Abstract
Flock house virus (FHV), a single-stranded RNA insect virus, has previously been reported to cross the kingdom barrier and replicate in barley protoplasts and in inoculated leaves of several plant species [Selling, B. H., Allison, R. F. & Kaesberg, P. (1990) Proc. Natl. Acad. Sci. USA 87, 434–438]. There was no systemic movement of FHV in plants. We tested the ability of movement proteins (MPs) of plant viruses to provide movement functions and cause systemic spread of FHV in plants. We compared the growth of FHV in leaves of nontransgenic and transgenic plants expressing the MP of tobacco mosaic virus or red clover necrotic mosaic virus (RCNMV). Both MPs mobilized cell-to-cell and systemic movement of FHV in Nicotiana benthamiana plants. The yield of FHV was more than 100-fold higher in the inoculated leaves of transgenic plants than in the inoculated leaves of nontransgenic plants. In addition, FHV accumulated in the noninoculated upper leaves of both MP-transgenic plants. RCNMV MP was more efficient in mobilizing FHV to noninoculated upper leaves. We also report here that FHV replicates in inoculated leaves of six additional plant species: alfalfa, Arabidopsis, Brassica, cucumber, maize, and rice. Our results demonstrate that plant viral MPs cause cell-to-cell and long-distance movement of an animal virus in plants and offer approaches to the study of the evolution of viruses and mechanisms governing mRNA trafficking in plants as well as to the development of promising vectors for transient expression of foreign genes in plants.
Keywords: nodavirus
Viruses whose host range spans two kingdoms are few. Most are insect-vectored plant viruses that multiply in a narrow insect host range (1). Flock house virus (FHV), a member of the insect/animal virus family called Nodaviridae (for reviews, see refs. 2–4), is a unique example of a virus that crosses the kingdom barrier. FHV was originally isolated from the New Zealand grass grub Costelytra zealandica (5). The range of insect hosts for FHV is not well studied. In the laboratory, FHV replicates in the larvae of the wax moth Galleria mellonella and cultured Drosophila melanogaster cells (4). In 1990, FHV was shown to replicate and produce infectious virions in barley protoplasts and the inoculated leaves of several other monocotyledon and dicotyledon plant species without producing symptoms (6). FHV thus replicates in both insects (2) and plants (6), although it is not transmitted to plants by its insect hosts (2).
The genome of FHV consists of two small messenger sense RNAs: RNA1 (3.1 kb; ref. 7) encodes the viral polymerase, and RNA2 (1.4 kb; ref. 8) encodes the virion capsid protein. RNA1 is capable of independent replication in insect host cells. Replication of RNA2 is RNA1-dependent, and the formation of progeny virus particles requires coinfection of cells by both RNAs (9). FHV infectivity titers can be quantified by a plaque assay on D. melanogaster cells (10).
Broad host range, simple genome organization, and asymptomatic growth in plants make FHV a powerful tool for the study of fundamental questions of virus–host interactions and the modification of gene expression in plants. A drawback to its use is that FHV does not encode a protein that enables it to move inside the plant. In all plants previously tested, FHV accumulated only in inoculated tissues or cells (6).
Here we report the results of experiments designed to determine whether FHV could be mobilized in plants with the use of plant viral movement proteins (MPs). We used genes encoding the 30-kDa protein of tobacco mosaic virus (TMV) and the 35-kDa protein of red clover necrotic mosaic virus (RCNMV) (dianthovirus), two of the most well-studied and widely used plant viral MPs, both expressed as transgenes in N. benthamiana plants (11–16). These plant viruses each encode a single, functionally homologous MP with conserved amino acid sequences (14, 17), and neither virus requires coat protein for cell-to-cell movement (16, 18). These MPs have been shown conclusively to bind RNA in vitro and to increase the size exclusion limit of plasmodesmata in mesophyl cells to allow cell-to-cell movement (19). Moreover, both TMV MP and RCNMV MP have been shown to complement the transport of other plant viruses belonging to different taxonomic groups, such as barley stripe mosaic virus (20, 21), potato virus X (22), and brome mosaic virus (23).
Our results show that MPs of both TMV and RCNMV initiate local as well as long-distance movement of FHV in MP transgenic plants. RCNMV MP is more effective in mobilizing movement of FHV to noninoculated leaves. We also show that FHV replicates in inoculated leaves of six more plant species, in addition to those reported previously (6). Newly discovered experimental hosts of FHV include alfalfa, Arabidopsis, Brassica, cucumber, rice, and sweet corn.
Materials and Methods
Plant Growth and Inoculation.
The plants tested for the growth of FHV were Nicotiana benthamiana, alfalfa (Medicago sativa L.), Arabidopsis (Arabidopsis thaliana), Brassica (Brassica rapa), cucumber (Cucumis sativus), rice (Oryza sativa L.), Golden Cross Bantam hybrid sweet corn (Zea mays L.), tomato (Lycopersicon esculentum L.), and potato (Solanum tuberosum L.). N. benthamiana seeds (wild-type and transgenic expressing TMV MP or RCNMV MP; refs. 15 and 16) were kindly provided by Roger Beachy (Donald Danforth Plant Science Center, St. Louis) and Steven Lommel (North Carolina State University), respectively. Other seeds were obtained from the Department of Horticulture and Department of Plant Pathology, University of Wisconsin–Madison.
