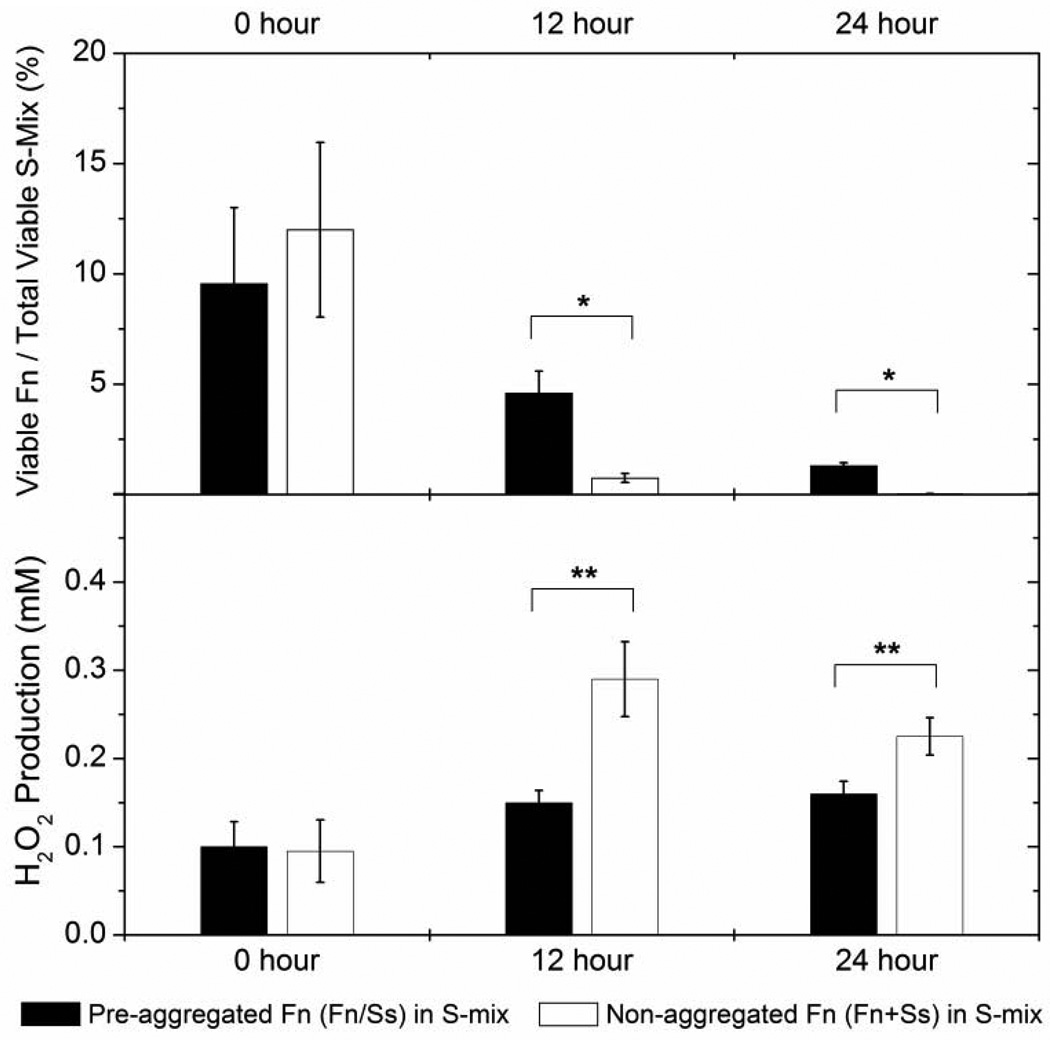
To further investigate the effect of co-aggregation with its partner on the survival of F. nucleatum within S-mix, co-aggregation buffer treated F. nucleatum mixtures with the salivary isolate S. sanguinis SI-101 were added to the S-mix in a 1:10 ratio. Co-cultures were incubated at 37°C, H2O2 concentration was measured periodically and viability of F. nucleatum was monitored every 12 hours. Colony counting data revealed that, when co-cultivated with S-mix, F. nucleatum cells alone suffered a drastic reduction in their viable count. The proportion relative to the total oral bacteria count reduced from around 10% to less than 0.5% within 24 hours of co-cultivation; while no significant change in the total bacterial count was apparent. Meanwhile, the viability decline of F. nucleatum forming aggregates with S. sanguinis within S-mix was significantly less compared to that of F. nucleatum cells alone (Fig. 5A). Quantitative H2O2 measurement revealed that co-cultivation of F. nucleatum alone with S-mix resulted in a much higher production of H2O2 by S-mix compared to the co-culture containing aggregates of F. nucleatum with S. sanguinis cells (Fig.5B).
