Abstract
Keloid represents overgrowth of granulation tissue, which is characterized by collection of atypical fibroblasts with excessive deposition of extracellular matrix components, after skin injury, but its etiology is still largely unknown. We recently performed genome-wide association study (GWAS) of keloid and identified NEDD4 to be one of candidate molecules associated with keloid susceptibility. Here we demonstrate a possible mechanism of NEDD4 involvement in keloid formation through enhancement of the proliferation and invasiveness of fibroblasts as well as upregulation of type 1 collagen expression. Activation of NEDD4 affected subcellular localization and protein stability of p27 which was implied its critical role in contact inhibition. It also induced accumulation of β-catenin in the cytoplasm and activated the TCF/β-catenin transcriptional activity. Furthermore, NEDD4 upregulated expressions of fibronectin and type 1 collagen and contributed to the excessive accumulation of extracellular matrix. Our findings provide new insights into mechanism developing keloid and can be applied for development of a novel treatment for keloid.
Keywords: NEDD4, keloid, contact inhibition, EMT
Introduction
Wound healing and tissue regeneration are important for maintenance of the tissue homeostasis and regulated very strictly by various factors. Dysregulation of this system in skin can lead to hyperproliferative scar formation such as hypertrophic scars and keloids. Keloid is clinically defined as skin trauma growing beyond the confines of original wounds and shows no tendency of regression over time.1,2) These lesions are often accompanied with severe pain, intolerable itching, and cosmetic disfigurement, which can significantly impair quality of life (QOL) of patients both physically and psycologically.3)
The formation of keloid is more common among individuals with a darker pigmented skin.1–4) Familial occurrence is also reported although most of the cases are observed sporadically. In a study of 14 pedigrees with familial keloid, the inheritance pattern is consistently autosomal dominant with incomplete clinical penetrance and variable expressions.5) These studies implicated that the involvement of genetic factors in the development of keloid formation. Although TGF-β and SMAD have been implicated their involvement in keloid pathogenesis,6–12) previous association studies failed to demonstrate any positive associations of polymorphisms in genes belonging to the TGF-β and SMAD families with keloid susceptibility.1,13–16) Despite of many challenges to elucidate the keloid pathogenesis, there have been no clear evidences explaining the molecular mechanism for keloid formation.
To clarify genetic factors associated with susceptibility to keloid and elucidate the molecular mechanisms for keloid formation, we recently performed a genome-wide association study (GWAS) for keloid disease and identified four susceptibility loci in the Japanese population.17) Among them, a SNP rs8032158, that is located in intron 5 of the neuronal precursor cell-expressed developmentally downregulated 4 (NEDD4) gene on chromosome 15, showed very significant association with keloid (P = 1.95 × 10−11; OR = 1.49 with 95%CI of 1.33–1.67), implying the possible involvement of NEDD4 in the keloid pathogenesis. NEDD4 is characterized as an E3 ubiquitin ligase with a HECT domain, involved in ubiquitin-mediated protein degradation.18) Ubiquitination regulates protein turnover in cells by regulating the degradation of specific proteins. The E3 ligases play a critical role in the ubiquitin conjugation cascade by recruiting ubiquitin-conjugating enzyme E2, recognizing specific substrates, and facilitating or directly catalyzing ubiquitin transfer to their target molecules.19,20) The relevance of the E3 ligases in several biological processes is demonstrated in vivo by the facts that their genetic alteration, abnormal expression, or dysfunction is often accompanied with pathological disorders including cancer.20)
Here we report the biological role of E3 ubiquitin ligase NEDD4 in keloid pathogenesis by regulation of fibroblast proliferation and migration through ubiquitination of PTEN. Loss of PTEN in fibroblasts is likely to induce cell cycle progression and cell migration through regulation of the Akt pathway. NEDD4-mediated Akt activation altered protein expression or subcellular localization of β-catenin and p27. Furthermore, NEDD4 in fibroblasts involved in the regulation of collagen and fibronectin expression. This aberrant regulation caused the accumulation of collagen and fibronectin in extracellular matrix. These results suggested that NEDD4 could contribute to the keloid formation and progression.
Materials and methods
Cell Lines.
NIH3T3 cells and keloid fibroblasts were purchased from the American Type Culture Collection (ATCC, Rockvill, MD). Normal Human Dermal Fibroblast (NHDF) and normal Human Epithelial Keratinocyte (NHEK) were purchased from Lonza (Walkersvill, MD). NIH3T3 cells and keloid fibroblasts were grown in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA; ATCC); the media were supplemented with 10% bovine serum (GIBCO, Grand Island, NY) for NIH3T3 cells, with 10% fetal bovine serum (Nichirei Bioscience Inc., Tokyo, Japan) and 1% antibiotic/antimycotic solution (Sigma-Aldrich, St. Louis, MO) for keloid fibroblasts. NHDF and NHEK were cultured with appropriate media according to the manufacturer’s recommendation. Cells were maintained at 37 ℃ in atmospheres of humidified air with 5% CO2.
Semi-quantitative RT-PCR.