Plants were grown from seeds planted in Scotts Redi-earth Plug and Seedling Mix and placed in growth chambers maintained at 24°C, with 12 h of light/24-h day provided by 40-W cool white fluorescent bulbs (215 μE/m2/s). Half-strength Hoagland's medium (24) was used as the source of nutrients.
Leaves of 2- to 3-week-old plants were dusted with carborundum powder and inoculated (with the use of cotton swabs) with 100 μl of a mixture containing bentonite (5 μg/μl) and FHV RNA (10 μg/μl) in 25 mM Tris⋅HCl, 25 mM Na2HPO4, 250 mM NaCl, and 5 mM EDTA (pH 7.5). After inoculation, leaves were rinsed with distilled water, and the plants were maintained as described above.
Virus and RNA Extraction.
We harvested inoculated leaves and noninoculated upper leaves by cutting the petioles with scissors. We rinsed the leaves with distilled water, air-dried them, and stored them at −80°C. For virus extractions, 100 mg of leaf tissues was homogenized in 1.5-ml Eppendorf tubes with the use of disposable plastic pestles (VWR Scientific). Isotonic buffer (100 mM NaCl/35 mM Pipes/10 mM KCl/1 mM MgCl2/1 mM CaCl2, pH 6.8) was added (1 ml), and the extracts were centrifuged (Eppendorf centrifuge, model 5415C) at 10,000 rpm for 8 min. The supernatant liquids were diluted for use in plaque assays (10). When not used immediately, the virus extracts were stored at −80°C.
RNA extractions were carried out according to a hot phenol procedure (25). Leaf samples (100 mg) were frozen in liquid N2 and homogenized to a powder by hand with a pestle in a chilled porcelain mortar. The homogenates were extracted with 0.5 ml of a 1:1 mixture of phenol (saturated phenol solution; Amresco, Solon, OH) and buffer (0.1 M LiCl/100 mM Tris⋅HCl/10 mM EDTA/1% SDS, pH 8.0) at 70°C, followed by another extraction of the aqueous phase with 0.25 ml of chloroform:isoamyl alcohol (24:1). RNAs were recovered by precipitation with the addition of one volume of 4 M LiCl, incubation overnight at 4°C, and centrifugation at 14,000 rpm. Pellets were resuspended in 200 μl of RNase-free water. We removed DNA by treatment with DNase I (RQ1 RNase-free DNase; Promega) at 1 unit of enzyme/μg of total nucleic acids (37°C for 30 min). The enzyme was inactivated by two extractions with phenol:chloroform (1:1), and the RNA was precipitated with ethanol in the presence of 0.3 M Na acetate (pH 5.2) at −20°C. RNA pellets were recovered by centrifugation for 15 min, washed with 70% ethanol, and dried in air at room temperature for 10 min. Pellets were resuspended in 40 μl of RNase-free water and quantified by UV absorption at 260 nm and 280 nm with the use of a Hitachi (model 2001) spectrophotometer.
Plaque Assay.
Plaque assays for the detection and quantitation of FHV in leaf homogenates were performed on monolayers of D. melanogaster cells as described (6, 10). Briefly, 95 μl of cells (8 × 106) grown in Schneider's insect cell medium (26) were added to 5 μl of virus extract (prepared as described above) and mixed with gentle shaking at room temperature for 1 h. Cells were then poured into tissue culture dishes (60- to 100-mm diameter Corning polystyrene cell culture dishes) and, after 1 h, when cells were attached to the dishes, medium was replaced with an overlay of 1% agarose (low melting point; GIBCO/BRL) containing Schneider's medium. Dishes were incubated in the dark at 26°C, and we visualized the plaques after 60 h by staining the cells with 0.5 ml of 3 mg/ml solution of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma) in isotonic buffer. Infectivity of plant homogenates was converted into plaque-forming units (pfu)/100 mg of leaf tissue.
Reverse Transcription–PCR (RT-PCR) Assay of FHV RNA.
Reverse transcription (RT) of FHV RNAs in total leaf RNA extracts was performed with Superscript II (GIBCO/BRL) to produce virus-specific cDNAs that were then amplified by PCR with the use of Taq DNA polymerase (Promega). FHV RNA primer pairs for amplifying full-length RNA2 (1.4 kb) and 1240 bases of RNA1 were designed from the known sequences (7, 8) and were synthesized at the Biotechnology Center, University of Wisconsin–Madison. For the first-strand cDNA reactions, total RNAs (1–5 μg) from buffer-inoculated, FHV RNA-inoculated, and non-inoculated plant leaves were used as templates. FHV RNA2 (1 μg) purified from virions grown in Drosophila cells was used as a positive control. RNA samples were combined with 10 pmol of reverse primers specific for FHV RNA1 (5′-CTG GCC AAT GGA CGC G-3′; complementary to nt 1225–1240; ref. 7) or RNA2 (5′-ACC TTA GTC TGT TGA C-3′; complementary to nt 1385–1400; ref. 8) and annealed at 65°C for 5 min. Two hundred units of Superscript II in a total volume of 20 μl was used for each reaction. Conditions were similar to the manufacturer's instructions, except that we added 2 units of RNasin (Promega) to each reaction to inhibit ribonuclease activity. Incubation was carried out at 42°C for 1 h. Reactions were terminated by heating at 95°C for 5 min. The reaction mixtures were then diluted to 50 μl, and a 2-μl sample was used for the PCR amplification by adding 10 pmol of forward primers specific for FHV RNA1 (5′-GTT TTC GAA ACA AAT-3′; nt 1–15; ref. 7) or RNA2 (5′-GTA AAC AAT TCC AAG-3′; nt 1–15; ref. 8) and 5 units of Taq polymerase. The cycling parameters for the PCR reactions (25 μl) were 1 min of denaturation at 94°C followed by 35 cycles at 94°C for 30 s, 55°C for 1 min, and 72°C for 1.5 min, followed by incubation at 72°C for 2 min. PCR products (10 μl) were analyzed on 1% agarose gels in 1× TBE buffer (89 mM Tris/89 mM boric acid/2.5 mM EDTA, pH 8.3).