Figure 5.
Survival of F. nucleatum aggregated and non-aggregated with S. sanguinis within S-mix. A: co-aggregation buffer treated (Fn/Ss) and non-treated (Fn+Ss) F. nucleatum and S. sanguinis SI-101 mixture were co-cultivated with S-mix in a 1:10 ratio in cell numbers. The viability of F. nucleatum (Fn) and S-mix was monitored every 12 hours. The data were expressed as percentage of viable count of F. nucleatum to the total viable count of S-mix. B: Quantitative measurement of H2O2 production. The H2O2 concentration within the co-cultures was monitored every 12 hours. Two independent experiments were performed in three replicates each and average values ± SD are shown. The asterisk indicates significant differences between two values (Student’s T-test P value < 0.05).
Discussion
The spatiotemporal model proposed that the establishment of the oral community is a highly coordinated and tightly regulated event [9], with adhesion of bacterial cells to each other being one of the crucial factors determining the presence of certain species within the structure of multispecies communities [10]. In addition, successful integration into a community requires that the incoming bacterial species are able to overcome the colonization resistance developed by the existing community [14,15,16,32]. In this study, we investigated community integration of F. nulceatum, a Gram-negative oral bacterial species that performs an important function during microbial community formation through its ability to function as a “bridging” organism connecting the early and late colonizing microorganisms [9]. This role as a bridging organism was recently supported by an in vivo study that confirmed the localization of F. nucleatum in the middle layer of tooth-attached human plaque samples [13]. In addition to its function in facilitating recruitment of late colonizing species which comprise a number of periodontal pathogens, F. nucleatum likely plays an active role in pathogenesis due to its ability to induce apoptosis in lymphocytes [33]. While these multifaceted tasks within the oral biofilm render F. nucleatum a key community member, the mechanism allowing it to overcome the integration resistance displayed by predominantly Gram-positive oral communities comprised of early colonizing species still remains to be investigated.
Previous studies have shown that the mere ability of a species to attach to members of an existing community is not sufficient to become part of a pre-existing biofilm. To successfully integrate into a community, an incoming bacterial species needs to overcome several “barriers”, one of them being the colonization resistance developed by the rest of the community [14]. The colonization resistance could be the competition for colonization sites, or more importantly, the inhibitory effect exerted by other microbes within the community via the production of inhibitory compounds [15,34]. The data presented here revealed that wild type F. nucleatum suffered a great loss in viability upon addition to cultivable mice oral microbial community (O-mix) (Fig. 1B). This reduced survival in the presence of the O-mix was due to contact induced production of H2O2, a mechanism found to be involved in the community resistance against integration of E. coli, a Gram-negative species that was not a commensal oral bacterium [15]. F. nucleatum is more sensitive to this bactericidal agent than the Gram-positive isolates present in the O-mix and therefore being killed at concentrations that do not affect the rest of the community (suppl. Table 2). Further analysis showed that, unlike E. coli which triggered the production of H2O2 by O-mix via its membrane associated LPS [15], the triggering component(s) of F. nucleatum are likely membrane associated proteins (Fig. 2) which yet remain to be identified.