Total RNAs from cells were extracted using RNeasy Kit (QIAGEN, Valencia, CA) according to manufacturer’s recommendation. Extracted RNAs were treated with RNase-Free DNase Set (QIAGEN) and reversely transcribed to single-stranded cDNAs using d(T)12-18 primer with Superscript II reverse transcriptase (Invitrogen). The RT-PCR exponential phase was determined to allow semi-quantitative comparisons among cDNAs developed from identical reactions. Each PCR regime involved a step of 95 ℃, 30 sec initial denaturation followed by 22 cycles (for ACTB), 25 cycles (for cyclinD1, COL1, COL3, fibronectin) and 30 cycles (for NEDD4, MMP7) of a step at 95 ℃ for 10 sec and 56 ℃ for 30 sec and a step of 72 ℃ for 30 sec (for ACTB, cyclinD1, MMP7, COL1, COL3, fibronectin) or 3 min (for NEDD4), on a Gene Amp PCR system 9600 (PE Applied Biosystems, Foster, CA). The PCR using the following primer sets; 5′-AAATGGCAACTTGCGCGGT-3′, 5′-CTAATCAACTCCATCAAAGCC-3′ for NEDD4; 5′-AATGTTAAACTCCCGCGTCA-3′ and 5′-TGGGGATCTCCATTTCCATA-3′ for MMP7; 5′-ATGGAACACCAGCTCCTGTG-3′ and 5′-TCAGATGTCCACGTCCC-3′ for cyclinD1; 5′-GTGCTAAAGGTGCCAATGGT-3′ and 5′-CTCCTCGCTTTCCTTCCTCT-3′ for COL1; 5′-TACGGCAATCCTGAACTTCC-3′ and 5′-GTGTGTTTCGTGCAACCATC-3′ for COL3; 5′-GGAGTTGATTATACCATCACTG-3′ and 5′-TTTCTGTTTGATCTGGACCT-3′ for fibronectin; 5′-TTGGCTTGACTCAGGATTTA-3′ and 5′-ATGCTATCACCTCCCCTGTG-3′ for ACTB.
Immunoblotting.
The cells were transfected with NEDD4 expression vector or control mock vector using Fugene 6 (Roche, Indianapolis, IN) according to the manufacturer’s recommended procedures. We synthesized siRNA duplex (sense; 5′-GUAUGAGUUCUUCCGAAGATT-3′, anti-sense; 5′-UCUUCGGAAGAACUCAUACTT-3′) for knockdown of NEDD4 and transfected with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s recommended procedures. Cells lysed in the RIPA buffer that contained 1 mM sodium orthovanadate, 10 mM sodium fluoride and protease inhibitor cocktail (Calbiochem, Darmstadt, Germany). For analysis of secreted fibronectin, conditioned culture media were collected after 96 or 120 h incubation and concentrated using UltraFree centrifugal filter and tube (Millipore, Bedford, MA). For collagen analysis, cells were directly added 5× sample buffer and collected. Protein from each sample was separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated with the indicated primary antibodies; anti-Flag, anti-ACTB (Sigma-Aldrich), anti-NEDD4, anti-PTEN, anti-β-catenin, anti-p27, anti-COL1A1, anti-fibronectin (Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-GSK3β (Ser9), anti-Akt, anti-phospho-Akt (Ser473) (Cell signaling, Beverly, MA) antibodies.
Cell viability assay.
The full length of NEDD4 cDNA (NP_006145) was prepared by PCR amplification and inserted into pcDNA3.1-myc-his vector (invitrogen). The plasmid was transfected into NHDF cells using Fugene6 (Roche) according to the manufacturer’s recommended procedures. For in vitro growth assay, 1 × 104 NHDF cells, in which NEDD4 was exogenously introduced, were seeded into each wells of a 48-well culture plate and incubated in their respective recommended media for 48 h. The growth curve of these established clones were measured using Cell-counting kit-8 (DOJINDO, Kumamoto, Japan).
Matrigel invasion assay.
NIH3T3 cells that were transfected with either plasmid designed to express NEDD4 (pcDNA3.1/NEDD4-Myc-His) or that with mock plasmid were grown to the near confluence condition in DMEM containing 10% BS. For Matrigel invasion assay, 1 × 104 cells were plated in the top chamber with Matrigel-coated membrane (BD Labware, Bedford, MA). Cells were plated in serum-free medium, and medium with serum was used as a chemoattractant in the lower chamber. The cells were incubated for 20 h and cells which did not invade were removed by a cotton swab. The cells invading through Matrigel were fixed and stained with Gimsa (Merck, Darmstadt, Germany) and counted.
Multi-layer cell culture and colony formation.
NIH3T3 cells were seeded onto 6-cm dish and transfected with control empty vector or NEDD4 expression vector. Cells were cultured until over 100% confluent condition, then photographed and stained with Giemsa staining solution (Merck) at 7 days after reaching to the confluent level.
Immunocytochemical staining.
Cells were transfected with NEDD4 expression vector (pCAGGS/NEDD4-HA) as described above and incubated for 48 h. After the incubation, the cells were fixed with methanol:acetone for 1 h at −20 ℃. Non-specific binding was blocked by treatment with PBS containing 3% BSA for 60 min at room temperature. The cells were incubated for 60 min at room temperature with each of the indicated primary antibodies (anti-HA (Roche), anti-β-catenin and anti-p27 as described above) diluted at 1:200 by PBS containing 3% BSA. After washing with PBS, the cells were stained by the FITC-conjugated secondary antibody (invitrogen) for 60 min at room temperature and visualized with Spectral Confocal Scanning Systems (Leica, Heidelberg, Germany).
Recombinant proteins and in vitro ubiquitination assay.