Sequence Analysis of RT-PCR Products.
RT-PCR reaction products (25 μl) were purified with the use of QIAquick PCR purification kits (Qiagen, Valencia, CA) according to the manufacturer's protocol. Typical sequencing reactions consisted of 0.5 μg of gel-purified RT-PCR product, 4 μl of Big Dye Terminator mix (PE Biosystems, Foster City, CA.), 4 μl of 2.5× dilution buffer containing 200 mM Tris⋅HCl (pH 9.0)/5 mM MgCl2, and 6 pmol of reverse primers specific for FHV RNA1 or RNA2 (above) in a final reaction volume of 20 μl. The PCR parameters were a 3-min denaturation step at 94°C followed by 35 cycles at 94°C for 30 s, 45°C for 20 s, and 60°C for 4 min, followed by incubation at 72°C for 7 min. Excess dye terminators were removed with the use of Amersham Pharmacia AutoSeq G50 columns (according to the manufacturer's instructions), and the samples were dried in a Speed-Vac (Savant). Sequencing gels were run at the Biotechnology Center, University of Wisconsin–Madison, with the use of an automated DNA sequencing instrument (PE Biosystems 377XL). Data were analyzed with abi (Version 3.3) sequence analysis software.
Results
Virus Yield in Inoculated Leaves of Wild-Type and Transgenic N. benthamiana Plants.
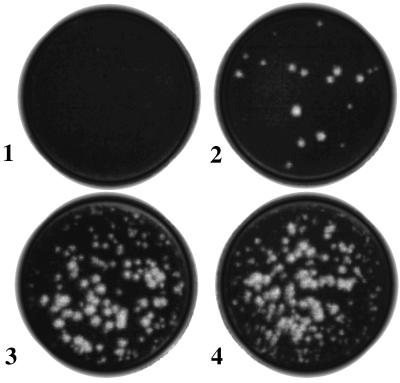
We attempted to determine whether viral MPs increased yield of FHV in inoculated leaves by comparing the infectivity titers of FHV recovered from plants expressing TMV MP or RCNMV MP with those recovered from nontransgenic control plants. Nontransgenic and transgenic plants were grown, inoculated with FHV RNA, and then processed under identical conditions. Plaque assays on monolayers of Drosophila cells were conducted with the use of diluted homogenates of leaves under conditions where infectivity due only to assembled virions would be detected. Results of a typical assay (Fig. 1) showed that nontransgenic leaf homogenates produced 17 plaques (equivalent to a virus titer of 3.4 × 105 pfu/100 mg tissue; plate 2) after 100-fold dilution, whereas leaf homogenates of TMV and RCNMV-MP transgenic plants each produced ≈200 plaques (2 × 107 pfu/100 mg tissue; plates 3 and 4) after 1,000-fold dilution. Buffer-inoculated leaf homogenates showed no plaques (plate 1). When half leaves were inoculated with FHV RNA, we detected similarly increased virus titer in the noninoculated half leaves of transgenic plants but not in those of nontransgenic control plants (data not shown). These results support the interpretation that MPs of both TMV and RCNMV mediated cell-to-cell movement of FHV in inoculated leaves and that the increase in virus titers was the result of virus spread beyond the localized sites of virus inoculation.
Figure 1.
Plaque assay showing an increase in FHV titers in inoculated leaves of MP-transgenic N. benthamiana plants. Leaves harvested 12 days after inoculation were homogenized and assayed for infectious FHV as described in the text. Five microliters of 1-ml homogenates derived from 100 mg of leaf tissues were used in each assay after appropriate dilutions (100-fold for nontransgenic and 1,000-fold for transgenic). Plates: 1, buffer-inoculated transgenic; 2, FHV RNA-inoculated nontransgenic; 3, FHV RNA-inoculated TMV-MP transgenic; 4, FHV RNA-inoculated RCNMV-MP transgenic leaf homogenates.
Detection of Infectious Virus in the Inoculated and Noninoculated Leaves of Transgenic N. benthamiana Plants.
