Abstract
A homology search of wheat chloroplast (ct) and mitochondrial (mt) genomes identified 54 ctDNA segments that have homology with 66 mtDNA segments. The mtDNA segments were classified according to their origin: orthologs (prokaryotic origin), xenologs (interorganellar DNA transfer origin) and paralogs (intraorganellar DNA amplification origin). The 66 mtDNA sequences with homology to ctDNA segments included 14 paralogs, 18 orthologs and 34 xenologs. Analysis of the xenologs indicated that the DNA transfer occurred unidirectionally from the ct genome to the mt genome. The evolutionary timing of each interorganellar DNA transfer that generated a xenolog was estimated. This analysis showed that 2 xenologs originated early in green plant evolution, 4 in angiosperm evolution, 3 in monocotyledon evolution, 9 during cereal diversification and 8 in the evolution of wheat. Six other xenologs showed recurrent transfer from the ct to mt genomes in more than one taxon. The two remaining xenologs were uninformative on the evolutionary timing of their transfer. The wheat mt nad9 gene was found to be chimeric, consisting of the cereal nad9 gene and its 291 bp 5′-flanking region that included a 58 bp xenolog of the ct-ndhC origin.
Keywords: organellar DNAs, recurrent ctDNA transfers, unidirectional transfer, timing of transfer, chimeric gene
Introduction
The cells of green plants contain three different genomes, nuclear, plastid (chloroplast) and mitochondrial. These genomes are autonomous genetic entities that are independent of each other and have their own genetic information. This autonomy, however, is not rigid, as interorganellar DNA transfer can occur between the three genomes,1) in addition to horizontal gene transfer from alien species.2)
Research into interorganellar DNA transfer in plants was initiated by the discovery of a common 12 kb DNA sequence present in the mitochondrial (mt) and chloroplast (ct) genomes of maize.3) Since then, other types of interorganellar DNA transfer have been identified, namely, ctDNA transfer to the mitochondria,4,5) nuclear DNA (abbreviated as ncDNA) transfer to the mitochondria,6) mtDNA transfer to the nucleus7) and ctDNA transfer to the nucleus.8) To date, no cases of mt or ncDNA transfer to the chloroplast have been reported.9)
The complete nucleotide sequences of the ct and mt genomes have been determined in a large number of green plants. Since the first description of the ct genomes of liverwort and tobacco,10,11) more than 100 species have now been sequenced. Similarly, the mt genomes of about 50 species have been described following the first report in liverwort.12) These sequence data now enable us to carry out genome-wide investigations of interorganellar DNA transfer during plant evolution.
We recently completed sequencing both the ct and mt genomes of wheat,13,14) and found that approximately 6% of the mtDNA sequences were homologous to those of the ctDNA.14) The present work was planned to clarify the nature and origin of the homologous DNA segments in the wheat ct and mt genomes. First, the homologous DNA sequences in the two organellar genomes were enumerated. Next, these sequences were classified by their origin into three categories, “paralog” (originating by amplification of a template sequence in an organellar genome), “ortholog” (descending from a common ancestral sequence) and “xenolog” (originating from interorganellar DNA transfer). The detailed sequence analysis of the xenologs confirmed nine cases of recurrent DNA transfer during wheat phylogeny, with the same region of the ct genome being transferred to different sites in the mt genome at different evolutionary times. No examples of transfer from the mt to the ct genome were found, confirming that interorganellar DNA transfer was unidirectional, i.e., only from the ct to mt genome. Finally, I estimated the age of the ctDNA transfers during wheat evolution based on the distribution of the same mtDNA segments transferred from the ct genome among divergent plant taxa: the oldest transfers occurred at an early stage of green plant emergence, the newest ones after divergence of the wheat lineage from other monocotyledons, and the remainder at intermediate evolutionary times. In one case, a xenolog contributed to the generation of a new nad9 gene in the wheat mt genome.
Materials and data analysis
Screening for homologous nucleotide sequences.
Homologous sequences between the subject and query DNA sequences were screened using the BLAST 2 version BLASTn 2.2.24+ program (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi?). Homologies between the wheat ct and mtDNAs were screened using the ct and mt genome sequences (Accession no. NC_002762 and Acc. no. NC_007579)15,14) as the subject and query sequences, respectively. To screen other genomes for homologies to the 52 wheat mtDNA sequences that had homology to ctDNA segments, the subject sequence was always the wheat mtDNA sequence and the query sequence was one of the following: the complete mt genome sequence of rice (Oryza sativa ssp. japonica, Acc. no. NC_011033),16) maize (Zea mays ssp. mays, Acc. no. NC_007982),17) sorghum (Sorghum bicolor, Acc. no. NC_008360),18) thale cress (Arabidopsis thaliana, Acc. no. NC_001284),19) sugar beet (Beta vulgaris ssp. vulgaris, Acc. no. NC_002511),20) tobacco (Nicotiana tabacum, Acc. no. NC_006581),21) a gymnosperm (Cycas taitungensis Acc. no. NC_010303),22) moss (Physcomitrella patens, Acc. no. NC_007945),23) liverwort (Marchantia polymorpha, Acc. no. NC_001660)12) and green alga (Chlorokybus atmophyticus, Acc. no. NC_009630),24) and the complete genome sequence of two prokaryotes, a cyanobacterium (Prochlorococcus marinus str. MIT 9312, Acc. no. CP000111)25) and an α-proteobacterium (Candidatus Pelagibacter ubique HTCC1062, Acc. no. CP000084).26)
Origin of homologous organellar DNA segments and principles for determining origin.
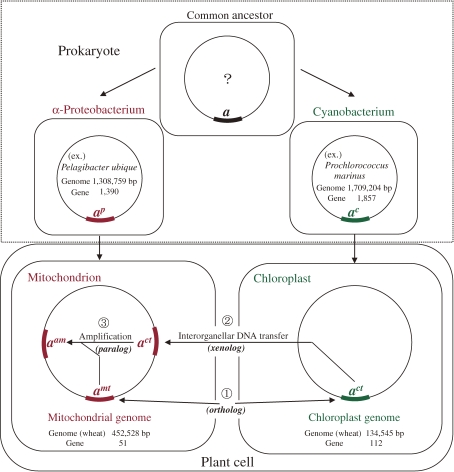
Sequence homology between ct and mtDNA segments may occur by three mechanisms, as diagrammatically shown in Fig. 1. We assume that present day bacteria were derived from a single ancestral prokaryote. In addition, the diagram assumes that the ct and mt genomes of green plants originated from a cyanobacterium and an α-proteobacterium, respectively.27–30) Here, I selected Prochlorococcus marinus (abbreviated as “Pm”) and Candidatus Pelagibacter ubique (abbreviated as “Pu”) as representative of the respective prokaryotes for the reasons given in our previous article.31)
Figure 1.
Three mechanisms for the origin of ctDNA-homologous mtDNA segments: (1) vertical transmission of a prokaryote DNA segment (orthology), (2) interorganellar DNA transfer from the ct to mt genome (xenology) and (3) amplification of an orthologous or xenologous DNA segment in the mitochondrion (paralogy). Circle: prokaryote genome and plant organellar genome. Thick arc on DNA molecule: homologous DNA segment descended from the prokaryote genome or by xenology or paralogy.
One of the mechanisms that produced homology between the ct and mtDNA segments is the vertical transmission of an ancestral prokaryote DNA segment (or gene) to both the ct and mt genomes via a cyanobacterium or an α-proteobacterium, respectively (“orthology”). The second mechanism is interorganellar DNA transfer between the ct and mt genomes (“xenology”).32) The third mechanism is amplification of a DNA segment of orthologous or xenologous origin in an organelle (“paralogy”).
Orthology between the wheat mt and ctDNA segments is identified by the occurrence of similar levels of sequence homology between the mt and the ct, Pm and Pu DNA segments, because the divergence times of the present day cyanobacterium, α-proteobacterium, chloroplast and mitochondrial genomes are assumed to be similar. Orthologous genes carried by corresponding mt and ctDNA segments provide supporting evidence for their orthology. Xenologous segments are identified by their high level of sequence homology between the ct and mtDNA segments compared to low or no homology to the corresponding DNA segments in the Pm and Pu genomes. This conclusion is based on the fact that interorganellar DNA transfer resulting in xenologous segments must have occurred after acquisition of the chloroplast and mitochondrion by the ancestral eukaryote through endosymbiosis of a cyanobacterium and α-proteobacterium. Paralogous segments are identified by high sequence homology between multiple DNA copies within an organellar genome, in which their amplification took place, than between them and the ct (or mt), Pm and Pu counterparts, because the amplification event is an evolutionarily recent event compared to the origins of orthologs and xenologs. It is not possible to determine which of the paralogous segments is the template and which the copy. For the sake of convenience, I assume that the paralog shown with the affix “a” on the mtDNA segment number (Table 2) is the template and the others the paralogs.
Table 2.