The full length cDNAs of human NEDD4 (900 amino acids, NP_006145) and human PTEN (403 amino acids, NP_000305) were prepared by PCR amplification and inserted into pGEX-6P-3 vector (GE Healthcare Bio-sciences, Piscataway, NJ). The recombinant NEDD4 or PTEN that was fused with glutathione S-transferase (GST) tag at N-terminus was expressed in Escherichia coli, BL21 codon plus (Stratagene, La Jolla, CA), and purified with Glutathione Sepharose 4B and PreScission protease (GE Healthcare Bio-sciences) under native condition according to supplier’s protocol. For in vitro ubiquitination assay, the NEDD4 and PTEN proteins were incubated with E1 and E2 (UbcH7) enzymes, and ubiquitin in the assay buffer of Ubiquitylation kit (ENZO Life Sciences, Farmingdale, NY). After 2 h of incubation at 30 ℃, the reactions were terminated by addition of SDS sample buffer.
Statistical analysis.
All values were presented as means ± s.d. Statistical significance was computed using Student’s t-test, and the level of significance was set at P < 0.05.
Results
NEDD4 contributed to loss of the cell–cell contact inhibition by promoted cell proliferation and invasiveness.
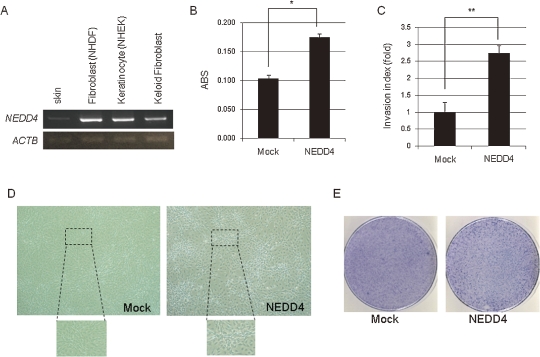
NEDD4 was reported its abundant expression in skeletal muscle, placenta, and cancer cell lines.21) Because NEDD4 was identified as a candidate gene susceptible to the keloid disease in our GWAS analysis,17) we suspected that genetic variations on this locus might contribute to its expression level. Hence, we first examined the expression of NEDD4 in fibroblasts, and found that NEDD4 was expressed in skin, normal fibroblast cell lines as well as keloid fibroblasts (Fig. 1A). To investigate the biological role of NEDD4 in keloid formation, we constructed a plasmid vector that was designed to express human NEDD4 and transfected it into normal human dermal fibroblast (NHDF) cells. The cells were incubated for 48 h and the number of the cells was measured by an MTT assay, suggesting that the over-expression of NEDD4 promoted the growth of the NHDF cells (Fig. 1B).
Figure 1.
Biological function of NEDD4 in human normal fibroblasts. (A) NEDD4 was expressed in normal human skin, normal human dermal fibroblasts (NHDF), human keratinocytes (NHEK) and keloid fibroblast, respectively. β-actin (ACTB) was used for quantitative normalization. (B) Cell viability assay showing that NEDD4 promoted cell proliferation. MTT assay of NHDF cells that were transfected with NEDD4 expression vector or the control mock vector. ABS on Y-axis means absorbance at 490 nm with that at 630 nm as a reference measured with a microplate reader. These experiments were carried out in triplicate (*P = 0.0001). (C) NEDD4 over-expressing NIH3T3 cells promoted cell invasion by Matrigel invasion assay. Invasion index means the average cell number of migration, counted by microscopic observation. These experiments were carried out in triplicate (**P = 0.0235). Error bars represent mean ± s.d. (D, E) NEDD4 introduced NIH3T3 cells were multi-layered after cells reached 100% confluent (D) and formed colonies (E), whereas these were not observed in control mock cells.
The pathological features of human keloid tissue are characterized by the increased density and proliferative rate of fibroblasts, and their aggressive invasion ability to the normal surrounding skin tissue.1) To examine a possible role of NEDD4 in the cellular invasiveness, we performed a Matrigel invasion assay using NIH3T3 fibroblast cells. As shown in Fig. 1C, NEDD4 over-expression drastically increased the invasive ability of NIH3T3 cells. Furthermore, NIH3T3 cells which exogenously expressed NEDD4 were multi-layered and formed colonies after they reached to the 100% confluence stage in the dish while we did not observe any colony formation in the control mock cells (Fig. 1D, E). These data imply that NEDD4 promoted keloid fibroblast proliferation and infiltration into the surrounding skin tissues, and deregulated the contact inhibition after cell–cell attachment.
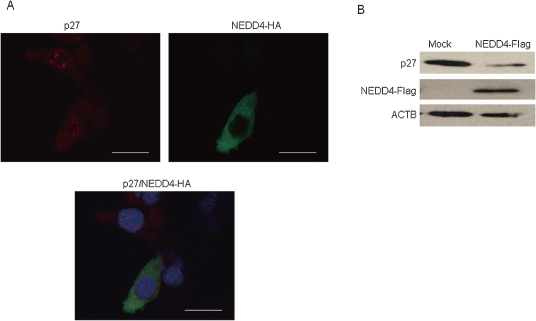
Over-expression of NEDD4 increased cytoplasmic p27 and promoted its degradation.