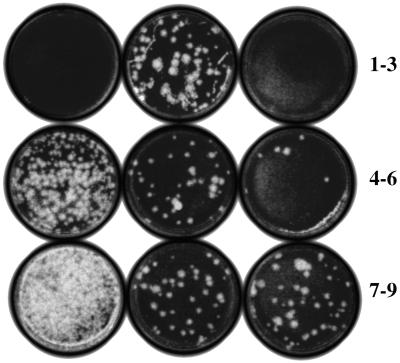
To test for and compare the systemic spread of FHV in nontransgenic and transgenic N. benthamiana plants, we prepared homogenates from upper noninoculated leaves and assayed for the presence of virus, following the procedure described above. Fig. 2 shows the results of a typical experiment. No plaques were found from assays of homogenates prepared from the buffer-inoculated leaf of a RCNMV-MP transgenic plant (plate 1). Homogenates from FHV RNA–inoculated leaves and noninoculated upper leaves of nontransgenic plants after 10-fold dilution showed 40 plaques (8 × 104 pfu/100 mg tissue; plate 2) and no plaques (plate 3), respectively, showing that FHV was present only in the inoculated leaves and not in the upper noninoculated leaves of nontransgenic plants. Homogenates from the inoculated leaves of TMV-MP transgenic plants produced 90 plaques (1.8 × 107 pfu/100 mg tissue; plate 4) after 1,000-fold dilution, which was more than 200-fold higher than the number of plaques found in the comparable leaves of nontransgenic plants (compare plates 2 and 4). The homogenates of the second and third upper noninoculated leaves after 1,000-fold dilution produced 30 and 5 plaques (6 × 106 and 1.0 × 106 pfu/100 mg tissue, respectively; plates 5 and 6), respectively, showing spread of FHV to those leaves. These virus titers are much lower (30% and 5%, respectively) compared with those in inoculated leaves of transgenic plants (1.8 × 107) but still significantly higher (about 75-fold in the second upper leaf and 13-fold in the upper third leaf, respectively) than was detected in the inoculated leaf of nontransgenic plants (8 × 104; plate 2).
Figure 2.
Plaque assay showing FHV accumulation in inoculated and noninoculated leaves of MP-transgenic N. benthamiana plants. Plaque assays, as described in Fig. 1, were conducted 14 days after inoculation with leaf homogenates after 10-fold (plates 1–3) and 1,000-fold (plates 4–9) dilutions. Plates: 1–3, nontransgenic plant, buffer-inoculated (1), FHV RNA-inoculated (2), and noninoculated upper second leaf (3); 4–6, TMV-MP transgenic plant, FHV RNA-inoculated (4) and noninoculated upper second (5) and third (6) leaves; 7–9, RCNMV-MP transgenic plant, FHV RNA-inoculated (7) and noninoculated upper second (8) and third (9) leaves.
Infectivity assays of leaf homogenates from inoculated leaves of RCNMV-MP transgenic plants after 1,000-fold dilution produced 120 plaques (2.4 × 107 pfu/100 mg tissue; plate 7). The second and third upper noninoculated leaves on these plants each produced 40 plaques (8.0 × 106 pfu/100 mg tissue; plates 8 and 9), showing spread of FHV to the upper noninoculated leaves. Once again, these virus titers were lower (33%) compared with that of the inoculated leaf of transgenic plants but still significantly higher (about 100-fold in both second and third upper leaves) than was detected in the inoculated leaf of nontransgenic plants. Notably, the virus titer in the third upper leaf was not lower than that in the second upper leaf of RCNMV-MP transgenic plants, in contrast with the case of TMV-MP transgenic plants. These results confirm that both TMV MP and RCNMV MP lead to at least a 100-fold increase in virus yield in the inoculated leaves and the accumulation as well of assembled virus particles in noninoculated leaves of MP transgenic N. benthamiana plants. We also attempted to determine whether the presence of the RCNMV MP transgene itself increased viral replication and accumulation by transfecting protoplasts isolated from leaves of transgenic and nontransgenic N. benthamiana plants with FHV RNA (equimolar mixture of FHV RNAs 1 + 2). We collected samples over a 48-h period and assayed viral titers over time by plaque assay as described (6, 10). The results showed that FHV viral titers are not higher in the RCNMV MP transgenic protoplasts than in nontransgenic protoplasts (data not shown; details can be found at http://www.plantpath.wisc.edu/jobob/addinfo.htm). These results demonstrate that neither the RCNMV MP transgene nor its gene product increased the virus titers in transfected, transgene-expressing cells.
Detection of Viral RNA Sequences in the Inoculated and Noninoculated Leaves of Transgenic N. benthamiana Plants.
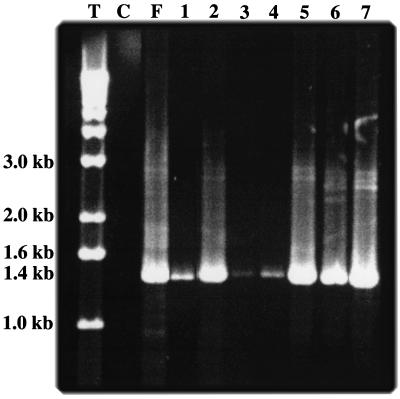
Total RNAs extracted from leaf samples collected 12 days after inoculation of TMV-MP transgenic plants were used as templates for RT-PCR to investigate the relationship between virus accumulation and FHV RNA accumulation in the inoculated and noninoculated leaves. Amplification with FHV RNA2-specific primers produced specific bands of expected sizes, although the yields were variable probably due to the presence of inhibitors of Taq polymerase in plant RNA preparations. As shown in Fig. 3, bands corresponding to FHV RNA2 were present in inoculated (lane 2) as well as noninoculated upper second and third leaves of TMV-MP transgenic plants (lanes 3 and 4). When total RNAs from leaves of nontransgenic plants were used as templates for RT-PCR, a band of much lower intensity was obtained from the inoculated leaf (lane 1), and no bands were obtained from upper noninoculated leaves (data not shown). This result shows that FHV RNA2 accumulates in the upper leaves of TMV-MP transgenic plants, although the amounts are significantly less than those in inoculated leaves of such plants.