Homologous segments in the wheat ct and mtDNAs arranged according to the ct-coordinates of the ctDNA segments
| CtDNA segment | MtDNA segment | Sequence homology (%)c) | ||||||
|---|---|---|---|---|---|---|---|---|
| Segm’t no. | Ct-coordinates | Size (bp) | Ct gene | Segm’t no. | Mt-coordinatesb) | Size (bp) | Mt genea) | |
| 1 | 6,696–6,749 | 54 | trnQ* | 1a | R 266,815–266,868 | 54 | trnQ-1* | 83.3 |
| ″ | ″ | ″ | ″ | 1b | R 174,918–174,971 | 54 | trnQ-2* | 83.3 |
| ″ | ″ | ″ | ″ | 1c | R 193,291–193,344 | 54 | trnQ-3* | 81.5 |
| 2 | 11,579–11,655 | 77 | trnS-2* | 2 | R 408,510–408,585 | 77 | trnS-2 | 81.6 |
| 3 | 18,282–18,754 | 473 | trnC | 3 | R 97,417–97,861 | 445 | trnC | 78.2 |
| 4 | 21,268–21,343 | 76 | rpoB* | 4 | 316,034–316,102 | 69 | - | 95.7 |
| 5 | 28,338–28,371 | 34 | rpoC2* | 5 | 68,014–68,047 | 34 | - | 91.2 |
| 6 | 33,284–33,319 | 36 | atpF 5′-ex* | 6 | 349,029–349,064 | 36 | - | 91.7 |
| 7 | 34,262–34,319 | 58 | atpF 3′-ex* | 7 | 104,644–104,702 | 59 | - | 86.2 |
| 8 | 34,309–34,366 | 58 | atpF 3′-ex* | 8 | 162,458–162,515 | 58 | - | 96.6 |
| 9 | 34,485–34,513 | 29 | atpF 3′-ex* | 9 | 388,154–388,182 | 29 | - | 100.0 |
| 10 | 34,918–35,245 | 328 | atpA* | 10 | 304,646–304,973 | 328 | - | 95.7 |
| 11 | 34,918–36,037 | 1,120 | atpA* | 11a | R 53,841–54,950 | 1,110 | - | 98.1 |
| ″ | ″ | ″ | ″ | 11b | R 390,809–391,918 | 1,110 | - | 98.1 |
| 12 | 35,099–35,864 | 766 | atpA* | 12 | 7,265–8,057 | 793 | atp1* | 67.7 |
| 13 | 35,542–36,108 | 567 | atpA* | 13 | 119,524–120,049 | 526 | - | 87.5 |
| 14 | 40,653–41,098 | 446 | psaA* | 14 | R 358,076–358,521 | 446 | - | 99.8 |
| 15 | 43,932–44,149 | 218 | - | 15 | R 378,941–379,150 | 210 | - | 89.5 |
| 16 | 44,967–45,160 | 194 | trnS-3 | 16 | R 383,409–383,603 | 195 | trnS-3 | 90.8 |
| 17 | 47,621–47,851 | 231 | trnL 3′-ex | 17 | R 383,203–383,422 | 220 | - | 80.5 |
| 18 | 48,013–48,128 | 116 | trnF | 18 | R 382,956–383,071 | 116 | trnF | 95.7 |
| 19 | 48,072–48,098 | 27 | trnF* | 19a | R 55,179–55,205 | 27 | - | 100.0 |
| ″ | ″ | ″ | ″ | 19b | 304,391–304,417 | 27 | - | 100.0 |
| ″ | ″ | ″ | ″ | 19c | R 392,147–392,173 | 27 | - | 100.0 |
| 20 | 48,073–48,119 | 47 | trnF* | 20 | 97,436–97,481 | 46 | trnC* | 84.8 |
| 21 | 49,041–49,072 | 32 | ndhJ* | 21 | 400,525–400,556 | 32 | - | 96.9 |
| 22 | 50,062–50,123 | 62 | ndhC* | 22 | R 212,481–212,538 | 58 | nad9* | 91.4 |
| 23 | 51,175–51,225 | 51 | trnV 3′-ex* | 23 | 249,838–249,890 | 53 | - | 88.7 |
| 24 | 52,035–52,107 | 73 | trnM | 24 | R 436,153–436,225 | 73 | trnM | 94.5 |
| 25 | 61,844–62,032 | 189 | psbF*, psbE* | 25 | 452,168–452,356 | 189 | - | 88.4 |
| 26 | 62,048–62,218 | 171 | psbE* | 26 | 986–1,157 | 172 | - | 90.7 |
| 27 | 63,336–63,441 | 106 | petL | 27 | R 79,301–79,405 | 105 | - | 96.2 |
| 28 | 63,846–63,927 | 82 | trnW | 28 | R 445,609–445,690 | 82 | trnW | 96.3 |
| 29 | 64,017–64,084 | 68 | trnP* | 29 | R 445,455–445,515 | 61 | - | 95.1 |
| 30 | 64,069–64,131 | 63 | trnP* | 30a | R 305,100–305,163 | 64 | trnP-1* | 82.8 |
| ″ | ″ | ″ | ″ | 30b | R 117,697–117,760 | 64 | trnP-2* | 82.8 |
| 31 | 64,246–64,290 | 45 | - | 31 | R 445,372–445,416 | 45 | - | 88.9 |
| 32 | 68,081–68,256 | 176 | clpP* | 32 | 380,710–380,885 | 176 | - | 95.5 |
| 33 | 75,509–75,643 | 135 | rps11* | 33 | R 324,416–324,550 | 135 | - | 94.1 |
| 34 | 76,715–76,833 | 119 | rps8* | 34 | R 99,254–99,373 | 120 | - | 86.7 |
| 35 | 77,024–77,395 | 372 | rpl14* | 35 | R 98,764–99,133 | 370 | - | 84.1 |
| 36 | 82,945–82,976 | 32 | trnI* | 36 | R 157,714–157,745 | 32 | trnI-p | 96.9 |
| 37 | 82,945–82,974 | 30 | trnI* | 37 | 184,631–184,660 | 30 | - | 83.3 |
| 38 | 83,963–84,016 | 54 | - | 38 | R 154,459–154,512 | 54 | - | 98.1 |
| 39 | 84,711–89,049 | 4,339 | Footnoted) | 39 | R 417,240–421,558 | 4,319 | - | 97.7 |
| 40 | 88,845–89,021 | 177 | - | 40 | 340,755–340,931 | 177 | rps12* | 72.3 |
| 41 | 91,016–92,532 | 1,472 | rrn16 | 41a | 300,896–302,827 | 1,932 | rrn18-1 | 75.6e) |
| ″ | ″ | ″ | ″ | 41b | R 393,737–395.668 | 1,932 | rrn18-2 | 75.6e) |
| ″ | ″ | ″ | ″ | 41c | R 56,769–58,700 | 1,932 | rrn18-3 | 75.6e) |
| 42 | 93,226–95,059 | 1,834 | Footnotef) | 42 | 74,171–76,003 | 1,833 | trnA | 99.8 |
| 43 | 94,995–97,615 | 2,621 | rrn23 | 43a | R 371,386–374,530 | 3,145 | rrn26-1 | 73.7g) |
| ” | ″ | ″ | ″ | 43b | R 259,648–262,792 | 3,145 | rrn26-2 | 73.7g) |
| 44 | 94,995–95,063h) | 69 | rrn23* | 44 | R 170,827–170,895 | 69 | rrn26-p* | 79.7 |
| 45 | 94,995–95,059 | 65 | rrn23* | 45 | 75,939–76,003 | 65 | rrn26-p* | 98.5 |
| 46 | 98,811–98,896 | 86 | trnN | 46 | R 428,633–428,718 | 86 | trnN | 98.8 |
| 47 | 98,831–98,888 | 58 | trnN* | 47a | R 270,543–270,601 | 59 | trnK-1* | 79.7 |
| ″ | ″ | ″ | ″ | 47b | R 442,708–442,766 | 59 | trnK-2* | 79.7 |
| ″ | ″ | ″ | ″ | 47c | R 178,647–178,705 | 59 | trnK-3* | 79.7 |
| 48 | 102,531–102,595 | 65 | ndhF* | 48 | 294,426–294,490 | 65 | nad5b* | 81.5 |
| 49 | 109,526–109,596 | 71 | ndhG* | 49 | R 146,989–147,055 | 67 | - | 89.6 |
| 50 | 110,973–111,387 | 415 | ndhA 3′-ex* | 50 | 242,709–243,106 | 398 | - | 91.7 |
| 51 | 111,103–111,141 | 39 | ndhA 3′-ex* | 51 | 16,001–16,039 | 39 | nad1c* | 82.1 |
| 52 | 111,319–111,361 | 43 | - | 52 | 43,065–43,107 | 43 | - | 81.4 |
| 53 | 111,324–111,355 | 32 | - | 53a | R 136,911–136,942 | 32 | - | 81.3 |
| ″ | ″ | ″ | - | 53b | 43,726–43,757 | 32 | - | 81.3 |
| 54 | 111,389–111,572 | 184 | - | 54 | R 343,407–343,586 | 180 | - | 90.6 |
| Total | - | 18,385 | - | Total | - | 27,733 | - | - |
a) Suffix “p” to the mt gene indicates a pseudogene. *: Part of the gene located.
b) “R” indicates the mtDNA segment reversed with respect to the corresponding ctDNA segment.
c) Sequence homology (%) = [No. matched nucleotides]/[No. nucleotides in mtDNA segment] × 100.
d) This ctDNA segment contains three ct genes, ndhB, rps7, and 3′-rps12 ex-2.
e) Homology of the 1,246 bp sequences conserved between the ct-rrn16 and mt-rrn18 genes.
f) This ctDNA segment contains three ct genes, trnI 3′-ex, trnA and rrn23*.
g) Homology of the 1,571 bp sequences conserved between the ct-rrn23 and mt-rrn26 genes.
h) The 69 bp 5′-end of no. 43 ctDNA segment.
Estimation of the significance of levels of homology between the subject and query sequences.
The 5% and 1% levels of significance for sequence homology were estimated for different size classes of the 52 wheat mtDNA segments of orthologous and xenologous origin. These estimates were based on their sequence homology to random DNA segments of the wheat ctDNA, the mtDNAs of five green plants (the monocotyledon Oryza sativa, the dicotyledon Arabidopsis thaliana, the gymnosperm Cycas taitungensis, the bryophyte Physcomitrella patens and the green alga Chlorokybus atmophyticus), and the genomic DNA of the prokaryote Prochlorococcus marinus. The percentage (“X”) sequence homology of random DNA segments from the seven listed species to wheat mtDNA segments showed a Poisson rather than a binomial distribution and was transformed to “X1/2” for its normalization. Using the normalized values, the mean and standard deviation were calculated for different size classes of wheat mtDNA segments, and these were used to estimate the 5% and 1% significance levels for sequence homology (Table 1 ; the 5% and 1% significance levels for sequence homologies that have been reconverted to percentages from the square root of “X”).
Table 1.
The upper limits of the 95% and 99% confidence intervals for sequence homology (%) of ctDNA-homologous mtDNA segments of wheat to random segments of wheat ctDNA, mtDNAs of five green plants and genomic DNA of a prokaryotea)
| Size class of wheat mt-segm’t (bp) | No. random DNA segm’ts examined | Mean sequence homology (%) | Upper confid. interval limit | |
|---|---|---|---|---|
| 95% level (%) | 99% level (%) | |||
| 20–40 | 439 | 42.1 | 66.0 | 75.4 |
| 41–70 | 631 | 26.9 | 43.4 | 49.6 |
| 71–100 | 246 | 22.0 | 38.4 | 44.0 |
| 101–150 | 350 | 14.8 | 25.5 | 28.9 |
| 151–200 | 510 | 9.4 | 15.1 | 16.9 |
| 201–300 | 192 | 7.9 | 12.7 | 14.3 |
| 301–500 | 455 | 4.7 | 8.3 | 9.4 |
| 501–1,000 | 124 | 3.2 | 8.0 | 9.5 |
| over 1,000 | 462 | 1.0 | 2.9 | 3.5 |
a) Five green plant and a prokaryote species were used in the analysis: Oryza sativa (monocotyledon), Arabidopsis thaliana (dicotyledon), Cycas taitungensis (gymnosperm), Physcomitrella patens (bryophyte), Chlorokybus atmophyticus (green alga), and Prochlorococcus marinus (prokaryote).
Results
Homologous DNA segments in wheat ct and mt genomes.
A BLAST search was carried out to identify homologous sequences in the wheat ct and mt genomes. The ct genome of angiosperms, including wheat, has a pair of large inverted repeats that are identical in their nucleotide sequences.13,33) In this analysis, only one of the two repeat copies was subjected to the homology search, together with two single copy regions of the ct genome. The ct and mtDNA segments with sequence homology are listed in Table 2. Hereafter, locations of the ct and mtDNA homologues in their respective genomes are shown by their ct- and mt-coordinates.
Homologous segments are arranged in Table 2 according to their locations in the ct genome, and are listed with their ct-coordinates and numbered in ascending order. Ct or mt genes, which were completely or partially located in the respective segments, are also shown. In total, 54 ctDNA segments ranging in size from 27 to 4,339 bp were found to be homologous to 66 mtDNA segments with sequence homologies of about 70% or higher.
The total length of mtDNA with homology to ctDNA was 27,608 bp (excluding 125 bp doubly counted), which corresponds to 6.1% of the wheat mt genome of 452,528 bp. The doubly counted mtDNA sequences were a 65 bp sequence of segment no. 45 that was part of segment no. 42, a 46 bp sequence of segment no. 20 that was part of segment no. 43, and a 14 bp terminal region in segments no. 16 and 17 that overlapped.
Identification of paralogs in multiple mtDNA segments with homology to the same ctDNA segment.
To discriminate paralogs from orthologous and xenologous mtDNA segments, I examined multiple mtDNA segments that showed homology to each other and to the same or partly overlapping ctDNA segments (Table 3).
Table 3.