To further investigate how NEDD4 promotes the cell proliferation and invasiveness of fibroblast cells, we examined molecules involved in the cell-cycle progression and the epithelial-mesenchymal transition (EMT). A cyclin-dependent kinase inhibitor p27 is known to be involved in contact inhibition by inducing the cell cycle arrest.22,23) Phosphorylation-dependent degradation of the p27 protein affected the contact inhibition process.24,25) To investigate roles of NEDD4 over-expression on p27 degradation and/or its subcellular localization, we performed immunocytochemical analysis and immunoblotting using NIH3T3 cells that were transfected with NEDD4 expression vector. We observed diffuse staining of p27 in the cytoplasm of the cells over-expressing NEDD4, while p27 was mainly stained in the nucleus of the parental NIH3T3 cells (Fig. 2A). We also detected the decrease of the p27 protein level in NEDD4-introduced cells (Fig. 2B), suggesting that NEDD4 over-expression induced translocation of p27 from nucleus to cytoplasm and enhanced its degradation.
Figure 2.
The regulation of p27 in NEDD4 over-expressing cells. (A) Immunocytochemical analysis reveals the diffuse subcellular localization of p27 in the cytoplasm in the cells in which NEDD4 was exogenously introduced while most of the p27 protein was localized in the nucleus in the control cells in which NEDD4 was not introduced. Nuclei were stained with DAPI. Bar = 17 µm. (B) The protein expression of p27 was decreased in NEDD4 over-expressed cells rather than mock cells.
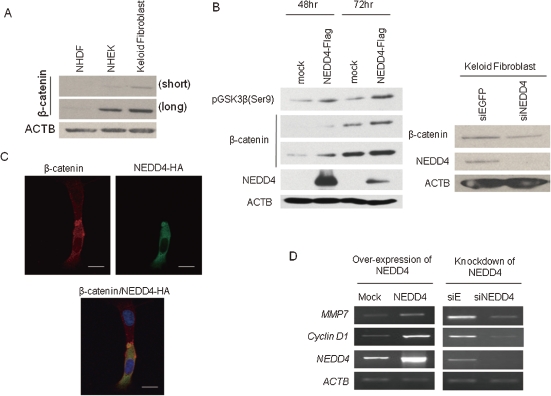
β-catenin signaling pathway was activated in NEDD4 exogenously expressed fibroblasts.
We also investigated the involvement of GSK3β/β-catenin signaling pathway because β-catenin is known to play a critical role in the EMT process and contact inhibition.26–28) β-catenin was reported to be accumulated in hyperplastic scars as well as keloid tissues.29) As shown in Fig. 3A, we indeed observed β-catenin accumulation in keloid fibroblasts. To clarify a possible role of NEDD4 in regulation of β-catenin, we examined the phosphorylation level of GSK3β and the amount of β-catenin protein in the NIH3T3 cells which were transfected with NEDD4 expression vector. Western blot analysis indicated that the level of GSK3β phosphorylation and the amount of β-catenin were significantly increased by exogenous induction of NEDD4. On the other hand, knockdown of NEDD4 expression in keloid fibroblasts by NEDD4-specific siRNA duplex reduced the amount of β-catenin (Fig. 3B). Immunocytochemical staining found β-catenin to be localized mainly in the cytoplasm in NEDD4 over-expressing cells (Fig. 3C). The cytoplasmic β-catenin is known to be translocated into the nucleus, binds to TCF/LEF-1 transcriptional factors,30) and induce the expression of several downstream genes such as cyclin D1 and MMP7 which are reported to be up-regulated during EMT.31,32) Hence, we performed semi-quantitative RT-PCR using cDNA isolated from NEDD4 over-expressing or NEDD4 knocked-down cells, and observed that the expression levels of MMP7 and cyclin D1 were reduced in the NHDF cells in which NEDD4 expression was knocked-down, while they were elavated in the cells in which NEDD4 was over-expressed (Fig. 3D).
Figure 3.
NEDD4 increased cytoplasmic β-catenin and upregulated β-catenin transcriptional activity. (A) Up-regulation of β-catenin in keloid fibroblasts. Endogenous β-catenin was detected by western blot analysis using total cell lysates. NHDF; normal human dermal fibroblast, NHEK; normal human epithelial keratinocyte, short; short exposure, long; long exposure. (B) Phospho-GSK3β and β-catenin were up-regulated in the cells that NEDD4 was transiently introduced. The cells were incubated for 48 h or 72 h after the transfection. Depletion of NEDD4 in keloid fibroblasts decreased the protein level of β-catenin. The cells were incubated for 48 h after the transfection of siRNA duplex. (C) Immunocytochemical staining detected β-catenin mainly at plasma membrane in the cells in which NEDD4 was not introduced while β-catenin was translocated to the cytoplasm in NEDD4 over-expressing cells. Bar = 12 µm. (D) Transcriptional changes of the genes regulated in the β-catenin/TCF were analyzed by semi-quantitative RT-PCR. Cells were incubated for 72 h (for transiently expressing) or 96 h (for knockdown). β-actin (ACTB) was used to quantify cDNA contents.
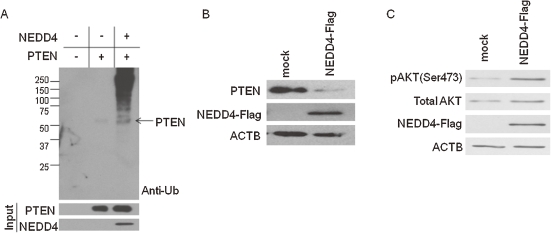
NEDD4 activated Akt signaling transduction through dysregulation of PTEN.