Figure 3.
FHV RNA accumulation in inoculated and noninoculated leaves of MP-transgenic N. benthamiana plants. FHV RNA2 (1 μg) purified from virions (positive control) or total RNAs (1–5 μg) extracted from leaves were used as templates for RT-PCR with FHV RNA2 specific primers, as described in the text. Products were analyzed by electrophoresis on 1% agarose gel. Lanes: M, 1-kb ladder; C, buffer-inoculated leaf (negative control); F, FHV RNA2; 1, nontransgenic plant, FHV RNA inoculated; 2–4, TMV MP transgenic plant, FHV RNA inoculated (2) and noninoculated upper second (3) and third (4) leaves; 5–7, RCNMV MP transgenic plant, FHV RNA inoculated (5) and noninoculated upper second (6) and third (7) leaves. The arrow shows the position of FHV RNA2 (1.4 kb).
In contrast to results from TMV-MP transgenic plants, the band intensities of FHV RNA2-specific RT-PCR products from both the second and third upper noninoculated leaves of RCNMV-MP transgenic plants (lanes 6 and 7) were as high as that from the inoculated leaf (lane 5). In addition, our preliminary results show low amounts of FHV RNA2 accumulation in flowers of the RCNMV MP transgenic plants (data not shown). Thus transgene expression of RCNMV MP appears to mobilize FHV infection to upper noninoculated leaves of N. benthamiana plants much more effectively than does expression of TMV MP. Moreover, although we observed a lower virus titer in the upper noninoculated leaves by plaque assay (Fig. 2), the quantity of FHV RNA2 estimated from RT-PCR reactions was approximately the same in all of the tested leaves of RCNMV-MP transgenic plants.
Amplification of FHV RNA1 (3.1 kb) by RT-PCR resulted in poor yields and almost undetectable bands corresponding to full-length FHV RNA1 in the noninoculated upper leaves. Inefficient polymerization in RT-PCR reactions and/or a low level of synthesis of FHV RNA1 in leaves at a later stage of FHV infection may have contributed to this effect. Therefore, for detection of FHV RNA1, we used PCR primers designed to amplify the first 1240 bases and obtained RT-PCR products corresponding to partial-length RNA1. Band intensities were comparable in the inoculated and upper noninoculated leaves of RCNMV-MP transgenic plants (data not shown). Authenticity of FHV RNA sequences in all of the bands was verified by elution of RT-PCR products from the gel and sequencing as described in Materials and Methods.
Detection of Infectious Virus in Inoculated Leaves of Alfalfa, Arabidopsis, Brassica, Cucumber, Rice, Maize, Tomato, and Potato.
We tested additional plant species for their ability to support FHV infection and replication. Nontransgenic and RCNMV-MP transgenic N. benthamiana plants were used as positive controls. Inoculated leaves were harvested and homogenized, and plaque assays were performed on Drosophila cell monolayers under conditions that were experimentally confirmed to detect only intact FHV virus particles. Results of one set of tests are shown in Table 1. Virus titers varied depending upon the plant species inoculated. Moreover, there were differences in the virus yield in different inoculated leaves of the same plant. Cucumber produced the highest yield, followed by sweet corn. Lower titers were obtained from rice, Brassica, alfalfa, and Arabidopsis. No plaques were detected when the leaf homogenates of potato and tomato were tested. No symptoms were detected in any inoculated leaves other than the slight tissue damage caused by rubbing of the leaves during inoculations, and FHV was not detected in any noninoculated leaf.
Table 1.
FHV yield in plants
| Plant | Mean number of plaques (± SD) | Infectivity titers, pfu/100 mg of leaf tissue |
|---|---|---|
| Dicotyledons | ||
| Alfalfa | 27 (6.3) | 5.4 × 104 |
| Arabidopsis | 60 (8.5) | 1.2 × 105 |
| Brassica | 140 (7.0) | 2.8 × 105 |
| Cucumber | 150 (7.9) | 3.0 × 106 |
| Tomato | 0 | 0 |
| Potato | 0 | 0 |
| N. benthamiana (nontransgenic) | 55 (14.1) | 1.1 × 105 |
| N. benthamiana (transgenic) | 130 (20.3) | 2.6 × 107 |
| Monocotyledons | ||
| Rice | 41 (20.1) | 8.2 × 104 |
| Maize (sweet corn) | 80 (23.3) | 1.6 × 106 |
Discussion
Our data provide conclusive evidence that FHV, a RNA insect virus capable of replicating in a wide variety of plants, can be induced to move systemically by MPs from at least two different plant viruses. We have also examined the host range of FHV and found that the virus can replicate in the inoculated leaves of six important plant species in addition to those reported by Selling et al. (6). A total of 11 plant species, including members of both Monocotyledonae and Dicotyledonae, are now known to be capable of supporting the replication, translation, and assembly of progeny FHV particles. This capability provides a unique opportunity to study evolutionary relationships and interactions between components of viruses in hosts from two different kingdoms. Moreover, asymptomatic growth of FHV in a wide variety of plants provides additional advantages over the plant viruses for the development of virus-based plant expression vectors.