Paralog identification based on sequence homology between the mtDNA segments that showed homology to the same ctDNA region
| Ct segment No. | Ct-coordinates | Mt segments compareda) | Size comp’d (bp) | No. matched nucleotides | % Homologyb) | Originc) |
|---|---|---|---|---|---|---|
| 1 | 6,696–6,749 | 1a/1b | 54 | 54 | 100.0 | Paralog |
| ″ | ″ | 1a, b/1c | 54 | 53 | 98.1 | Paralog |
| 7,8 | 34,262–34,366 | 7/8 | 58 | 8 | 13.8 | - |
| 10–13 | 34,918–36,108 | 10/11a,b | 328 | 327 | 99.7 | Paralogd) |
| ″ | ″ | 10/12 | 147 | 88 | 59.9 | - |
| ″ | ″ | 11a/11b | 1,110 | 1,110 | 100.0 | Paralog |
| ″ | ″ | 11a,b/12 | 766 | 518 | 67.6 | - |
| ″ | ″ | 11a,b/13 | 496 | 348 | 70.2 | - |
| ″ | ″ | 12/13 | 323 | 81 | 25.1 | - |
| 18–20 | 48,013–48,128 | 18/19a,b,c | 27 | 26 | 96.3 | - |
| ″ | ″ | 18/20 | 47 | 38 | 80.9 | - |
| ″ | ″ | 19a/19b/19c | 27 | 27 | 100.0 | Paralog |
| ″ | ″ | 19a,b,c/20 | 26 | 17 | 65.4 | - |
| 29,30 | 64,017–64,131 | 29/30a,b | 16 | 15 | 87.5 | - |
| ″ | 64,069–64,131 | 30a/30b | 64 | 64 | 100.0 | Paralog |
| 36,37 | 82,945–82,976 | 36/37 | 30 | 21 | 70.0 | - |
| 39,40 | 84,711–89,049 | 39/40 | 177 | 45 | 25.4 | - |
| 41 | 91,016–92,532 | 41a/41b/41c | 1,932 | 1,932 | 100.0 | Paralog |
| 43–45 | 94,995–97,615 | 43a/43b | 3,145 | 3,145 | 100.0 | Paralog |
| ″ | ″ | 43a,b/44 | 69 | 69e) | 100.0 | Paralog |
| ″ | ″ | 43a,b/45 | 65 | 50e) | 76.9 | - |
| ″ | ″ | 44/45 | 65 | 50 | 76.9 | - |
| 46,47 | 98,811–98,896 | 46/47a,b,c | 58 | 47 | 81.0 | - |
| ″ | ″ | 47a/47b/47c | 59 | 59 | 100.0 | Paralog |
| 50–53 | 110,973–111,387 | 50/51 | 39 | 15 | 38.5 | - |
| ″ | ″ | 50/52 | 43 | 34 | 79.1 | - |
| ″ | ″ | 50/53a,b | 32 | 26 | 81.3 | - |
| ″ | ″ | 52/53a,b | 32 | 20 | 65.2 | - |
| ″ | ″ | 53a/53b | 32 | 32 | 100.0 | Paralog |
a) MtDNA segment numbers are taken from Table 2. Numerator and denominator indicate the DNA segments compared.
b) [No. of matched nucleotides]/[Number of nucleotides compared] × 100.
c) Definition of paralog is given in the text.
d) Segment no. 10 (size 328 bp) is 99.7% homologous to the 328 bp 5′-end of segments no. 11a and 11b, indicating paralogy of the former to the 5′-end of the latter two segments.
e) The 69 bp 5′-end of segments no. 43a and 43b is completely homologous to segment no. 44 and the 65 bp 5′-end of 43a and 43b is partially homologous to segment no. 45.
Two doublet mtDNA segments (nos. 30a, b; and 53a, b) and six triplet segments (nos. 1a, b, c; 10, 11a, b; 19a, b, c; 41a, b, c; 43a, b, 44; and 47a, b, c) showed complete or almost complete homology (98.1–100.0%) between the twin or triplet segments, whereas their homologies to the corresponding ctDNA segments were generally lower (75.6–98.1%; Table 2) with the exception of triplet no. 19a, b, c, which showed complete homology between the triplets and the ctDNA counterpart.
With respect to the assumed paralogy between the 1a, 1b and 1c mtDNA segments, the 1c segment showed a single base substitution compared to the la/1b pair; the latter pair showed the same 9 base mismatches to the ctDNA homologue, whereas the 1c segment had an additional mismatch. These findings indicate that the 1c segment underwent a single base substitution after its amplification in the mt genome. Two triplet segments, nos. 10, 11a and 11b, and nos. 43a, 43b and 44 were exceptional. The 328 bp no. 10 segment was 99.7% homologous to the 328 bp 5′-end of the 11a and 11b segments that were completely homologous to each other. Similarly, the 69 bp no. 44 segment was completely homologous to the 69 bp 5′-end of the 47a and 47b segments, which were also completely homologous to each other. Apparently, both the no. 10 and no. 44 segments are partial copies of their counterpart templates.
All of the data support the assumption that the paralogs identified in the present study originated by amplification of their founder copy in the wheat mitochondria.
Determination of orthology between the wheat mt and ctDNA segments.
Orthology between the mtDNA segment and its ctDNA homologue was examined by comparing sequence homologies between homologous segments in the mt and ct genomes of wheat and the Pm and Pu prokaryotic genomes. The criterion for identification of orthology was high sequence homology of the mtDNA segment to all of the corresponding homologues in the wheat ct, Pm and Pu genomes. The presence of genes with the same or similar function was regarded as supportive evidence. As for the paralogs, orthology was examined only for segment with the affix “a” from each paralog group. All the orthologous segments deduced from this homology search are shown in Table 4.
Table 4.
Assumed orthologs in the wheat mt and ct genomes and Pm and Pu genomes, based on the sequence homology of the wheat mtDNA segments to the homologous segments in wheat ct, Pm and Pu genomesa) and the genes located in those segments of the four genomesb)
| Wheat mtDNA segment | Wheat ct genome | Pm genome | Pu genome | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Segment no. | Size (bp) | Gene | Match. nts. | Homol. (%) | Gene | Match. nts. | Homol. (%) | Gene | Match. nts. | Homol. (%) | Gene |
| 1a | 54 | trnQ-1* | 45 | 83.3 | trnQ* | 46 | 85.2 | trnQ* | 41 | 75.9 | trnQ* |
| 2 | 76 | trnS-2* | 62 | 81.6 | trnS-2* | 21 | 27.3 | trnS* | 55 | 71.4 | trnS* |
| 12 | 793 | atp1* | 537 | 67.7 | atpA* | 461 | 66.0 | atpA* | 564 | 71.1 | atpA* |
| 18 | 116 | trnF | 111 | 95.7 | trnF | 62 | 53.4 | trnF* | 43 | 37.1 | trnF* |
| 19a | 27 | - | 27 | 100.0 | trnF* | 22 | 81.5 | trnF* | 22 | 81.5 | trnF* |
| 20 | 46 | trnC* | 39 | 84.8 | trnF* | 43 | 93.5 | trnC* | 39 | 84.8 | trnC* |
| 24 | 73 | trnM | 69 | 94.5 | trnM | 40 | 54.8 | trnT* | 25 | 34.2 | aldA* |
| 28 | 82 | trnW | 79 | 96.3 | trnW | 57 | 69.5 | trnW* | 40 | 48.8 | trnW* |
| 30a | 64 | trnP-1* | 53 | 82.8 | trnP* | 46 | 71.9 | trnP-1* | 51 | 79.7 | trnP* |
| 31 | 45 | - | 40 | 88.9 | - | 30 | 66.7 | (Note 1) | 21 | 46.7 | (Note 2) |
| 40 | 177 | rps12* | 128 | 72.3 | - | 103 | 58.2 | rps12* | 54 | 30.5 | rps12* |
| 41ac) | 1,932 | rrn18-1 | 927 | 75.6 | rrn16 | 947 | 77.2 | rrn16 | 968 | 79.0 | rrn16 |
| 43ad) | 3,145 | rrn26-1 | 1,160 | 73.7 | rrn23 | 1,202 | 73.0 | rrn23 | 1,600 | 75.8 | rrn23 |
| 45 | 65 | rrn26-p | 64 | 98.5 | rrn23* | 56 | 86.2 | rrn23* | 53 | 81.5 | rrn23* |
| 46 | 86 | trnN | 85 | 98.8 | trnN | 61 | 70.9 | trnN | 53 | 61.6 | trnN* |
| 47a | 59 | trnK-1* | 47 | 79.7 | trnN* | 45 | 76.3 | trnN* | 50 | 84.7 | trnN* |
| 48e) | 65 | nad5b* | 53 | 81.5 | ndhF* | 53 | 81.5 | ? | 54 | 83.1 | nuoL* |
| 51e) | 39 | nad1c* | 32 | 82.1 | ndhA 3′-ex* | 27 | 69.2 | ndh SU* | 21 | 53.8 | ? |
a) Sequence homology (%) = [No. nucleotides matched between the wheat mtDNA segment and the DNA segment in comparison]/[No. nucleotides in the wheat mtDNA segment] × 100.
b) Genes reported in wheat mt, wheat ct, Pm and Pu genomes by Ogihara et al.,14,15) Copeland et al.25) and Giovannoni et al.,26) respectively. *: Part of the gene located. “-“, “?” and “-p” indicate no gene reported, undesignated gene located, and pseudogene, respectively.
c) Homology of the 1,226 bp conserved regions of the no. 41a mtDNA segment to those of the wheat ct, Pm and Pu genomes.31)
d) Homology of the 1,573–2,110 bp conserved regions of the no. 43a mtDNA segment to those of the wheat ct, Pm and Pu genomes.31)
e) nad, ndh and nuo: All are NADH dehydrogenase genes.
Note 1: cytidylate kinase gene, Note 2: D amino acid oxidase gene.
Eleven mtDNA segments showed high sequence homology (ca. 70% or higher) to sequences in all genomes of wheat ct, Pm and Pu. Additionally, five mtDNA segments, nos. 2, 18, 28, 40 and 51, showed high sequence homologies (ca. 60% or higher) to ct sequences and also to either Pm or Pu sequences. In addition to their high sequence homologies, most of the wheat mtDNA segments and their corresponding wheat ct and prokaryote DNA segments included complete or partial sequences of orthologous genes (Table 4). These observations support my conclusion on the orthology of the 16 wheat mtDNA segments and their ctDNA counterparts. Orthology of two other mtDNA segments, nos. 24 and 31, was not supported by the presence of orthologous gene sequences. However, they did show high sequence homology to the wheat ct genome and to one or other of the prokaryote genomes. They also had significant and high sequence homology to the mtDNAs of the 10 green plant species examined later (ref. Table 6). Based on these findings, I believe these sequences are of orthologous origin.
Table 6.