NEDD4 is characterized as an E3 ubiquitin ligase with a HECT domain, involved in ubiquitin-mediated protein degradation.18) Recently, NEDD4 was demonstrated to directly ubiquitinate PTEN (phosphatase and tensin homolog deleted on chromosome 10) and regulate its stability.33,34) p27 and β-catenin are known to be targets of the Akt protein which is regulated by PTEN. Interestingly, germline PTEN mutation associated with multiple hamartoma syndrome, also known as PTEN hamartoma tumor syndrome (PHTS), and the patient of PHTS tends to form keloids.35,36) Hence, we performed in vitro ubiquitination assay using human NEDD4 and PTEN recombinant proteins. We incubated them with E1 and E2 enzymes, and examined the ubiquitination of PTEN by anti-Ub antibody. The ubiquitination of PTEN was detected in the presence of the NEDD4 recombinant protein, while it was not without NEDD4 (Fig. 4A). Subsequently, we also found that the drastic reduction of the PTEN protein level in NEDD4 over-expressing cells, compared with the parental cells, by western blot analysis (Fig. 4B).
Figure 4.
NEDD4 activated Akt signaling pathway through diminished PTEN protein level in fibroblasts. (A) In vitro ubiquitination assay of PTEN by NEDD4. NEDD4 ubiquitinated PTEN directly. (B) NEDD4 ubiquitinated PTEN directly and promoted the protein degradation. Immunoblot analysis for endogenous PTEN in NEDD4 over-expressing NIH3T3 cells. Control or NEDD4 expression vector were transfected and incubated for 48 h. β-actin (ACTB) was blotted as the loading control. (C) Over-expression of NEDD4 in NIH3T3 cells enhanced the phosphorylation level of Akt. Control or NEDD4 expression vector were transfected and incubated for 48 h. Phosphor-Akt and total Akt were detected by immunoblotting. β-actin (ACTB) was blotted as the loading control.
The activated/phosphorylated Akt was reported to phosphorylate many substrates including p27 and GSK3β, and to promote growth and migration of cells.37–39) Since PTEN is known to negatively regulate the PI3K/Akt pathway,37,40) we examined the phosphorylation level of Akt in the NEDD4 over-expressing NIH3T3 cells. The phosphorylated Akt level was increased in the NEDD4 over-expressing cells, in concordance with the reduction of the amount of PTEN protein (Fig. 4C). Therefore, we suspected that NEDD4 enhances proliferative and invasive properties of cells through dysregulation of the PTEN/PI3K/Akt signaling pathway.
NEDD4 regulated the expression of fibronectin and type 1 collagen in ECM of fibronectin.
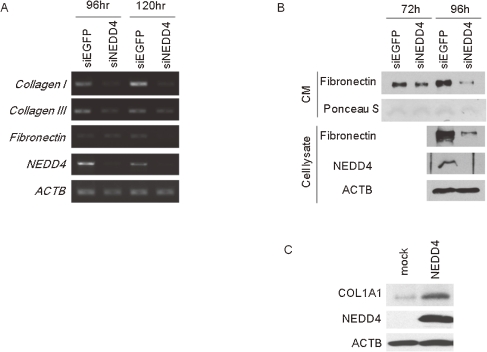
The excessive deposition of collagen in extracellular matrix (ECM) is another clinical characteristic of keloid disease.1) To elucidate this accumulation of collagen and other ECM-related molecules, we examined the expression levels of ECM components for their relation to the NEDD4. As shown in Fig. 5A, RT-PCR experiments revealed that expression of type I and III collagen was decreased in NHDF cells in which NEDD4 was knocked down, compared with control cells. The transcriptional level of fibronectin was also decreased in NEDD4-depleted cells (Fig. 5A). In addition, by western blot analysis using the conditioned medium, we found that the secreted fibronectin was drastically decreased in NEDD4-knocked down NHDF cells as similar to the intracellular fibronectin (Fig. 5B). Subsequently, we investigated the expression of collagen in NEDD4 over-expressing cells by western blot analysis. As shown in Fig. 5C, type1 collagen (COL1A1) was upregulated in the NEDD4 transiently expressed NIH3T3 cells.
Figure 5.
Depletion of NEDD4 decreased the expression of collagen and fibronectin. (A) Type 1 or 3 collagen and fibronectin expression was diminished in NEDD4 knocked down NHDF cells by RT-PCR. (B) The extracellular fibronectin was detected by immunoblot analysis using cell conditioned culture media. The protein level of fibronectin also decreased in NEDD4 depleted NHDF cells both extra- and intracellular compartment. CM; conditioned media. (C) The collagen expression was analyzed 72h after transfection of expression vector. The protein level of collagen was increased in NEDD4 over-expressing NIH3T3 cells than mock cells.
Discussion
In this study, we investigated for biological functions of NEDD4, which was identified as one of the genes susceptible to keloid through our recent GWAS for keloid disease.17) Keloid fibroblasts are characterized by its hyperproliferative and invasive nature.1) Since our data indicated that NEDD4 over-expression in fibroblasts promoted proliferation and invasiveness of the cells through dysregulation of the PTEN/Akt pathway, NEDD4 is very likely to be involved in the abnormal process of the wound healing.