Because growth of FHV in N. benthamiana was reported earlier (6) and transgenic N. benthamiana plants expressing genes encoding TMV and RCNMV MP were available (15, 16), we used this plant species as a model to test for mobility. TMV MP mobilized FHV mostly within the inoculated leaves, whereas RCNMV caused both local and systemic spread of FHV. It is not yet known whether and how TMV and RCNMV MPs interact with FHV RNAs and/or coat protein to affect movement. We also do not know why RCNMV was functionally superior to TMV MP in mobilizing FHV to upper noninoculated leaves, although cell-to-cell movement of FHV with either MP was comparable. This superiority could be due to a difference in the level of expression of RCNMV MP and TMV MP in different parts of transgenic plants. Alternatively, it could be due to similarity between nodavirus and dianthovirus (the group to which RCNMV belongs) genome strategies. FHV and RCNMV possess bipartite genomes with RNAs of similar molecular weight (7, 8, 27, 28). FHV and RCNMV RNAs lack significant similarity in primary sequences but may share functional similarities based on secondary or tertiary structures.
The mechanism by which RCNMV MP supports long-distance movement of FHV through N. benthamiana plants is unusual and needs further investigation. Normally, long-distance movement of plant viruses through vascular tissues requires the presence of plant viral coat proteins and yet unknown host factors (17, 29, 30). However, long-distance movement of viruses in plants is an area where our knowledge is incomplete, and plant virologists appreciate that different viruses solve the movement problem in different ways, depending on the cell types infected. For example, host and viral factors independent of coat proteins determine long-distance movement of bipartite geminiviruses (31). Hordeiviruses, such as barley stripe mosaic virus, also apparently do not require coat protein for systemic infection (32). A chimeric virus, in which the TMV MP gene was replaced by the RCNMV MP gene, systemically infected both N. benthamiana and tobacco, although tobacco is not a systemic host of RCNMV (14). Evidence for a paralog of the RCNMV 35-kDa MP in Cucurbita maxima has been found (33). This protein has been shown to move RNA within the phloem, the long-distance transport system that delivers nutrients and hormones to plant tissues and organs.
All of the foregoing evidence suggests that plants possess the tools for intercellular trafficking of ribonucleoprotein complexes, including those of viruses, through vascular systems and that the appropriate combination of MP and coat protein, not necessarily coded by the same family of viruses, may be sufficient to carry out such movement functions. Our finding that FHV can be mobilized to move systemically in RCNMV-MP transgenic plants provides an experimental system in which it may be possible to determine the molecular requirements for long-distance virus movement in the absence of symptoms that may complicate studies in other systems. It is possible that whereas RCNMV MP was effective in moving FHV in N. benthamiana plants, MP of a different plant virus specific for monocotyledonous (e.g., brome mosaic virus or maize dwarf mosaic virus) plants will be more suitable for moving FHV in monocotyledonous hosts.
The infectivity titers of FHV in noninoculated leaves were significantly less (5–30%) compared with those found in the inoculated leaves of the same plants. These lower infectivity titers may be due to fewer cells containing infectious FHV particles in systemically infected leaves or lower virus titer achieved per cell. Both possibilities could result from a difference in expression of MPs in upper, younger leaves, leading to uneven movement of FHV particles or a nucleoprotein complex through the plant. Degradation of FHV nucleoprotein complexes during movement through the vascular system or a lower level of FHV polymerase activity in the upper leaves could also explain the observed results. Even with plant viruses that move systemically, final distribution of virus throughout the plant depends on plant species, age of the plant, method of inoculation, and temperature, and uneven distribution is fairly common (1). Optimum conditions for growth and movement of FHV in a plant host have not been determined and could be different from what we used in our experiments. Interestingly, synthesis of RCNMV RNAs was also at a lower level in systemic leaves of RCNMV MP-transgenic N. benthamiana plants than that found in leaves inoculated with RCNMV RNAs (figure 2 in ref. 16).
Synthesis of FHV in 11 of 13 plant species tested so far, including members of both Monocotyledonae and Dicotyledonae, is particularly interesting. FHV yields were high in inoculated leaves of cucumber and sweet corn, indicating that FHV can use the cellular environment of monocotyledon and dicotyledon plants with comparable efficiency. Why FHV is capable of replication and assembly in plants and how plant cells mimic the functions provided by animal cells in the FHV infection cycle remain unclear (see the discussion in ref. 6). Several plant viruses belonging to the family of Reoviridae, Rhabdoviridae, and Bunyaviridae are known to replicate in insects, but, unlike the case with FHV, they are all transmitted to plants by the insect hosts. It seems very probable from their genome structures that these viruses originated in their animal/insect hosts. However, they are not single-stranded, positive-sense RNA viruses, and they are more complex in structure than FHV. They also cannot be mechanically transmitted to plants (1). On the other hand, FHV is not known to be transmitted to plants by its insect hosts, and FHV RNAs can be mechanically transmitted to plant hosts.