Sequence homology of wheat ctDNA-homologous mtDNA segments to the wheat ct genome, mt genomes of 10 green plant taxa and two prokaryote genomesa) and their presumed origins
| MtDNA segment | Sequence homology (%) of wheat mtDNA segment to: | Deduced origin of ctDNA- homologous wheat mtDNA | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wheat ct genome | Mt genome of ten green plant species; | Prokaryote genome | |||||||||||||
| No. | Size (bp) | Monocotyledon | Dicotyledon | Gymno- sperm | Bryophyta | Green alga | Cyano- bacterium (Pm) | α-Proteo- bacterium (Pu) | |||||||
| Rice | Maize | Sorghum | Arabi- dopsis | Sugar beet | Tobacco | Moss | Liver- wort | ||||||||
| 1a | 54 | 83** | 98** | 100** | 100** | 98** | 100** | 100** | 96** | 91** | 85** | 93** | 85** | 76** | Prokaryoteb) |
| 2 | 77 | 82** | 100** | 100** | 100** | 97** | 100** | 97** | 97** | 96** | 96** | 37 | 27 | 71** | ″ |
| 12 | 793 | 67** | 100** | 99** | 99** | 94** | 94** | 94** | 91** | 86** | 80** | 76** | 60** | 74** | ″ |
| 18 | 116 | 96** | 98** | 97** | 98** | 38* | 44** | 44** | 45** | 43** | 41** | 37** | 53** | 37** | ″ |
| 19a | 27 | 100** | 100** | 100** | 100** | 85** | 81** | 81** | 74* | 81** | 81** | 70* | 82** | 82** | ″ |
| 20 | 46 | 85** | 100** | 100** | 100** | 63** | 50** | 37 | 46* | 57** | 48* | 48* | 93** | 85** | ″ |
| 24c) | 73 | 95** | 100** | 100** | 100** | 100** | 100** | 100** | 96** | 58** | 52** | 58** | 55** | 34 | ″ |
| 28 | 82 | 96** | 99** | 28 | 99** | 99** | 96** | 96** | 40* | 40* | 49** | 56** | 70** | 49** | ″ |
| 30a | 64 | 83** | 100** | 100** | 100** | 100** | 100** | 100** | 95** | 95** | 80** | 53** | 72** | 80** | ″ |
| 31c) | 45 | 89** | 96** | 53* | 100** | 47* | 49* | 56** | 58** | 53** | 62** | 67** | 67** | 47* | ″ |
| 40 | 177 | 72** | 100** | 99** | 100** | 93** | 96** | 95** | 91** | 84** | 85** | 72** | 58** | 31** | ″ |
| 41a | 1,932 | 76** | 86** | 98** | 99** | 91** | 91** | 85** | 78** | 70** | 69** | 58** | 77** | 77** | ″ |
| 43a | 3,145 | 76** | 99** | 99** | 99** | 84** | 90** | 80** | 73** | 69** | 67** | 54** | 78** | 76** | ″ |
| 45 | 65 | 98** | 77** | 85** | 77** | 78** | 78** | 95** | 77** | 82** | 82** | 74** | 86** | 82** | ″ |
| 46 | 86 | 99** | 100** | 100** | 100** | 88** | 87** | 87** | 55** | 47** | 67** | 71** | 71** | 62** | ″ |
| 47a | 59 | 80** | 100** | 100** | 100** | 100** | 100** | 100** | 98** | 93** | 93** | 86** | 76** | 85** | ″ |
| 48 | 65 | 82** | 100** | 100** | 100** | 94** | 95** | 94** | 94** | 62** | 62** | 78** | 82** | 83** | ″ |
| 51 | 39 | 82** | 95** | 97** | 97** | 97** | 95** | 97** | 95** | 87** | 38 | 87** | 69* | 54 | ″ |
| 52 | 43 | 81** | 100** | 100** | 100** | 98** | 100** | 100** | 95** | 93** | 81** | 65** | 49* | 40 | Green plant |
| 53a | 32 | 81** | 100** | 100** | 100** | 94** | 94** | 94** | 94** | 94** | 88** | 88** | 0 | 0 | ″ |
| 16 | 195 | 91** | 95** | 13 | 99** | 84** | 75** | 72** | 42** | 15 | 18* | 17* | 38** | 32** | Angiosperm |
| 29 | 61 | 95** | 100** | 38 | 100** | 41 | 87** | 95** | 30 | 26 | 34 | 38 | 39 | 41 | ″ |
| 36 | 32 | 97** | 100** | 100** | 100** | 91** | 75** | 91** | 53 | 63 | 56 | 63 | 56 | 53 | ″ |
| 37 | 30 | 83** | 87** | 87** | 87** | 83** | 70* | 83** | 40 | 50 | 53 | 43 | 63 | 53 | ″ |
| 17 | 220 | 80** | 97** | 99** | 96** | 12 | 15** | 13* | 12 | 10 | 10 | 10 | 19** | 12 | Monocotyledon |
| 22 | 58 | 91** | 97** | 100** | 100** | 47* | 33 | 40 | 34 | 29 | 26 | 31 | 34 | 34 | ″ |
| 34 | 120 | 87** | 100** | 51** | 97** | 18 | 19 | 27* | 23 | 18 | 20 | 20 | 22 | 25 | ″ |
| 3 | 445 | 78** | 62** | 39** | 39** | 7 | 7 | 5 | 7 | 6 | 6 | 6 | 15** | 12** | Cereald) |
| 4 | 69 | 96** | 93** | 30 | 93** | 33 | 35 | 46* | 35 | 29 | 39 | 36 | 38 | 43 | ″ |
| 21 | 32 | 97** | 91** | 59 | 53 | 38 | 59 | 69* | 63 | 47 | 50 | 47 | 53 | 53 | ″ |
| 33 | 135 | 94** | 17 | 21 | 97** | 17 | 21 | 18 | 13 | 13 | 13 | 16 | 20 | 16 | ″ |
| 35 | 370 | 84** | 96** | 6 | 95** | 6 | 5 | 7 | 6 | 5 | 6 | 6 | 40** | 9* | ″ |
| 38 | 54 | 98** | 41 | 96** | 37 | 35 | 31 | 43 | 35 | 44* | 30 | 33 | 39 | 30 | ″ |
| 39 | 4,319 | 98** | 3 | 98** | 1 | 1 | 34** | 37** | 1 | 1 | 29** | 3 | 6 | 4 | ″ |
| 42 | 1,833 | 100** | 3 | 64** | 34** | 4 | 9* | 12** | 2 | 3 | 3 | 3 | 14** | 10** | ″ |
| 49 | 67 | 90** | 22 | 31 | 100** | 30 | 31 | 24 | 28 | 37 | 28 | 24 | 30 | 27 | ″ |
| 5 | 34 | 91** | 44 | 62 | 62 | 50 | 62 | 50 | 53 | 50 | 44 | 50 | 59 | 56 | Wheat |
| 6 | 36 | 92** | 39 | 47 | 44 | 64 | 44 | 56 | 64 | 47 | 53 | 53 | 64 | 56 | ″ |
| 7 | 59 | 86** | 27 | 34 | 44* | 34 | 42 | 42 | 46* | 27 | 27 | 32 | 34 | 39 | ″ |
| 9 | 29 | 100** | 52 | 62 | 69* | 62 | 62 | 55 | 48 | 55 | 62 | 69* | 69* | 69* | ″ |
| 15 | 210 | 90** | 14* | 14* | 15** | 11 | 10 | 9 | 34** | 10 | 10 | 15** | 14* | 12 | ″ |
| 32 | 176 | 95** | 27** | 14 | 16* | 13 | 19** | 13 | 16* | 14 | 12 | 13 | 28* | 20** | ″ |
| 50 | 398 | 92** | 9* | 12** | 9* | 8 | 9* | 9* | 9* | 8 | 8 | 7 | 40** | 11** | ″ |
| 54 | 180 | 91** | 16* | 15 | 13 | 18** | 17** | 10 | 16* | 16* | 17** | 11 | 18** | 17** | ″ |
| 8 | 58 | 97** | 98** | 98** | 98** | 43 | 64** | 40 | 38 | 31 | 40 | 41 | 53** | 48* | Recurrent transfer |
| 14 | 446 | 100** | 4 | 7 | 8 | 4 | 6 | 64** | 4 | 4 | 6 | 4 | 32** | 6 | ″ |
| 23 | 53 | 89** | 49* | 36 | 89** | 38 | 38 | 25 | 79** | 34 | 32 | 53* | 43 | 43 | ″ |
| 25 | 189 | 88** | 13 | 14 | 14 | 12 | 9 | 88** | 11 | 8 | 14 | 10 | 13 | 12 | ″ |
| 26 | 172 | 91** | 15 | 17** | 13 | 12 | 12 | 88** | 16* | 17** | 16* | 15 | 27** | 14 | ″ |
| 27 | 105 | 96** | 21 | 19 | 17 | 23 | 20 | 88** | 21 | 16 | 22 | 18 | 23 | 21 | ″ |
| 11 | 1,110 | 98** | 61** | 47** | 47** | 26** | 26** | 46** | 25** | 46** | 30** | 26** | 53** | 24** | Undetermined |
| 13 | 526 | 87** | 88** | 20** | 20** | 28** | 20** | 19** | 16** | 16** | 23** | 26** | 28** | 12** | ″ |
| Reference: Wheat ct-rbcL | |||||||||||||||
| rbcL | 1,434 | 2e) | 96** | 92** | 94** | 34** | 1 | 2 | 75** | 1 | 2 | 1 | 81** | 2 | Recurrent transfer |
a) Sequence homology (%) = [No. matched nucleotides]/[Wheat mt-segment size (bp)] × 100. Significant sequence homology at the 5% and 1% levels of probability are shown by * and **, respectively.
b) Synonymous to the orthologous origin.
c) The indicated origin is inconsistent with the genes located.
d) “Cereal” means the occurrence of ctDNA transfer in cereals after divergence of the cereal from other groups of monocotyledon.
e) No sequence homology between the wheat ct-rbcL and any wheat mt segments.
Xenologous mtDNA segments and origin of a chimeric mt gene.
As stated above, I identified 14 paralogous and 18 orthologous segments (Tables 3 and 4) among the 66 mtDNA segments with homology to ctDNA sequences (Table 2). By default, the remaining 34 segments were putatively of xenologous origin (Table 5).
Table 5.