NEDD4 is an E3 ubiquitin ligase containing a HECT domain and PTEN was recently identified as one of its substrates.33) PTEN was first identified as a tumor suppressor gene on human chromosome 10q23 and later its frequent inactivation in various types of human cancer was reported.41,42) The functional significance of PTEN is also supported by its germline mutations in patients with a group of autosomal dominant syndromes characterized by developmental disorders, multiple hamartomas and neurological deficits that are referred as PHTS.35,36,43) Cowden disease, Bannayan–Riley–Ruvalcaba syndrome (BRRS), Proteus syndrome and Proteus-like syndrome that belong to PHTS are characterized by occurrence of multiple tumor-like masses.35) Patients with PHTS are well known to have high risk to develop keloid disease after surgical operations or injuries,36,43) implying a potential role of the PTEN pathway in keloid formation. PTEN is a plasma membrane lipid phosphatase that antagonizes phophatidylinositol 3-kinase (PI3K)/Akt signaling.44) Loss of PTEN activity leads to Akt activation that can promote cell survival, proliferation, and invasion through phosphorylation of many substrates including MDM2, GSK3, and p27.37,44)
During wound healing process, p27 as well as GSK3β/β-catenin are considered to be important molecules for the cell–cell contact inhibition22,28) that is a mechanism inhibiting the cell motility and proliferation when cells reach to the high density and/or establish cell–cell contacts. This contact inhibition process is implicated to be critically important for development and tissue regeneration. Failure of the contact inhibition is usually associated with abnormal growth of cells and the appearance of multilayered foci in culture, which are often observed in the malignant transformation process.45,46) The contact inhibition consists of two important processes, EMT and cell-cycle regulation. EMT is critical for appropriate tissue remodeling and cell migration during embryonic development, and is re-engaged in adults during wound healing and tissues regeneration. During EMT, β-catenin is up-regulated and located in the cytoplasm and/or the nucleus.26–28) The nuclear β-catenin binds to T-cell factor/lymphoid enhancer binding factor (TCF/LEF) transcription factors and activates transcription of their target genes.30–32,47) β-catenin is known to be accumulated during fibroproliferation, and was shown its elevated expression in aggressive fibromatosis and keloid tissues.29,48) Cytoplasmic β-catenin is degraded after it was phosphorylated by GSK3β, which was shown to be phosphorylated and inactivated by Akt. Hence, the activation of Akt is considered to induce β-catenin stabilization in cytoplasm. Akt is also known to phosphorylate p27, a cyclin-dependent kinase inhibitor, that arrests the cell cycle at G0/G1 phase.22,49) Several studies implicated that the level of p27 protein was increased in the cell-cycle arrested cells by contact inhibition.22,23,50,51) On the other hand, down-regulation of p27 was indicated to lead to failure of contact inhibition.24,25)
Our study implicated that the fibroblast cells, in which NEDD4 was exogenously over-expressed, decreased the amount of p27 and accumulated cytoplasmic β-catenin through dysregulation of PTEN. This abnormal regulation contributed to promote the cell proliferation or invasiveness. Furthermore, NEDD4 expression led to expression of ECM components and induced the accumulation of collagen or fibronectin. These results demonstrated that NEDD4 over-expressing cells acquired similar phenotypes as keloid fibroblasts. While TGF-β/SMAD signaling is known as the regulator of collagen expression, the mechanisms of accumulation of collagen in the patients of keloid disease are still not fully understood. Our results provide the clue for the clarification of the formation of keloid tissues. We are still puzzled in the relationship between NEDD4 expression and predisposed individuals of keloid and the direct regulator of ECM compounds in NEDD4 signal transduction. Although the expression level in keloid fibroblasts seemed to be lower than that in NHDF in Fig. 1A, these in vitro expression levels may not reflect exactly the in vivo pathological conditions. We need to examine the expression of NEDD4 using the clinical samples of an active stage of keloid disease, but biopsy samples at such stage is extremely difficult to obtain because biopsy itself becomes the stimuli for cell growth and may make the keloid disease condition worse. Although further investigation is required, our study first implies that NEDD4 is functionally related to keloid formation and suggest that the NEDD4 is a potential target for treatment and/or prevention of the keloid disease.
Acknowledgments
We would like to thank Miss Mami U for her outstanding technical assistance. This work was conducted as a part of the BioBank Japan Project that was supported by the Ministry of Education, Culture, Sports, Science and Technology of the Japanese government.