To facilitate their integration into existing microbiota, bacteria have evolved diverse strategies to overcome community based colonization resistances. For example, instead of competing for abiotic colonization sites, some bacteria adhered to other resident bacteria to achieve the first step in integration [35,36]; in other cases, certain bacterial species neutralized the inhibitory compound, or converted it to substances that are toxic to competing organisms [11,37,38]. Our data indicate that F. nucleatum overcomes community resistance by adhering to oral streptococci, a strategy resembling the one employed by many bacterial symbionts or pathogens to evade the detection by host defense systems. For example, as a commensal bacterium of the human gastrointestinal tract, Bacteroides fragilis is able to decorate its surface capsule polysaccharides and glycoproteins with L-fucose, an abundant surface molecule of intestinal epithelial cells, thus evading being recognized as “foreign” [39]. Many bacterial pathogens carry complement regulator-binding proteins, such as the factor H-binding protein (fHbp) in Neisseria meningitides [40], the outer surface protein E (OspE) in Borrelia burgdorferi [41], and the pneumococcal surface protein C (PspC) in Streptococcus pneumoniae [42]. Through these proteins, pathogens are able to recruit and “disguise” themselves with host complement regulators, and are recognized by the host as self and protected from complement-mediated killing.
Our study provided an intriguing addition to the strategies used by bacterial species to survive and integrate themselves into a community. Adherence of F. nucleatum to S. sanguinis significantly reduced the induction of H2O2 production in the O-mix (Fig. 3), likely by “masking” the triggering surface component(s) as a result of binding between the partner species. This “masking” effect was lost in the F. nucleatum mutant lacking radD which encodes an outer membrane protein mediating the adhesion between F. nucleatum and early colonizers, including S. sanguinis (Fig.3). At the same time, binding to S. sanguinis increased the resistance F. nucleatum to H2O2 (Fig.4). It has been shown that adhesion with Actinomyces naeslundii protected Streptococcus gordonii from oxidative damage due to catalase production by A. naeslundii [43]. It is unlikely that a similar mechanism could be responsible for the protective effect we observed in this study since no catalase activity has been reported in S. sanguinis. However, increasing evidence suggests that binding between two oral bacterial species could result in different gene expression pattern which could enhance survival within a multispecies community. For example, Streptococcus gordonii up-regulated several genes when co-aggregated with Actinomyces oris compared with co-culture [44]; while a proteomics analysis of Porphyromonas gingivalis revealed that, when co-aggregated with F. nucleatum and S. gordonii, 403 proteins were down regulated and 89 proteins were up-regulated compared to the cultures of P. gingivalis alone [45]. When adhered to F. nucleatum or S. gordonii, many DNA repair-related proteins are down regulated in P. gingivalis, while PGN0090, a phoH family protein with proposed function in oxidation protection increased in abundance [45]. Furthermore, a recent microarray analysis comparing planktonic cells of F. nucleatum with those autoaggregated by the presence of saliva showed that nearly 100 genes were differentially regulated after 60 min of aggregation [46]. Auto-aggregation was suggested to activate a transcriptional response that could prepare cells for growing in the high cell density environment of oral biofilms. Collectively, these studied showed that there is a far more complex cellular response after cell-cell contact or aggregate formation. The change in post-aggregational gene expression could prepare bacterial species to better cope with the environmental stress experienced within the multispecies communities.
The data obtained from human salivary microbiota (S-mix) study (Fig. 5) further supported our hypothesis that adherence with early colonizer, such as S. sanguinis could facilitate F. nucleatum integration into Gram-positive bacteria dominated supragingival microbial community. Although binding between F. nucleatum and S. sanguinis was achieved under artificial condition in our in vitro systems, the physical association of F. nucleatum and other bacteria, including streptococci in vivo has been well documented [13,47]. Within dental plaque, F. nucleatum is often found in “corncob” formations with Streptococci, a structure that was originally regarded to serve as a connecting link between gram-positive dominated supragingival and gram-negative dominated subgingival plaque [47]. Our study suggested that adherence of F. nucleatum to Streptococci facilitates integration into oral communities that are dominated by gram-positive bacteria (supragingival or initial subgingival communities). Being decorated with streptococci could help F. nucleatum evade detection by oral species which respond to the presence of F. nucleatum alone by the production of H2O2. Adherence to its partner species could also trigger a specific cellular response in F. nucleatum cells and result in increased resistance to environmental stress, such as the exposure to H2O2.