Assumed xenologous mtDNA segments identified by sequence homology between wheat mt and ctDNA segments and between the wheat mt and genomic DNA segments of two prokaryotes
| Wheat mtDNA segment | Wheat ct genome | Pm genome | Pu genome | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Segment no. | Size (bp) | Genea) | Match. nts. | Homol. (%) | Genea) | Match. nts. | Homol. (%) | Genea) | Match. nts. | Homol. (%) | Genea) |
| 3 | 445 | trnC | 348 | 78.2 | trnC | 68 | 15.3 | trnC | 54 | 12.1 | trnC* |
| 4 | 69 | - | 66 | 95.7 | rpoB* | 26 | 37.7 | (Note 1) | 30 | 43.5 | cgtA* |
| 5 | 34 | - | 31 | 91.2 | rpoC2* | 20 | 58.8 | (Note 2) | 19 | 55.9 | (Note 3) |
| 6 | 36 | - | 33 | 91.7 | atpF 5′-ex* | 23 | 63.9 | peptidase M* | 20 | 55.6 | phoR* |
| 7 | 59 | - | 50 | 86.2 | atpF 3′-ex* | 20 | 33.9 | (Note 4) | 23 | 39.0 | aceE* |
| 8 | 58 | - | 56 | 96.6 | “ | 31 | 53.4 | psb(?)* | 28 | 48.3 | gcvP* |
| 9 | 29 | - | 29 | 100.0 | “ | 20 | 69.0 | (Note 5) | 20 | 69.0 | priA* |
| 11a | 1,110 | atp1* | 1,089 | 98.1 | atpA* | 589 | 53.1 | atpA* | 264 | 23.8 | atpA* |
| 13 | 526 | atp1* | 460 | 87.5 | atpA* | 149 | 28.3 | atpA* | 64 | 12.2 | atpA* |
| 14 | 446 | - | 445 | 99.8 | psaA* | 143 | 32.1 | psaA* | 26 | 5.8 | glxB* |
| 15 | 210 | - | 188 | 89.5 | - | 29 | 13.8 | (Note 6) | 26 | 12.4 | ? |
| 16 | 195 | trnS-3 | 177 | 90.8 | trnS-3 | 75 | 38.5 | trnS | 63 | 32.3 | trnS |
| 17 | 220 | - | 177 | 80.5 | trnL 3′-ex | 41 | 18.6 | trnL* | 27 | 12.3 | tolB* |
| 21 | 32 | - | 31 | 96.9 | ndhJ* | 17 | 53.1 | (Note 7) | 17 | 53.1 | (Note 8) |
| 22 | 58 | nad9* | 53 | 91.4 | ndhC* | 18 | 31.0 | ? | 20 | 34.5 | nuoD* |
| 23 | 53 | - | 47 | 88.7 | trnV 3′-ex* | 23 | 43.4 | ATPase* | 23 | 43.4 | ? |
| 25 | 189 | - | 167 | 88.4 | psbF*, psbE* | 24 | 12.7 | ? | 22 | 11.6 | (Note 9) |
| 26 | 172 | - | 156 | 90.7 | psbE* | 47 | 27.3 | petA* | 24 | 14.0 | (Note 10) |
| 27 | 105 | - | 101 | 96.2 | petL | 24 | 22.9 | dioxygenase* | 22 | 21.0 | - |
| 29 | 61 | - | 58 | 95.1 | trnP* | 24 | 39.3 | ? | 25 | 41.0 | ? |
| 32 | 176 | - | 168 | 95.5 | clpP* | 49 | 27.8 | clpP* | 36 | 20.5 | tauC* |
| 33 | 135 | - | 127 | 94.1 | rps11* | 27 | 20.0 | psudogene | 21 | 15.6 | metS* |
| 34 | 120 | - | 104 | 86.7 | rps8* | 26 | 21.7 | (Note 11) | 30 | 25.0 | rsbU |
| 35 | 370 | - | 311 | 84.1 | rpl14* | 148 | 40.0 | rpl14* | 34 | 9.2 | (Note 12) |
| 36 | 32 | trnI-p | 31 | 96.9 | trnI* | 18 | 56.3 | trnS* | 17 | 53.1 | (Note 13) |
| 37 | 30 | - | 25 | 83.3 | trnI* | 19 | 63.3 | α-amylase* | 16 | 53.3 | braC* |
| 38 | 54 | - | 53 | 98.1 | - | 21 | 38.9 | (Note 14) | 16 | 29.6 | hom* |
| 39 | 4,319 | - | 4,220 | 97.7 | (Note 15) | 247 | 5.7 | rps7 | 179 | 4.1 | rps12* |
| 42 | 1,833 | trnA | 1,830 | 99.8 | (Note 16) | 263 | 14.3 | trnA | 191 | 10.4 | trnA* |
| 49 | 67 | - | 60 | 89.6 | ndhG* | 20 | 29.9 | trnL* | 18 | 26.9 | mrsA* |
| 50 | 398 | - | 365 | 91.7 | ndhA 3′-ex* | 161 | 40.5 | ndh SU* | 43 | 10.8 | nuoH* |
| 52 | 43 | - | 35 | 81.4 | - | 21 | 48.8 | ? | 17 | 39.5 | ycjp* |
| 53a | 32 | - | 26 | 81.3 | - | 0 | 0.0 | - | 0 | 0.0 | - |
| 54 | 180 | - | 163 | 90.6 | - | 33 | 18.3 | ? | 30 | 16.7 | thiD* |
a) Gene located in the respective DNA segment. Genes for wheat mt and ct genomes and those for Pm and Pu genomes are reported by Ogihara et al.,14,15) Copeland et al.25) and Giovannoni et al.,26) respectively. Some Pm and Pu genes are indicated by their products, for which gene symbols have not been designated. -p: pseudogene, -: no gene located, ?: unidentified gene located
Notes 1–16: (1) carbon storage regulator, (2) exopolyphosphatase, (3) peptidase M50, (4) outer envelope membrane, (5) generic methyl transferase, (6) galactose epimerase, (7) glycosyl transferase, (8) α/β hydrolase fold, (9) branched-chain amino acid transporter protein, (10) secretary autotransporter, (11) Mg-transporter, (12) monomeric sarcosine oxidase, (13) glutamine amidotransferase class-I, (14) gene for RNA methyltransferase, (15) three genes, ndhB, rps7 and 3′-rps12 ex-2, and (16) three genes, trnI 3′-ex, trnA and rrn23.
Three of the 34 candidate xenologs, namely, nos. 3, 16 and 42 segments, contained complete gene sequences of trnC, trnS-3 and trnA, respectively; all three genes are known to be of ct genome origin.17)
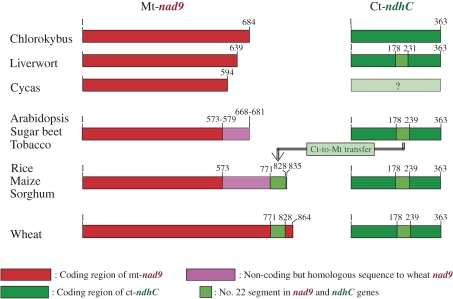
The xenology of the no. 22 mtDNA segment to its counterpart ctDNA segment needs special mention because they carried partial sequences of the active mt-nad9 and ct-ndhC genes, respectively; both genes encode an NADH dehydrogenase subunit. Neither of the two prokaryote genomes had sequences with significant homology to the no. 22 segment, indicating the non-orthologous origin of these segments in the mt and ct genomes. Homologues of the no. 22 mtDNA segment were found only in the monocotyledon species (ref. Table 6), whereas nad9 and ndhC were found, respectively, in the mt and ct genomes of all green plants (Fig. 2).
Figure 2.
Structures of the mt-nad9 and ct-ndhC genes of ten green plants, showing interorganellar transfer of segment no. 22 DNA from the ct to mt genome in a common ancestor of the cereal and the origin of the wheat mt-nad9 gene by expansion of its coding region to include the 5′-flanking region where segment no. 22 was transferred. Numbers given in the figure: nucleotide position of the respective borders. Double-lined arrow: transfer of segment no. 22 in the ct-ndhC to the 5′-flanking region of mt-nad9 in a common ancestor of the cereal. Nucleotide 1 of each gene is the third nucleotide of its stop codon.
The mt-nad9 gene had a nearly constant size, between 573 and 579 bp, in all angiosperms except wheat; in three green plants of lower taxa, i.e., gymnosperm, bryophyte and green alga, the gene size varied from 594 to 684 bp. By contrast, the nad9 gene of wheat was 864 bp.34) Another remarkable feature was that the 5′-flanking regions of the nad9 gene in angiosperms (other than wheat) were homologous to the 5′-end of the wheat nad9 coding region (colored pink in Fig. 2) but varied in size between 95 and 262 bp.
With respect to the ct-ndhC gene, all green plants, except for Cycas for which the ct genome sequence is not yet available, carried the gene, which had an invariant size of 363 bp. All of these species, except for the green alga, possessed a no. 22 ctDNA segment of 54–62 bp in the ndhC coding region. A 58 bp homologue of this segment was found in the mt genomes of all four cereals but not in other plants, including three dicotyledon species. This fact indicated that the ct-to-mt genome transfer of the no. 22 segment occurred in the common ancestor of cereals. The mt site of its transfer was the 5′-flanking region of the cereal mt-nad9, the site being incorporated into the coding region of nad9 in wheat by its expansion to the 5′-flanking region. This is a possible case of the origination of a functional chimeric mt gene from a native and a xenologous mtDNA segment, suggesting an evolutionary role for the xenologous DNA sequence in organellar genome evolution. Similar chimeric mt genes, consisting of a native and xenologous mtDNA segment, have been reported previously for the male-sterile cytoplasm of several crop species. An example of this is the mt T-orf25 gene of the maize mt genome, associated with T-type male-sterile cytoplasm, that consists of partial sequences of the mt-rrn26 and ct-trnR genes.35,36)
Recurrent interorganellar DNA transfers occur unidirectionally from the ct to mt genome.
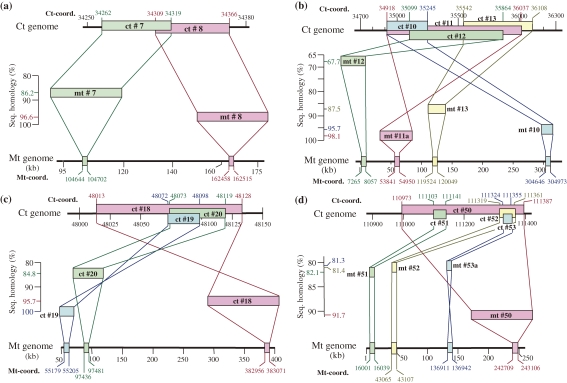
Four cases of recurrent ctDNA transfer from the same ctDNA regions to the mt genome were analyzed in detail (Fig. 3a–d). The first example involved the ctDNA segments no. 7 (ct-coordinates 34,262–34,319) and no. 8 (34,309–34,366), which overlapped with each other in an 11 bp sequence (ct-coordinates, 34,309–34,319) and had their mtDNA homologues in two different regions of the mt genome, mt-coordinates 104,644–104,702 for mtDNA segment no. 7 and 162,458–162,515 for no. 8 (Fig. 3a). The homologies of the no. 7 and no. 8 mtDNA segments to their ctDNA counterparts were 86.2% and 96.6%, respectively (Table 2). These observations indicate that the ctDNA transfer occurred twice at different times in evolution with no. 7 being transferred earlier than no. 8; each segment was transferred from the same region of the ct genome to different sites on the mt genome. The alternative explanation of transfer in the opposite direction, i.e., mt genome to ct genome, seems unlikely, because the probability of two separate mtDNA segments transferring to the same ctDNA site would be expected to be very low as the site choice is non site-specific.37) The order of transfer, i.e., no. 7 being earlier and no. 8 later, was supported by nucleotide differences in their overlapping 11 bp ctDNA sequences: the no. 7 mtDNA segment had three base substitutions (mt-coordinates, 104,693 (T-to-G), 104,694 (C-to-A) and 104,697 (A-to-T)), whereas the no. 8 mtDNA segment did not have any base substitution in this sequence.
Figure 3.
Four cases of recurrent ctDNA transfer from the same regions of the wheat ct genome to different sites in the wheat mt genome. Ct and mtDNA segments involved in each event are drawn, respectively, in the top and bottom of each figure by their positions in the ct and mt genomes, which are shown by their ct- and mt-coordinates. Sequence homologies between the corresponding ct and mtDNA segments are shown in the scale drawn in the space between the ct and mt genomes and the transferred mtDNA segments are drawn in positions that fit their sequence homologies to the corresponding ctDNA segments. (a) Segments no. 7 and 8 originated from the ct-coordinates, 34262–34366. (b) Segments no. 10–13 originated from the ct-coordinates, 34918–36108. (c) Segments no. 18–20 originated from the ct-coordinates, 48013–48128. (d) Segments no. 50–53 originated from the ct-coordinates, 110973–111387.
The second example involved the ctDNA segments nos. 10, 11, 12 and 13, which were located within a 1,191 bp ctDNA region (ct-coordinates 34,918–36,108). These segments were transferred to different sites in the mt genome. Ct- and mt-coordinates of all the ct and mtDNA segments involved and the sequence homologies observed between the template ct and copied mtDNA segments are shown in Fig. 3b. Three mtDNA segments, nos. 12, 13, and 11a, were transferred in succession in the order listed (oldest to most recent) as indicated by their homologies of 67.7, 87.5 and 98.1% to the corresponding ctDNA segments. The no. 10 mtDNA segment was a partial paralog of the no. 11a mtDNA segment, as previously described (Table 3). Nos. 11a and 12 mtDNA segments shared a 766 bp sequence with the same ctDNA segment; no. 11a mtDNA segment showed nine base substitutions and six deletions (four 1 bp deletions and one each of 2 bp and 4 bp), while the no. 12 mtDNA segment had 217 base substitutions and 12 deletions of variable sizes (three 1 bp, two 2 bp, two 3 bp, four 6 bp and one 14 bp) in their common sequence. Similarly, nos. 11a and 13 mtDNA segments shared a common 496 bp sequence with the same ctDNA segment. In this region, no. 11a had eight base substitutions and three deletions (two 1 bp and one 2 bp), whereas no. 13 mtDNA segment possessed 52 base substitutions and 18 deletions of variable size (eleven 1 bp, three 2 bp, two 4 bp, one each of 5 and 9 bp deletions). Those findings indicated that transfer from the ct to mt genome occurred earliest for no. 12, followed by no. 13 and then no. 11a, in complete agreement with the transfer order suggested by overall sequence homology to the corresponding ctDNA segments.