The small bipartite genome organization and the presence of subgenomic RNA suggest a close evolutionary relationship of FHV and other nodaviruses with certain plant viruses. A recent report shows evidence that a nanovirus (single-stranded DNA virus of plants) switched to a vertebrate host and then evolved into a circovirus (circular, single-stranded DNA virus of vertebrates) after recombination with an RNA calcivirus (34). Part of the nanovirus DNA, including the origin of replication, is postulated to have been transferred to the vertebrate genome when the vertebrate was exposed to sap from a nanovirus-infected plant host. Thereafter, recombination took place in the vertebrate host. We speculate that FHV too may have evolved from such recombination events between plant viruses and viruses of insects that feed on plants.
Although FHV is not known to infect animal cells other than those of insects, nodamura virus, another member of the family Nodaviridae, multiplies in insect cells (such as wax moth larvae, mosquitoes, and honey bees; refs. 35 and 36) as well as in mammalian cells (such as baby hamster kidney cells and baby mice; ref. 4). At least 25 isolates of nodaviruses infecting fish have been reported (37). Replication of FHV in yeast has also been reported (38). All of these results indicate the adaptability of nodaviruses in different hosts and show FHV to be a simple model system for the study of evolution and replication of viruses in different hosts.
Symptomless growth and systemic spread in the presence of plant MPs makes FHV an ideal candidate for vector development for gene expression in plants. Plant viruses are generally narrow in their host range, and vectors currently developed from them show variability in expression and often produce undesirable symptoms (39–41). Growth of FHV in both monocotyledons and dicotyledons and in economically important plants like rice, corn, cucumber, and Brassica are particularly promising. FHV growth in Arabidopsis provides opportunities for using FHV-based vectors for the study of the host genes responsible for replication and movement of FHV in plants as well as for applications in functional genomics.
Acknowledgments
We thank Lisa Mueller, Melissa Lehti, Violet Best, and Russell Spear for technical assistance; Dr. Mark Kainz (Colgate University) for technical advice; and Dr. Roger Beachy (Donald Danforth Plant Science Center, St. Louis, MO) and Dr. Steven Lommel (North Carolina State University) for providing transgenic N. benthamiana seeds. We are grateful to Dr. Tim Sit (North Carolina State University) and Drs. Thomas German and Jo Handelsman (University of Wisconsin–Madison) for helpful discussions and reviews of earlier drafts of the manuscript. This research was funded by University-Industry Relations, University of Wisconsin–Madison; Mycogen Seeds; the McKnight Foundation; and the United States Department of Agriculture (Grant 94-34190-1204).
Abbreviations
- FHV
flock house virus
- TMV
tobacco mosaic virus
- RCNMV
red clover necrotic mosaic virus
- MP
movement protein
- pfu
plaque-forming units
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mathews R E F. Plant Virology. 3rd ed. New York: Academic; 1991. [Google Scholar]
- 2.Hendry D A. In: Viruses of Invertebrates. Kurstak E, editor. New York: Dekker; 1991. pp. 227–276. [Google Scholar]
- 3.Schneemann A, Reddy V, Johnson J E. Adv Virus Res. 1998;50:381–446. doi: 10.1016/s0065-3527(08)60812-x. [DOI] [PubMed] [Google Scholar]
- 4.Ball L A, Johnson K L. In: The Insect Viruses. Miller L K, Ball L A, editors. New York: Plenum; 1998. pp. 225–267. [Google Scholar]
- 5.Scotti P D, Dearing S, Mossop D W. Arch Virol. 1983;75:181–189. doi: 10.1007/BF01315272. [DOI] [PubMed] [Google Scholar]
- 6.Selling B H, Allison R F, Kaesberg P. Proc Natl Acad Sci USA. 1990;87:434–438. doi: 10.1073/pnas.87.1.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasmahapatra B, Dasgupta R, Ghosh A, Kaesberg P. J Mol Biol. 1985;182:183–189. doi: 10.1016/0022-2836(85)90337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dasgupta R, Sgro J Y. Nucleic Acids Res. 1989;17:7525–7526. doi: 10.1093/nar/17.18.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher T M, Friesen P D, Rueckert R R. J Virol. 1983;46:481–489. doi: 10.1128/jvi.46.2.481-489.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selling B H, Rueckert R R. J Virol. 1984;51:251–253. doi: 10.1128/jvi.51.1.251-253.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deom C M, Schubert K R, Wolf S, Holt C A, Lucas W J, Beachy R N. Proc Natl Acad Sci USA. 1990;87:3284–3288. doi: 10.1073/pnas.87.9.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Citovsky V. Philos Trans R Soc London B. 1999;354:637–643. doi: 10.1098/rstb.1999.0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Z, Kim K H, Giesman-Cookmeyer D, Lommel S A. Virology. 1993;192:27–32. doi: 10.1006/viro.1993.1004. [DOI] [PubMed] [Google Scholar]