Due to the limitation of culture-dependent methods, the results we obtained using an in vitro system can not entirely represent the real situation in the oral cavity. We would expect a far more complex process during the integration of F. nucleatum into the oral community, which might involve extensive bacterial inter-species and bacteria-host interactions. Nevertheless, our results provide important insights into one of the possible mechanisms by which the “bridging” bacterium, F. nucleatum integrates into supragingival or initial subgingival communities dominated by Gram-positive species.
Acknowledgement
The study was supported by US National Institutes of Health (NIH) Grants DE020102 and DE021108 to R. Lux and W. Shi.
References
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 4.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahlén G. Role of suspected periodontopathogens in microbiological monitoring of periodontitis. Adv Dent Res. 1993;7:163–174. doi: 10.1177/08959374930070020701. [DOI] [PubMed] [Google Scholar]
- 6.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 7.Nishihara T, Koseki T. Microbial etiology of periodontitis. Periodontology 2000. 2004;36:14–26. doi: 10.1111/j.1600-0757.2004.03671.x. [DOI] [PubMed] [Google Scholar]
- 8.Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems1. Annu Rev Microbiol. 2000;54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 9.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, et al. Communication among oral bacteria. Microbiol Mol Biol Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell–cell distance. Nat Rev Micro. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 11.Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolenbrander PE, Egland PG, Diaz PI, Palmer RJ., Jr Genome–genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 2005;13:11–15. doi: 10.1016/j.tim.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Zijnge V, van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, et al. Oral biofilm architecture on natural teeth. PLoS ONE. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vollaard E, Clasener H. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–414. doi: 10.1128/aac.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X, Tian Y, Guo L, Lux R, Zusman D, et al. Oral-derived bacterial flora defends its domain by recognizing and killing intruders—a molecular analysis using Escherichia coli as a model intestinal bacterium. Microb Ecol. 2010b;60:655–664. doi: 10.1007/s00248-010-9708-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X, Tian Y, Guo L, Ano T, Lux R, et al. In vitro communities derived from oral and gut microbial floras inhibit the growth of bacteria of foreign origins. Microb Ecol. 2010a;60:665–676. doi: 10.1007/s00248-010-9711-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun J, Kim K, Lee J-H, Choi Y. The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol. 2010;10:101. doi: 10.1186/1471-2180-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott DR, Wilson M, Buckley CMF, Spratt DA. Cultivable oral microbiota of domestic dogs. J Clin Microbiol. 2005;43:5470–5476. doi: 10.1128/JCM.43.11.5470-5476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Citron Diane M. Update on the taxonomy and clinical aspects of the genus Fusobacterium. Clin Infect Dis. 2002;35:S22–S27. doi: 10.1086/341916. [DOI] [PubMed] [Google Scholar]
- 20.Guggenheim B, Giertsen E, Schüpbach P, Shapiro S. Validation of an in vitro biofilm model of supragingival plaque. J Dent Res. 2001;80:363–370. doi: 10.1177/00220345010800011201. [DOI] [PubMed] [Google Scholar]
- 21.Hintao J, Teanpaisan R, Chongsuvivatwong V, Ratarasan C, Dahlen G. The microbiological profiles of saliva, supragingival and subgingival plaque and dental caries in adults with and without type 2 diabetes mellitus. Oral Microbiol Immunol. 2007;22:175–181. doi: 10.1111/j.1399-302X.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- 22.Palmer RJJ, Diaz PI, Kolenbrander PE. Biocomplexity in the oral cavity – the basics of structure in supragingival bacterial communities. Biofilms. 2004;1:329–335. [Google Scholar]