The third example involved ctDNA segments nos. 18, 19 and 20 located within a 118 bp region (ct-coordinates, 48,013–48,130) and their homologous mtDNA segments, which were distributed across a wide range of the mt genome (Fig. 3c). Transfer of the no. 20 mtDNA segment seemed to have occurred earliest, followed by those of no. 18 and no. 19a in that order, as suggested by their homologies of 84.8, 95.7 and 100.0%, respectively, to their ctDNA homologues. In their 26 bp common sequence (ct-coordinates 48,073–48,098), no. 19a showed no base changes, no. 18 had a single base substitution, and no. 20 had five base substitutions, compared to their ctDNA homologue; these findings confirmed the above transfer order.
The fourth example involved ctDNA segments nos. 50, 51, 52 and 53, which were located within a 415 bp region (ct-coordinates 110,973–111,387) of the ct genome, whereas their mt homologues were scattered through a 227,106 bp region of the mt genome (Fig. 3d). The sequence homologies of the mtDNA segments to the corresponding ctDNA segments were 81.3% (no. 53a), 81.4% (no. 52), 82.1% (no. 51) and 91.7% (no. 50), suggesting the transfer of nos. 53a, 52 and 51 occurred at nearly the same time, while that of no. 50 was a more recent event. Amplification of the paralogous sequence no. 53b took place soon after the transfer of no. 53a to the mt genome, because the nucleotide sequences of these two mtDNA segments were the same and both mtDNA sequences showed the same six nucleotide substitutions to the ctDNA homologue.
In addition to these, members of five groups of mtDNA segments, namely, nos. 29 and 30, nos. 36 and 37, nos. 39 and 40, nos. 42, 43a and 45, and nos. 46 and 47, shared a common 16 to 177 bp stretch of DNA in the corresponding ctDNA sequences, in spite of their separate locations in the mt genome. These are all cases of recurrent ctDNA transfer from the same regions of the ct genome to different sites in the mt genome; detailed descriptions of each will be omitted due to the limitation in space.
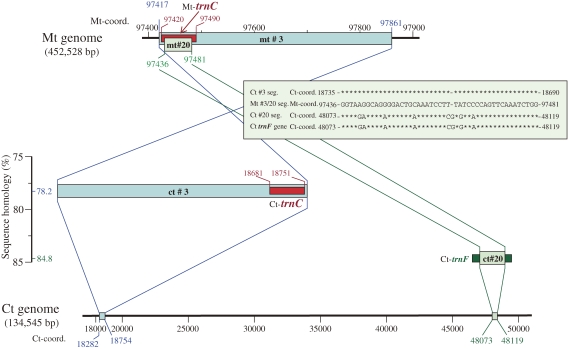
In contrast, a dual transfer of an mtDNA segment from a single mt genomic region (mt-coordinates 97,417–97,861) to two separate sites in the ct genome was suspected in only one case (Fig. 4). The no. 20 mtDNA segment is a part of the no. 3 mtDNA segment, as was clear from their mt-coordinates (97,436–97,481 and 97,417–97,861, respectively), whereas their ctDNA homologues were found in two different sites of the ct genome (ct-coordinates 18,282–18,754 and 48,073–48,119; Fig. 4).
Figure 4.
Map and sequence relationships between segments no. 3 and 20 in the wheat mt and ct genomes. The no. 3 mtDNA segment is a transferant of the no. 3 ctDNA segment that carries the ct-trnC gene, whereas the no. 20 ctDNA segment is not a transferant of the no. 3 mtDNA segment but is the 47 bp central portion of ct-trnF gene. The mt-trnC is the ct genome-derived trnC gene.14) The 46 bp nucleotide sequence of the no. 3 ctDNA segment, the corresponding 46 bp sequence of the nos. 3 and 20 mtDNA segments, the 47 bp nucleotide sequences of the no. 20 ctDNA segment, and the 47 bp central portion of ct-trnF gene are shown in the inserted box.
Detailed analyses of the nucleotide sequences of the two mt and two ctDNA segments involved in this suspected event, however, clearly indicated that this is not the case. Although sequence homology between the no. 3 mt and ctDNA segments was as low as 78.2%, their 71 bp regions (mt-coordinates 97,420–97,490 and ct-coordinates 18,751–18,681, in reversed direction) showed 100% sequence homology. The mtDNA region is the site of the active mt-trnC gene of chloroplast origin,14) while the ctDNA region is the site of the native ct-trnC gene.15) This suggests that the no. 3 mtDNA segment was derived by transfer of the no. 3 ctDNA segment, and not by a transfer in the opposite direction. On the other hand, the no. 20 ctDNA segment (47 bp) showed complete nucleotide matching over its entire region to the corresponding 47 bp region of the native ct-trnF gene (see insert in Fig. 4). This indicates that the no. 20 ctDNA segment is a part of the ct-trnF gene. The observed homology of 84.8% between the no. 20 mt and ctDNA segments was due to homology between the mt-trnC (of chloroplast origin) and ct-trnF genes (Fig. 4). Thus, the sequence homology detected between the no. 20 ct and mtDNA segments was not due to the transfer of the latter mtDNA segment to the ct genome, but rather was produced by the partial sequence homology that exists between the mt-trnC and ct-trnF genes.
Phylogenetic origin of ctDNA transfer to the mt genomes in green plant evolution.
A search of the distribution of ctDNA-homologous mtDNA sequences among the mt genomes of a wide range of green plants, including the reference prokaryote genomes, will provide insights into the phylogenetic origin of ctDNA transfer in plant evolution. For this purpose, the mtDNAs of 10 green plant species of diverse taxa and the two prokaryote reference genomes were BLAST searched for sequences with homology to the 52 wheat mtDNA segments of orthologous or xenologous origin. To confirm the validity of the present method, the presence of homologous sequences to the wheat ct-rbcL gene in the mt genomes and prokaryote genomes of all those species was investigated and the results were compared to those of Cummings et al.38) who investigated this distribution among a large number of angiosperm species. The results are shown in Table 6, in which homology between the wheat mtDNA segments and their ctDNA counterparts are also shown.
The 5% and 1% levels of significance for sequence homology estimated in Table 1 are shown in Table 6 by one and two asterisks, respectively. All 18 wheat mtDNA segments identified to be of orthologous origin (ref. Table 4) had counterpart segments with a significant level of sequence homology in the mt genomes of a minimum of nine of the ten examined green plants and one or both of the two prokaryote genomes, supporting the interpretation of their prokaryote origin.
Two mtDNA segments, nos. 52 and 53a, maintained a high level of sequence homology to the mtDNA segments of all 10 green plants, with no or a low level of homology to prokaryote genomes. This finding indicated that these two mtDNA segments were transferred from the ct genome at an early stage of green plant evolution. Four wheat mtDNA segments, nos. 16, 29, 36 and 37, appeared to have been transferred at an early stage of angiosperm evolution, because their homologues were found in almost all of the monocotyledons and dicotyledons examined, but not in the lower plant taxa, including the gymnosperm. Three other mtDNA segments, nos. 17, 22 and 34, showed highly significant sequence homology to all monocotyledons examined, but not to other plant taxa, suggesting they were transferred at an early stage of monocotyledon evolution. Nine mtDNA segments, nos. 3, 4, 21, 33, 35, 38, 39, 42 and 49, showed highly significant homology to the mtDNAs in various combinations of two or three monocotyledons, including wheat, suggesting their transfer occurred during divergence of the cereal species. Our previous phylogenetic studies on the organellar genomes indicated that genetic distances between the organellar genomes of wheat, rice and maize were similar, suggesting that the phylogenetic divergence of their organellar genomes occurred at almost the same time.31) As a result, I could not conclude with any certainty whether the observed ctDNA transfer occurred in a common ancestor of the two to three cereal species or if it occurred independently in these species. Transfer of eight mtDNA segments, nos. 5, 6, 7, 9, 15, 32, 50 and 54, undoubtedly occurred only once in the wheat lineage.
Six ctDNA segments, nos. 8, 14, 23, 25, 26 and 27, appeared to have been transferred recurrently to the mitochondrion during green plant evolution: no. 8 in the sugar beet and cereal lineages; nos. 14, 25, 26 and 27 in the tobacco and wheat lineages; and no. 23 in the Cycas, sorghum and wheat lineages. Transfer of the ctDNA segments nos. 25, 26 and 27 merit special mention. Their transfer to the wheat mt genome occurred independently at different times, because their sequence homologies to the corresponding mtDNA homologues were 88.4, 90.7 and 96.2% for nos. 25, 26 and 27, respectively (Table 2). In contrast, in tobacco, they were transferred to the mt genome in a 2,726 bp ctDNA segment, in which they were completely linked, including their intervening sequences. Sequence homology of the 2,726 bp tobacco ctDNA segment to its mtDNA counterpart (2,703 bp) was 94.2%; this homology is slightly higher than the 91.0% homology of the three wheat mtDNA segments as a group to their counterpart ctDNA segments, indicating that transfer of this ctDNA region in tobacco was more recent than those of the wheat ctDNA segments. The two remaining mtDNA segments, nos. 11 and 13, showed a low but significant level of homology to the mt or genomic DNAs of all species examined. Thus, the timing of their transfer could not be estimated with any certainty.
In addition, wheat ct-rbcL homologues were detected in the mt genomes of rice, maize, sorghum, Arabidopsis (partial sequence) and Cycas, but missing in the mt genomes of wheat, sugar beet, tobacco, moss, liverwort and green alga (bottom line, Table 6). These findings, for the most part, agree with those of Cummings et al.38) with regard to the presence of the rbcL homologue in rice, maize and Arabidopsis and absence in wheat and tobacco; one exception was sorghum in which the homologue was found to be present in this study but absent in the earlier one. The rbcL homologue was reported recently in the mt genome of Cycas, whereas it was missing from the mt genomes of moss, liverwort and green alga. These results confirmed recurrent ct-to-mt genome transfer of the rbcL gene during the evolution of seed plants.
Based on the results described above, the origin of the ctDNA-homologous mtDNA segments in wheat is summarized in Table 7. In total, 14 paralogous, 18 orthologous and 34 xenologous mtDNA segments were identified; these occupied 1.96, 1.48 and 2.68% of the entire wheat mt genome, respectively, their total amounting to 6.13%.
Table 7.