- 14.Giesman-Cookmeyer D, Silver S, Vaewhongs A A, Lommel S A, Deom C M. Virology. 1995;213:38–45. doi: 10.1006/viro.1995.1544. [DOI] [PubMed] [Google Scholar]
- 15.Deom C M, Oliver M J, Beachy R N. Science. 1987;237:389–394. doi: 10.1126/science.237.4813.389. [DOI] [PubMed] [Google Scholar]
- 16.Vaewhongs A A, Lommel S A. Virology. 1995;212:607–613. doi: 10.1006/viro.1995.1518. [DOI] [PubMed] [Google Scholar]
- 17.Melcher U. J Gen Virol. 2000;81:257–266. doi: 10.1099/0022-1317-81-1-257. [DOI] [PubMed] [Google Scholar]
- 18.Lazarowitz S G, Beachy R N. Plant Cell. 1999;11:535–548. doi: 10.1105/tpc.11.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara T, Geisman-Cookmeyer D, Ding B, Lommel S A, Lucas W J. Plant Cell. 1993;5:1783–1794. doi: 10.1105/tpc.5.12.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solovyev A G, Zelenina D A, Savenkov E I, Grdzelishvili V Z, Morozov S Y, Maiss E, Casper R, Atabekov J G. Intervirology. 1997;40:1–6. doi: 10.1159/000150514. [DOI] [PubMed] [Google Scholar]
- 21.Solovyev A G, Zelenina D A, Savenkov E I, Grdzelishvili V Z, Morozov S Y, Lesemann D E, Maiss E, Casper R, Atabekov J G. Virology. 1996;217:435–441. doi: 10.1006/viro.1996.0137. [DOI] [PubMed] [Google Scholar]
- 22.Morozov S Y, Fedorkin O N, Juttner G, Schiemann J, Baulcombe D C, Atabekov J G. J Gen Virol. 1997;78:2077–2083. doi: 10.1099/0022-1317-78-8-2077. [DOI] [PubMed] [Google Scholar]
- 23.Rao A L N, Cooper B, Deom C M. Phytopathology. 1998;88:666–672. doi: 10.1094/PHYTO.1998.88.7.666. [DOI] [PubMed] [Google Scholar]
- 24.Hoagland D R, Snyder W C. Proc Am Soc Hort Sci. 1933;30:288. [Google Scholar]
- 25.Verwoerd T C, Dekker B M M, Hoekema A. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider I. J Exp Zool. 1964;156:91–104. doi: 10.1002/jez.1401560107. [DOI] [PubMed] [Google Scholar]
- 27.Xiong Z, Lommel S A. Virology. 1991;182:388–392. doi: 10.1016/0042-6822(91)90687-7. [DOI] [PubMed] [Google Scholar]
- 28.Lommel S A, Weston-Fina M, Xiong Z, Lomonossoff G P. Nucleic Acids Res. 1988;16:8587–8602. doi: 10.1093/nar/16.17.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrington J C, Kasschau K D, Mahajan S K, Schaad M C. Plant Cell. 1999;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson R S, Van Bel A J E. Prog Bot. 1998;59:476–533. [Google Scholar]
- 31.Gardiner W, Sunter G, Brand L, Elmer J S, Rogers S G, Bisaro D M. EMBO J. 1988;7:899–904. doi: 10.1002/j.1460-2075.1988.tb02894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petty I T, Jackson A O. Virology. 1990;179:712–718. doi: 10.1016/0042-6822(90)90138-h. [DOI] [PubMed] [Google Scholar]
- 33.Xoconostle-Cazares B, Xiang Y, Ruiz-Medrano R, Wang H L, Monzer J, Yoo B C, McFarland K C, Franceschi V R, Lucas W J. Science. 1999;283:94–98. doi: 10.1126/science.283.5398.94. [DOI] [PubMed] [Google Scholar]
- 34.Gibbs M J, Weiller G F. Proc Natl Acad Sci USA. 1999;96:8022–8027. doi: 10.1073/pnas.96.14.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey L, Newman J F E, Porterfield J S. J Gen Virol. 1975;26:15–20. doi: 10.1099/0022-1317-26-1-15. [DOI] [PubMed] [Google Scholar]
- 36.Tesh R B. J Gen Virol. 1980;48:177–182. [Google Scholar]
- 37.Nishizawa T, Furuhashi M, Nagai T, Nakai T, Muroga K. Appl Environ Microbiol. 1997;63:1633–1636. doi: 10.1128/aem.63.4.1633-1636.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price B D, Rueckert R R, Ahlquist P. Proc Natl Acad Sci USA. 1996;93:9465–9469. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yusibov V, Shivprasad S, Turpen T H, Dawson W, Koprowski H. Curr Top Microbiol Immunol. 1999;240:81–94. doi: 10.1007/978-3-642-60234-4_4. [DOI] [PubMed] [Google Scholar]
- 40.Gopinath K, Wellink J, Porta C, Taylor K M, Lomonossoff G, van Kammen A. Virology. 2000;267:159–173. doi: 10.1006/viro.1999.0126. [DOI] [PubMed] [Google Scholar]
- 41.Chapman S, Kavanagh T, Baulcombe D. Plant J. 1992;2:549–557. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]