- 23.Zaura E, Keijser B, Huse S, Crielaard W. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haffajee AD, Socransky SS, Patel MR, Song X. Microbial complexes in supragingival plaque. Oral Microbiol Immunol. 2008;23:196–205. doi: 10.1111/j.1399-302X.2007.00411.x. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan CW, Lux R, Haake SK, Shi W. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mole Microbiol. 2009;71:35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan CW, Lux R, Huynh T, Jewett A, Shi W, et al. Fusobacterium nucleatum apoptosis-inducing outer membrane protein. J Dent Res. 2005;84:700–704. doi: 10.1177/154405910508400803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human Lymphocytes. Infect Immun. 2010;78:4773–4778. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Y, He X, Torralba M, Yooseph S, Nelson KE, et al. Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol Oral Microbiol. 2010;25:357–367. doi: 10.1111/j.2041-1014.2010.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cisar JO, Kolenbrander PE, McIntire FC. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun. 1979;24:742–752. doi: 10.1128/iai.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreth J, Merritt J, Shi W, Qi F. Competition and Coexistence between Streptococcus mutans and Streptococcus sanguinis in the Dental Biofilm. J Bacteriol. 2005;187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uehara Y, Kikuchi K, Nakamura T, Nakama H, Agematsu K, et al. H2O2 produced by viridans group Streptococci may contribute to inhibition of methicillin-resistant Staphylococcus aureus colonization of oral cavities in newborns. Clin Infec Dis. 2001;32:1408–1413. doi: 10.1086/320179. [DOI] [PubMed] [Google Scholar]
- 32.Waaij Dvd, Berghuis-de Vries JM, Lekkerkerk-van der Wees JEC. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) 1971;69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan CW, Lux R, Huynh T, Jewett A, Shi W, et al. Fusobacterium nucleatum Apoptosis-inducing outer membrane protein. J Dent Res. 2005;84:700–704. doi: 10.1177/154405910508400803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liévin V, Peiffer I, Hudault S, Rochat F, Brassart D, et al. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut. 2000;47:646–652. doi: 10.1136/gut.47.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamont R, Jenkinson H. Adhesion as an ecological determinant in the oral cavity. In: Kuramitsu KK, Ellen RP, editors. Oral bacterial ecology: the molecular basis. Norfolk: Horizon Scientific Press; 2000. pp. 131–168. [Google Scholar]
- 36.Kolenbrander PE, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi N. Acid-neutralizing activity during amino acid fermentation by Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol. 2003;18:109–113. doi: 10.1034/j.1399-302x.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 38.Tong H, Chen W, Merritt J, Qi F, Shi W, et al. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol. 2007;63:872–880. doi: 10.1111/j.1365-2958.2006.05546.x. [DOI] [PubMed] [Google Scholar]
- 39.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 40.Pizza M, Donnelly J, Rappuoli R. Factor H-binding protein, a unique meningococcal vaccine antigen. Vaccine. 2008;26:I46–I48. doi: 10.1016/j.vaccine.2008.11.068. [DOI] [PubMed] [Google Scholar]
- 41.Hellwage J, Meri T, Heikkilä T, Alitalo A, Panelius J, et al. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J Biol Chem. 2001;276:8427–8435. doi: 10.1074/jbc.M007994200. [DOI] [PubMed] [Google Scholar]
- 42.Janulczyk R, Iannelli F, Sjöholm AG, Pozzi G, Björck L. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J Biol Chem. 2000;275:37257–37263. doi: 10.1074/jbc.M004572200. [DOI] [PubMed] [Google Scholar]
- 43.Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66:637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakubovics NS, Gill SR, Iobst SE, Vickerman MM, Kolenbrander PE. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J Bacteriol. 2008;190:3646–3657. doi: 10.1128/JB.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuboniwa M, Hendrickson E, Xia Q, Wang T, Xie H, et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98. doi: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merritt J, Niu G, Okinaga T, Qi F. Autoaggregation response of Fusobacterium nucleatum. Appl Environ Microbiol. 2009;75:7725–7733. doi: 10.1128/AEM.00916-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lancy PJ, Dirienzo JM, Appelbaum B, Rosan B, Holt SC. Corncob formation between Fusobacterium nucleatum and Streptococcus sanguis. Infect Immun. 1983;40:303–309. doi: 10.1128/iai.40.1.303-309.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]