Summary of the origins of the 66 wheat ctDNA-homologous mtDNA segments
| MtDNA segment | Nature | Size (bp) | Genea) (ct/mt/Pm/Pu) | Originb) |
|---|---|---|---|---|
| 1a | Ortholog | 54 | trnQ/trnQ-1/trnQ/trnQ | Prokaryote |
| 1b | Paralog | 54 | - | (Copy of #1a) |
| 1c | Paralog | 54 | - | (Copy of #1a) |
| 2 | Ortholog | 77 | trnS-2/trnS-2/trnS/trnS | Prokaryote |
| 3 | Xenolog | 445 | - | Cereal |
| 4 | Xenolog | 69 | - | Cereal |
| 5 | Xenolog | 34 | - | Wheat |
| 6 | Xenolog | 36 | - | Wheat |
| 7 | Xenolog | 59 | - | Wheat |
| 8 | Xenolog | 58 | - | Recurrent (beta & cereal) |
| 9 | Xenolog | 29 | - | Wheat |
| 10 | Paralog | 328 | - | (Partial copy of #11a)c) |
| 11a | Xenolog | 1,110 | - | (Undetermined) |
| 11b | Paralog | 1,110 | - | (Copy of #11a) |
| 12 | Ortholog | 793 | aptA*/atp1*/atpA*/atpA* | Prokaryote |
| 13 | Xenolog | 526 | - | (Undetermined) |
| 14 | Xenolog | 446 | - | Recurrent (wheat & tobacco) |
| 15 | Xenolog | 210 | - | Wheat |
| 16 | Xenolog | 195 | - | Angiosperm |
| 17 | Xenolog | 220 | - | Monocotyledon |
| 18 | Ortholog | 116 | trnF/trnF/trnF*/trnF* | Prokaryote |
| 19a | Ortholog | 27 | -/trnF*/trnF*/trnF* | Prokaryote |
| 19b | Paralog | 27 | - | (Copy of #19a) |
| 19c | Paralog | 27 | - | (Copy of #19a) |
| 20 | Ortholog | 46 | trnC/trnF/trnC/trnC | Prokaryote |
| 21 | Xenolog | 32 | - | Cereal |
| 22 | Xenolog | 58 | - | Monocotyledon |
| 23 | Xenolog | 53 | - | Recurrent (cycas & cereal) |
| 24 | Ortholog | 73 | trnM/trnM/trnT/aldA | Prokaryote |
| 25 | Xenolog | 189 | - | Recurrent (wheat & tobacco) |
| 26 | Xenolog | 172 | - | Recurrent (wheat & tobacco) |
| 27 | Xenolog | 105 | - | Recurrent (wheat & tobacco) |
| 28 | Ortholog | 82 | trnW/trnW/trnW/trnW | Prokaryote |
| 29 | Xenolog | 61 | - | Angiosperm |
| 30a | Ortholog | 64 | trnP/trnP-1/trnP-1/trnP | Prokaryote |
| 30b | Paralog | 64 | - | (Copy of #30a) |
| 31 | Ortholog | 45 | -d) | Prokaryote |
| 32 | Xenolog | 176 | - | Wheat |
| 33 | Xenolog | 135 | - | Cereal |
| 34 | Xenolog | 120 | - | Monocotyledon |
| 35 | Xenolog | 370 | - | Cereal |
| 36 | Xenolog | 32 | - | Angiosperm |
| 37 | Xenolog | 30 | - | Angiosperm |
| 38 | Xenolog | 54 | - | Cereal |
| 39 | Xenolog | 4,319 | - | Cereal |
| 40 | Ortholog | 177 | -/rps12/rps12/rps12 | Prokaryote |
| 41a | Ortholog | 1,932 | rrn16/rrn18-1/rrn16/rrn16 | Prokaryote |
| 41b | Paralog | 1,932 | - | (Copy of #41a) |
| 41c | Paralog | 1,932 | - | (Copy of #41a) |
| 42 | Xenolog | 1,833 | - | Cereal |
| 43a | Ortholog | 3,145 | rrn23/rrn26-1/rrn23/rrn23 | Prokaryote |
| 43b | Paralog | 3,145 | - | (Copy of #43a) |
| 44 | Paralog | 69 | - | (Partial copy of #43a)e) |
| 45 | Ortholog | 65 | rrn23/rrn26*/rrn23*/rrn23* | Prokaryote |
| 46 | Ortholog | 86 | trnN/trnN/trnN/trnN | Prokaryote |
| 47a | Ortholog | 59 | trnN*/trnK-1*/trnN*/trnN* | Prokaryote |
| 47b | Paralog | 59 | - | (Copy of #47a) |
| 47c | Paralog | 59 | - | (Copy of #47a) |
| 48 | Ortholog | 65 | ndhF/nad5b/-/nuoL | Prokaryote |
| 49 | Xenolog | 67 | - | Cereal |
| 50 | Xenolog | 398 | - | Wheat |
| 51 | Ortholog | 39 | ndhA ex/nad1c/ndhSU/nuoH | Prokaryote |
| 52 | Xenolog | 43 | - | Green plant |
| 53a | Xenolog | 32 | - | Green plant |
| 53b | Paralog | 32 | - | (Copy of #53a) |
| 54 | Xenolog | 180 | - | Wheat |
| Total | Paralogs | 8,892 | 1.96%f) | - |
| ″ | Orthologs | 6,711 | 1.48%f) | - |
| ″ | Xenologs | 12,130 | 2.68%f) | - |
| Grand total | 27,733 | 6.13%f) | - | |
a) Orthologous genes found in the four genomes are indicated (ref. Table 5). *: Part of the gene sequence.
b) Origin of each paralog is shown as a copy of its postulated template in parentheses. Monocotyledons analyzed are all cereals: therefore, “monocotyledon” is synonymous to cereal. “Recurrent” origin means probable independent occurrence of the ctDNA transfer in the two or more species indicated in parentheses. “Cereal” means the occurrence of the ctDNA transfer in a common lineage of two or three cereal species.
c) Paralog of the 328 bp 5′-end of no. 11a mtDNA segment.
d) No genes in the wheat mt and ct genomes, and the cytidylate kinase and D amino acid oxidase gene in the Pm and Pu genome, respectively.
e) Paralog of the 69 bp 5′-end of no. 43a mtDNA segment.
f) The proportions (%) of the total mt genome (452,528 bp).
Discussion
Investigation of organellar genomes for interorganellar DNA transfer.
Due to the rapid progress in DNA sequencing technology, the number of plant species for which complete nucleotide sequencing of organellar genomes has been accomplished has greatly expanded since 2000. However, sequence information is available on both the ct and mt genomes from only about ten plant species. As a consequence, systematic and genome-wide analysis of interorganellar DNA transfer is still limited.
Previous studies did not detect ctDNA-homologous mtDNA sequences in the liverwort12) and moss.23) In contrast, these sequences have been identified in vascular plants and shown to represent a significant proportion of the whole mt genome, e.g., 4.4% in Cycas,22) ca. 1% in Arabidopsis,19) 2.1% in sugar beet,20) 2.5% in tobacco,21) 3.6% in rape,39) 4.4% in maize17) and 6.3% in rice.16) In most of these studies, the nature of the sequence homology was not analyzed in depth and the proportions reported will almost certainly include paralogs and orthologs, thus resulting in an overestimate of the proportions of xenologs.
In wheat, we previously estimated that the ctDNA-homologous mtDNA sequences represented 5.8% of the entire mt genome;14) here, however, this proportion was estimated as 6.13%. The discrepancy may be due to the use of different BLASTn versions; the previous study used version 2.2.15, the present investigation version 2.2.24+. In the present study, I also classified the homologous sequences into three categories, paralogous, orthologous and xenologous (= ctDNA-derived mtDNA sequences), and estimated that these occupied 1.96, 1.48 and 2.68%, respectively, of the entire mt genome (Table 7). To my knowledge, this is the first time that a precise estimation of the total sizes and proportions of these three types of ctDNA-homologous sequences in the mt genome have been estimated.
Is interorganellar DNA transfer between the ct and mt genome bi- or unidirectional?
In his review on organellar DNA transfer to the nucleus, Leister9) showed that interorganellar DNA transfer has occurred between the nucleus, chloroplast and mitochondrion in four of the six possible directions; the two exceptions were from the nucleus to the chloroplast and from the mitochondrion to the chloroplast.
One of the limiting factors in analysis of interorganellar DNA transfer between the ct and mt genomes is that it is difficult to determine the direction of transfer when it has occurred only once from one site of the ct genome to one site of the mt genome, or vice versa. However, if a transfer has occurred recurrently from a single site of the ct or mt genome to multi-sites of the other organellar genome, we can unequivocally determine the direction of transfer (Fig. 3). In this study, every case of recurrent transfer occurred from the ct to mt genome, and never in the opposite direction. Since recurrent transfers are derived by accumulation of single transfer events that occurred at different times (Fig. 3a–d), it is reasonable to assume that the direction of single transfers is also in most cases, if not all, from the ct to the mt genome. This interpretation is supported by the fact that some chloroplast tRNA genes have been transferred into the mitochondrial genome and function there in many plants; this is known to have occurred in sugar beet,20) rice,16) rapeseed,39) maize,17) tobacco21) and wheat.14) Thus, I conclude that interorganellar DNA transfer between the ct and mt genomes occurs in only one direction, from the ct to mt genome; this conclusion is in agreement with that of two previous reviews on interorganellar gene transfers.9,32)
Evolutionary timing of ctDNA transfers to the mt genome in green plants.
As long ago as 1988, Nugent and Palmer40) reported a Southern blot analysis in Brassicaceae that showed ct-to-mt transfer of several ct genes. Transfer of the 3′-end of psaA and rbcL occurred in a common ancestor of six crucifer species belonging to five genera, Brassica, Raphanus, Crambe, Capsella and Arabidopsis. This transfer was followed by that of the 5′-end of psaA, rpoB and other genes in a common ancestor of four species of Brassica, Raphanus and Crambe, after the divergence of two genera, Capsella and Arabidopsis. The latest transfer occurred in the lineage of Crambe abyssinica, after divergence of all other species. Similarly, Cummings et al.38) investigated ct-to-mt transfer of the rbcL gene using Southern blot hybridization, and found that transfer of the gene occurred on at least five occasions during angiosperm evolution, more specifically, twice in Poaceae and Brassicaceae and once in Convolvulaceae. Recently, Wang et al.41) used ct and mt genome sequences to perform the first systematic survey of ctDNA transfers to the mt genome in 11 plant species: two species of alga, one species each of liverwort, moss and gymnosperm, and three monocotyledon and three dicotyledon angiosperm species. They focused only on organellar genes and used common gene order and boundaries present in the ct and mt genomes as the criterion for identification of ct gene transfer to the mt genome. The reason for adopting this criterion was a concern that dependence on sequence similarity might be misleading because of high base substitution and recombination rates in ctDNA-derived mtDNA sequences. They successfully identified the ct-to-mt genome transfer of seven ct gene clusters, consisting of two to five genes, and estimated the evolutionary timing of these transfers. Thus, transfer of the trnV-trnM-atpE-atpB-rbcL cluster occurred at the emergence of the seed plant lineage, while the gene clusters psaA-psaB, rps19-trnH-rpl2-rpl23 and psbE-psbF occurred in the common ancestor of six angiosperm species, and three other ct gene clusters in different branches of angiosperm divergence.
The aims and materials of the present investigation were very similar to those of Wang et al.41) with the following exceptions: (1) I surveyed sequence homologies between all the ct and mtDNA segments, irrespective of the presence/absence of genes, to obtain an entire picture of ctDNA transfers; (2) the ctDNA-homologous mtDNA segments were classified with respect to origin as orthologous (prokaryote origin), xenologous (ctDNA origin) and paralogous (amplification of orthologs or xenologs) using prokaryotic genome sequences as a reference; (3) the dynamics of recurrent ct-to-mt transfers were analyzed in detail with respect to the transfer of ctDNA segments in the same ct genome region to different sites of the mt genome, in order to assess directionality of the transfer; and (4) the evolutionary timing of transfer of most of the xenologs to the mt genome was estimated, based on the results of a homology search between the ctDNA-derived wheat mtDNA segments and the mt genomes of ten green plant species and two reference prokaryote genomes. These analyses identified new aspects of the dynamics and phylogenetic origins of interorganellar DNA transfers.
Mechanism of interorganellar DNA transfer.
On the basis of current ideas on the origin of green plants, we assume that the mt and ct genomes were derived by endosymbiosis of a Rickettsiales species of α-proteobacterium and a cyanobacterium into a hypothetical protoeukaryotic cell.1,26,29,30) During the evolution of green plants, both endosymbionts transferred a large part (more than 90%) of their genetic information to the nucleus (ref. Fig. 1),1) probably because of the advantages for maintenance and transmission of genetic information in the nuclear genome compared to the organelles. There are two contrasting views regarding the vehicle of organellar DNA transfer to the nucleus:1) first, transfer of bulk organellar DNAs;9,42,43) second, transfer through cDNA intermediates.8,44,45) The first view is supported by the fact that organelle-derived ncDNA segments possess organellar DNA-specific intergenic spacers and non-coding regions. Additionally, large organelle-derived ncDNA sequences of over 100 kb contain organellar DNA-derived introns, trn genes and non-coding sequences. By contrast, the second view is favored by the fact that organelle-derived ncDNA does not possess mt gene-specific introns and RNA editing sites. Our understanding of the mechanisms of ct-to-mt DNA transfer has deepened little since Schuster and Brennicke6,32) who proposed cDNAs as the transfer vehicle; these cDNAs were postulated to be produced by reverse transcription of RNA intermediates. Currently, there is an urgent need to elucidate the mechanism for unidirectional DNA transfer from the ct to mt genomes.