Abbreviations
- COL1A1
type 1 collagen
- ECM
extracellular matrix
- EMT
ephithelial mesenchymal transition
- GSK3β
glycogen synthase kinase 3 beta
- NEDD4
neuronal precursor cell-expressed developmentally downregulated 4
- NHDF
normal human dermal fibroblast
- PTEN
phosphatase and tensin homolog deleted on chromosome10
References
- 1).Seifert O., Mrowietz U. (2009) Keloid scarring: bench and bedside. Arch. Dermatol. Res. 301, 259–272 [DOI] [PubMed] [Google Scholar]
- 2).Tuan T.L., Nichter L.S. (1998) The molecular basis of keloid and hypertrophic scar formation. Mol. Med. Today 4, 19–24 [DOI] [PubMed] [Google Scholar]
- 3).Kose O., Waseem A. (2008) Keloids and hypertrophic scars: are they two different sides of the same coin? Dermatol. Surg. 34, 336–346 [DOI] [PubMed] [Google Scholar]
- 4).Shih B., Bayat A. (2010) Genetics of keloid scarring. Arch. Dermatol. Res. 302, 319–339 [DOI] [PubMed] [Google Scholar]
- 5).Marneros A.G., Norris J.E., Olsen B.R., Reichenberger E. (2001) Clinical genetics of familial keloids. Arch. Dermatol. 137, 1429–1434 [DOI] [PubMed] [Google Scholar]
- 6).Babu M., Diegelmann R., Oliver N. (1992) Keloid fibroblasts exhibit an altered response to TGF-beta. J. Invest. Dermatol. 99, 650–655 [DOI] [PubMed] [Google Scholar]
- 7).Bettinger D.A., Yager D.R., Diegelmann R.F., Cohen I.K. (1996) The effect of TGF-beta on keloid fibroblast proliferation and collagen synthesis. Plast. Reconstr. Surg. 98, 827–833 [DOI] [PubMed] [Google Scholar]
- 8).Lee T.Y., Chin G.S., Kim W.J., Chau D., Gittes G.K., Longaker M.T. (1999) Expression of transforming growth factor beta 1, 2, and 3 proteins in keloids. Ann. Plast. Surg. 43, 179–184 [PubMed] [Google Scholar]
- 9).Fujiwara M., Muragaki Y., Ooshima A. (2005) Upregulation of transforming growth factor-beta1 and vascular endothelial growth factor in cultured keloid fibroblasts: relevance to angiogenic activity. Arch. Dermatol. Res. 297, 161–169 [DOI] [PubMed] [Google Scholar]
- 10).Bock O., Yu H., Zitron S., Bayat A., Ferguson M.W., Mrowietz U. (2005) Studies of transforming growth factors beta 1–3 and their receptors I and II in fibroblast of keloids and hypertrophic scars. Acta Derm. Venereol. 85, 216–220 [DOI] [PubMed] [Google Scholar]
- 11).Phan T.T., Lim I.J., Aalami O., Lorget F., Khoo A., Tan E.K., Mukhopadhyay A., Longaker M.T. (2005) Smad3 signalling plays an important role in keloid pathogenesis via epithelial-mesenchymal interactions. J. Pathol. 207, 232–242 [DOI] [PubMed] [Google Scholar]
- 12).Gao Z., Wang Z., Shi Y., Lin Z., Jiang H., Hou T., Wang Q., Yuan X., Zhao Y., Wu H., Jin Y. (2006) Modulation of collagen synthesis in keloid fibroblasts by silencing Smad2 with siRNA. Plast. Reconstr. Surg. 118, 1328–1337 [DOI] [PubMed] [Google Scholar]
- 13).Bayat A., Bock O., Mrowietz U., Ollier W.E., Ferguson M.W. (2002) Genetic susceptibility to keloid disease and transforming growth factor beta 2 polymorphisms. Br. J. Plast. Surg. 55, 283–286 [DOI] [PubMed] [Google Scholar]
- 14).Bayat A., Bock O., Mrowietz U., Ollier W.E., Ferguson M.W. (2003) Genetic susceptibility to keloid disease and hypertrophic scarring: transforming growth factor beta1 common polymorphisms and plasma levels. Plast. Reconstr. Surg. 111, 535–543; discussion 544–536 [DOI] [PubMed] [Google Scholar]
- 15).Bayat A., Bock O., Mrowietz U., Ollier W.E., Ferguson M.W. (2004) Genetic susceptibility to keloid disease: transforming growth factor beta receptor gene polymorphisms are not associated with keloid disease. Exp. Dermatol. 13, 120–124 [DOI] [PubMed] [Google Scholar]
- 16).Bayat A., Walter J.M., Bock O., Mrowietz U., Ollier W.E., Ferguson M.W. (2005) Genetic susceptibility to keloid disease: mutation screening of the TGFbeta3 gene. Br. J. Plast. Surg. 58, 914–921 [DOI] [PubMed] [Google Scholar]
- 17).Nakashima M., Chung S., Takahashi A., Kamatani N., Kawaguchi T., Tsunoda T., Hosono N., Kubo M., Nakamura Y., Zembutsu H. (2010) A genome-wide association study identifies four susceptibility loci for keloid in Japanese population. Nat. Genet. 42, 768–771 [DOI] [PubMed] [Google Scholar]
- 18).Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 19).Mani A., Gelmann E.P. (2005) The ubiquitin-proteasome pathway and its role in cancer. J. Clin. Oncol. 23, 4776–4789 [DOI] [PubMed] [Google Scholar]
- 20).Bernassola F., Karin M., Ciechanover A., Melino G. (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 [DOI] [PubMed] [Google Scholar]
- 21).Anan T., Nagata Y., Koga H., Honda Y., Yabuki N., Miyamoto C., Kuwano A., Matsuda I., Endo F., Saya H., Nakao M. (1998) Human ubiquitin-protein ligase Nedd4: expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes. Genes Cells 3, 751–763 [DOI] [PubMed] [Google Scholar]
- 22).Polyak K., Kato J.Y., Solomon M.J., Sherr C.J., Massague J., Roberts J.M., Koff A. (1994) p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 8, 9–22 [DOI] [PubMed] [Google Scholar]
- 23).Levenberg S., Yarden A., Kam Z., Geiger B. (1999) p27 is involved in N-cadherin-mediated contact inhibition of cell growth and S-phase entry. Oncogene 18, 869–876 [DOI] [PubMed] [Google Scholar]
- 24).Motti M.L., Califano D., Baldassarre G., Celetti A., Merolla F., Forzati F., Napolitano M., Tavernise B., Fusco A., Viglietto G. (2005) Reduced E-cadherin expression contributes to the loss of p27kip1-mediated mechanism of contact inhibition in thyroid anaplastic carcinomas. Carcinogenesis 26, 1021–1034 [DOI] [PubMed] [Google Scholar]