Acknowledgments
I wish to acknowledge kind help of Yukiko Yamazaki and Masahiro Sugiura providing useful information on the plant organellar genome database and tobacco organellar genomes, respectively.
References
- 1).Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 [DOI] [PubMed] [Google Scholar]
- 2).Richardson A.O., Palmer J.D. (2006) Horizontal gene transfer in plants. J. Exp. Bot. 58, 1–9 [DOI] [PubMed] [Google Scholar]
- 3).Stern D.B., Lonsdale D.M. (1982) Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature 299, 698–702 [DOI] [PubMed] [Google Scholar]
- 4).Stern D.B., Palmer J.D. (1984) Extensive and widespread homologies between mitochondrial DNA and chloroplast DNA in plants. Proc. Natl. Acad. Sci. U.S.A. 81, 1946–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Stern D.B., Palmer J.D. (1986) Tripartite mitochondrial genome of spinach: physical structure, mitochondrial gene mapping, and locations of transposed chloroplast DNA sequences. Nucleic Acids Res. 14, 5651–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Schuster W., Brennicke A. (1987) Plastid, nuclear and reverse transcriptase sequences in the mitochondrial genome of Oenothera: is genetic information transferred between organelles via RNA? EMBO J. 6, 2857–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Kobayashi Y., Knoop V., Fukuzawa H., Brennicke A., Ohyama K. (1997) Interorganellar gene transfer in bryophytes: the functional nad7 gene is nuclear encoded in Marchantia polymorpha. Mol. Gen. Genet. 256, 589–592 [DOI] [PubMed] [Google Scholar]
- 8).Millen R.S., Olmstead R.G., Adams K.L., Palmer J.D., Lao N.T., Heggie L., Kavanagh T.A., Hibberd J.M., Gray J.C., Morden C.W., Calie P.J., Jermiin L.S., Wolfe K.H. (2001) Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Leister D. (2005) Origin, evolution and genetic effects of nuclear insertions of organellar DNA. Trends Genet. 21, 655–663 [DOI] [PubMed] [Google Scholar]
- 10).Ohyama K., Fukuzawa H., Kohchi T., Shirai H., Sano T., Sano S., Umesono K., Shiki Y., Takeuchi M., Chang Z., Aota S., Inokuchi H., Ozeki H. (1986) Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322, 572–574 [Google Scholar]
- 11).Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K., Ohto C., Torazawa K., Meng B.Y., Sugita M., Deno H., Kamogashira T., Yamada K., Kusuda J., Takaiwa F., Kato A., Tohdoh N., Shimada H., Sugiura M. (1986) The complete nucleotide sequences of tobacco chloroplast genome: its gene organization and expression. EMBO J. 5, 2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Oda K., Yamato K., Ohta E., Nakamura Y., Takemura M., Nozato N., Akashi K., Kanegae T., Ogura Y., Kohchi T., Ohyama K. (1992) Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA, a primitive form of plant mitochondrial genome. J. Mol. Biol. 223, 1–7 [DOI] [PubMed] [Google Scholar]
- 13).Ogihara Y., Isono K., Kojima T., Endo A., Hanaoka M., Shiina T., Terachi T., Utsugi S., Murata M., Mori N., Takumi S., Ikeo K., Gojobori T., Murai R., Murai K., Matsuoka Y., Ohnishi Y., Tajiri H., Tsunewaki K. (2000) Chinese Spring wheat (Triticum aestivum L.) chloroplast genome: Complete sequence and contig clones. Plant Mol. Biol. Rep. 18, 243–253 [Google Scholar]
- 14).Ogihara Y., Yamazaki Y., Murai K., Kanno A., Terachi T., Shiina T., Miyashita N., Nasuda S., Nakamura C., Mori N., Takumi S., Murata M., Futo S., Tsunewaki K. (2005) Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 33, 6235–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Ogihara Y., Isono K., Kojima T., Endo A., Hanaoka M., Shiina T., Terachi T., Utsugi S., Murata M., Mori N., Takumi S., Ikeo K., Gojobori T., Murai R., Murai K., Matsuoka Y., Ohnishi Y., Tajiri H., Tsunewaki K. (2002) Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol. Genet. Genomics 266, 740–746 [DOI] [PubMed] [Google Scholar]
- 16).Notsu Y., Masood S., Nishikawa T., Kubo N., Akiduki G., Nakazono M., Hirai A., Kadowaki K. (2002) The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics 268, 434–445 [DOI] [PubMed] [Google Scholar]
- 17).Clifton S.W., Minx P., Fauron C.M., Gibson M., Allen J.O., Sun H., Thompson M., Barbazuk W.B., Kanuganti S., Tayloe C., Meyer L., Wilson R.K., Newton K.J. (2004) Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 136, 3486–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Allen, J.O., Minx, P., Fauron, C.M., Oddiraju, S., Clifton, S.W. and Newton, K.J. (unpubl.) GenBank NC_008360 (Submission 06-Sept-2006) (Sorghum bicolor mitochondrial genome).
- 19).Unseld M., Marienfeld J.R., Brandt P., Brennicke A. (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15, 57–62 [DOI] [PubMed] [Google Scholar]
- 20).Kubo T., Nishizawa S., Sugawara A., Itchoda N., Estiati A., Mikami T. (2000) The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNACys (GCA). Nucleic Acids Res. 28, 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Sugiyama Y., Watase Y., Nagase M., Makita N., Yagura S., Hirai A., Sugiura M. (2005) The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genomics 272, 603–615 [DOI] [PubMed] [Google Scholar]
- 22).Chaw S.M., Shih A.C., Wang D., Wu Y.W., Liu S.M., Chou T.Y. (2008) The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol. Biol. Evol. 25, 603–615 [DOI] [PubMed] [Google Scholar]
- 23).Terasawa K., Odahara M., Kabeya Y., Kikugawa T., Sekine Y., Fujiwara M., Sato N. (2007) The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol. Biol. Evol. 24, 699–709 [DOI] [PubMed] [Google Scholar]
- 24).Turmel M., Otis C., Lemieux C. (2007) An unexpectedly large and loosely packed mitochondrial genome in the charophycean green alga Chlorokybus atmophyticus. BMC Genomics 8, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Copeland, A., Lucas, S., Lapidus, A., Barry, K., Detter, J.C., Glavina, T., Hammon, N., Israni, S., Pitluck, S., Thiel, J., Schmutz, J., Larimer, F., Land, M., Kyrpides, N., Lykidis, A. and Richardson, P. (unpubl.) Complete sequence of Prochlorococcus marinus str. MIT 9312 (database accession no. CP000111, submitted Aug. 2007).
- 26).Giovannoni S.J., Tripp H.J., Givan S., Podar M., Vergin K.L., Baptista D., Bibbs L., Eads J., Richardson T.H., Noordewier M., Rappe M.S., Short J.M., Carrington J.C., Mathur E.J. (2005) Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245 [DOI] [PubMed] [Google Scholar]
- 27).Gray M.W., Burger G., Lang B.F. (2001) The origin and early evolution of mitochondria. Genome Biol. 2, 1018.1–1018.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., Leister D., Stoebe B., Hasegawa M., Penny D. (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Knoop V. (2004) The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr. Genet. 46, 123–139 [DOI] [PubMed] [Google Scholar]
- 30).Fitzpatrick D.A., Creevey C.J., McInerney J.O. (2006) Genome phylogenies indicate a meaningful α-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol. Biol. Evol. 23, 74–85 [DOI] [PubMed] [Google Scholar]
- 31).Tsunewaki K., Matsuoka Y., Yamazaki Y., Ogihara Y. (2008) Evolutionary dynamics of wheat mitochondrial gene structure with special remarks on the origin and effects of RNA editing in cereals. Genes Genet. Syst. 83, 301–320 [DOI] [PubMed] [Google Scholar]
- 32).Schuster W., Brennicke A. (1988) Interorganellar sequence transfer: plant mitochondrial DNA is nuclear, is plastid, is mitochondrial. Plant Sci. 54, 1–10 [Google Scholar]
- 33).Palmer, J.D. (1991) Plastid chromosomes: structure and evolution. In The Molecular Biology of Plastids (eds. Bogorad, L. and Vasil, I.K.). Academic Press, Inc., New York, pp. 5–53. [Google Scholar]
- 34).Lamattina L., Gonzalez D., Gualberto J., Grienenberger J.M. (1993) Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem. 217, 831–838 [DOI] [PubMed] [Google Scholar]
- 35).Dewey R.E., Levings C.S., III, Timothy D.H. (1986) Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell 44, 439–449 [DOI] [PubMed] [Google Scholar]
- 36).Stamper S.E., Dewey R.E., Bland M.M., Levings C.S., III (1987) Characterization of the gene urf13-T and an unidentified reading frame, ORF25, in maize and tobacco mitochondria. Curr. Genet. 12, 457–463 [DOI] [PubMed] [Google Scholar]
- 37).Nakazono M., Hirai A. (1993) Identification of the entire set of transferred chloroplast DNA sequences in the mitochondrial genome of rice. Mol. Gen. Genet. 236, 341–346 [DOI] [PubMed] [Google Scholar]
- 38).Cummings M.P., Nugent J.M., Olmstead R.G., Palmer J.D. (2003) Phylogenetic analysis reveals five independent transfers of the chloroplast gene rbcL to the mitochondrial genome in angiosperms. Curr. Genet. 43, 131–138 [DOI] [PubMed] [Google Scholar]
- 39).Handa H. (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 3, 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Nugent J.M., Palmer J.D. (1988) Location, identity, amount and serial entry of chloroplast DNA sequences in crucifer mitochondrial DNAs. Curr. Genet. 14, 501–509 [DOI] [PubMed] [Google Scholar]
- 41).Wang D., Wu Y.W., Shih A.C., Wu C.S., Wang Y.N., Chaw S.M. (2007) Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol. Biol. Evol. 24, 2040–2048 [DOI] [PubMed] [Google Scholar]
- 42).Lin X.Y., Kaul S., Rounsley S., Shea T.P., Benito M., Town C.D., Fujii C.Y., Mason T., Bowman C.L., Barnstead M., Feldblyum T.V., Buell C.R., Ketchum K.A., Lee J., Ronning C.M., Koo H.L., Moffat K.S., Cronin L.A., Shen M., Pai G., Van Aken S., Umayam L., Tallon L.J., Gill J.E., Adams M.D., Carrera A.J., Creasy T.H., Goodman H.M., Somerville C.R., Copenhaver G.P., Preuss D., Nierman W.C., White O., Eisen J.A., Salzberg S.L., Fraser C.M., Craig Venter J. (1999) Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402, 761–768 [DOI] [PubMed] [Google Scholar]
- 43).Henze K., Martin W. (2001) How do mitochondrial genes get into the nucleus? Trends Genet. 17, 383–387 [DOI] [PubMed] [Google Scholar]
- 44).Nugent J.M., Palmer J.D. (1991) RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell 66, 473–481 [DOI] [PubMed] [Google Scholar]
- 45).Adams K.L., Palmer J.D. (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 29, 380–395 [DOI] [PubMed] [Google Scholar]