- 25).Carrano A.C., Eytan E., Hershko A., Pagano M. (1999) SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1, 193–199 [DOI] [PubMed] [Google Scholar]
- 26).Kim K., Lu Z., Hay E.D. (2002) Direct evidence for a role of beta-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol. Int. 26, 463–476 [DOI] [PubMed] [Google Scholar]
- 27).Liebner S., Cattelino A., Gallini R., Rudini N., Iurlaro M., Piccolo S., Dejana E. (2004) Beta-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J. Cell Biol. 166, 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Orford K., Orford C.C., Byers S.W. (1999) Exogenous expression of beta-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J. Cell Biol. 146, 855–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Sato M. (2006) Upregulation of the Wnt/beta-catenin pathway induced by transforming growth factor-beta in hypertrophic scars and keloids. Acta Derm. Venereol. 86, 300–307 [DOI] [PubMed] [Google Scholar]
- 30).Huber O., Korn R., McLaughlin J., Ohsugi M., Herrmann B.G., Kemler R. (1996) Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech. Dev. 59, 3–10 [DOI] [PubMed] [Google Scholar]
- 31).Tetsu O., McCormick F. (1999) Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 [DOI] [PubMed] [Google Scholar]
- 32).Brabletz T., Jung A., Dag S., Hlubek F., Kirchner T. (1999) Beta-catenin regulates the expression of the matrix metalloproteinase-7 in human colorectal cancer. Am. J. Pathol. 155, 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Wang X., Trotman L.C., Koppie T., Alimonti A., Chen Z., Gao Z., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C., Pandolfi P.P., Jiang X. (2007) NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Fouladkou F., Landry T., Kawabe H., Neeb A., Lu C., Brose N., Stambolic V., Rotin D. (2008) The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc. Natl. Acad. Sci. U. S. A. 105, 8585–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Eng C. (2003) PTEN: One gene, many syndromes. Hum. Mutat. 22, 183–198 [DOI] [PubMed] [Google Scholar]
- 36).Malamitsi-Puchner A., Dimitriadis D., Bartsocas C., Wiedemann H.R. (1990) Proteus syndrome: course of a severe case. Am. J. Med. Genet. 35, 283–285 [DOI] [PubMed] [Google Scholar]
- 37).Scheid M.P., Woodgett J.R. (2001) PKB/AKT: functional insights from genetic models. Nat. Rev. Mol. Cell Biol. 2, 760–768 [DOI] [PubMed] [Google Scholar]
- 38).Vivanco I., Sawyers C.L. (2002) The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer 2, 489–501 [DOI] [PubMed] [Google Scholar]
- 39).Shin I., Yakes F.M., Rojo F., Shin N.Y., Bakin A.V., Baselga J., Arteaga C.L. (2002) PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat. Med. 8, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 40).Bunney T.D., Katan M. (2010) Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat. Rev. Cancer 10, 342–352 [DOI] [PubMed] [Google Scholar]
- 41).Steck P.A., Pershouse M.A., Jasser S.A., Yung W.K., Lin H., Ligon A.H., Langford L.A., Baumgard M.L., Hattier T., Davis T., Frye C., Hu R., Swedlund B., Teng D.H., Tavtigian S.V. (1997) Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 15, 356–362 [DOI] [PubMed] [Google Scholar]
- 42).Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S.I., Puc J., Miliaresis C., Rodgers L., McCombie R., Bigner S.H., Giovanella B.C., Ittmann M., Tycko B., Hibshoosh H., Wigler M.H., Parsons R. (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 [DOI] [PubMed] [Google Scholar]
- 43).Viljoen D.L., Nelson M.M., de Jong G., Beighton P. (1987) Proteus syndrome in southern Africa: natural history and clinical manifestations in six individuals. Am. J. Med. Genet. 27, 87–97 [DOI] [PubMed] [Google Scholar]
- 44).Salmena L., Carracedo A., Pandolfi P.P. (2008) Tenets of PTEN tumor suppression. Cell 133, 403–414 [DOI] [PubMed] [Google Scholar]
- 45).Fagotto F., Gumbiner B.M. (1996) Cell contact-dependent signaling. Dev. Biol. 180, 445–454 [DOI] [PubMed] [Google Scholar]
- 46).Abercrombie M. (1979) Contact inhibition and malignancy. Nature 281, 259–262 [DOI] [PubMed] [Google Scholar]
- 47).Jho E.H., Zhang T., Domon C., Joo C.K., Freund J.N., Costantini F. (2002) Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Cheon S.S., Cheah A.Y., Turley S., Nadesan P., Poon R., Clevers H., Alman B.A. (2002) beta-Catenin stabilization dysregulates mesenchymal cell proliferation, motility, and invasiveness and causes aggressive fibromatosis and hyperplastic cutaneous wounds. Proc. Natl. Acad. Sci. U. S. A. 99, 6973–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Nelson P.J., Daniel T.O. (2002) Emerging targets: molecular mechanisms of cell contact-mediated growth control. Kidney Int. 61, S99–S105 [DOI] [PubMed] [Google Scholar]
- 50).Deleu L., Fuks F., Spitkovsky D., Horlein R., Faisst S., Rommelaere J. (1998) Opposite transcriptional effects of cyclic AMP-responsive elements in confluent or p27KIP-overexpressing cells versus serum-starved or growing cells. Mol. Cell. Biol. 18, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Dietrich C., Wallenfang K., Oesch F., Wieser R. (1997) Differences in the mechanisms of growth control in contact-inhibited and serum-deprived human fibroblasts. Oncogene 15, 2743–2747 [DOI] [PubMed] [Google Scholar]